Abstract
The coffee crops are exposed to different pathogens that directly affect yield. These include nematodes, which attack the roots of plants and compromise their physiological development. Given the losses caused by this pathogen and the lack of information on spatial distribution in infested areas, it is important to adopt technologies that enable crops under different management systems to be monitored during their growth cycle. The remote sensing associated with machine learning algorithms is presented as a potential tool for monitoring agricultural crops. The present study assesses different machine learning algorithms, using radiometric values of multispectral images as input datasets, and identifies the best algorithms, to estimate the physiological agronomic parameters in coffee crops submitted to 11 treatments for nematode management. Based on the association between the images taken by a low-cost camera (bands: (R) red, (G) green and (B) blue) mounted on a remotely piloted aircraft (RPA), machine learning algorithms (Random Forest (RF) and support-vector machines (SVM)), the results made it possible to estimate with satisfactory accuracy (root mean square error (RMSE) less than 26.5% the main physical parameters of coffee plants: chlorophyll, plant height, branch length, number of branches and number of nodes per branch. With Planet satellite-derived multispectral bands, the SVM algorithm estimated plant canopy diameters with an RMSE of 7.74%. Based on the spatial distribution maps of the physical parameters, the application machine learning methods offered an opportunity to better use remote sensing data for monitoring coffee crop growth conditions and accurately guiding several management techniques.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the world’s largest producer and exporter of coffee, Brazil plays an important role in coffee production. According to the International Coffee Organization (ICO, 2021), in the 2019/2020 growing season the country produced 63,400 million bags of coffee, 40,511 million of which were exported. Given the economic importance of coffee farming, a wide range of management techniques are used to directly influence aspects such as yield, production costs and quality. Additionally, much like other crops, coffee is exposed to different physiological disturbances, as well as pests and diseases that affect the plants and compromise yields or make it impossible to economically exploit the crop.
One example are nematodes, plant pathogens that live and develop in the soil and attack the roots of coffee plants, compromising their physiological development in the field and hampering growth and production. Visible symptoms on plants parasitized by nematodes include leaf yellowing, foliar necrosis, leaf drop, nutrient deficiencies, dry branches, a general plant decline and can even cause plant death (Ferraz, 2008; Villain et al., 2013). Several methods have been applied to reduce nematode populations in different crops, including the use of cultivars resistant to certain nematode species and chemical control. Biological control with bionematicides, using fungi and rhizobacteria, has been effectively applied in coffee crops to reduce and manage nematode populations in the soil (Campos & Silva, 2008).
The high cost of laboratory testing to quantify levels of nematode infestation in the soil and the losses caused by these pathogens has demonstrated the need for new methods to detect and quantify them in coffee crops (Martins et al., 2017). In this respect, it is important to consider that despite the scarcity of data on the spatial distribution, management and surrounding environments of infested areas, it is vital to monitor the crop throughout its growth cycle in order to obtain high yields and quality crops (Alves et al., 2016).
In light of the need to obtain more information on agricultural systems, technological advances have led to the use of remote sensors to estimate yield components (Zerbato et al., 2016). These sensors measure the radiation reflected from targets, which can be used to obtain information on the type of crop planted and its phenological or nutritional status, and to identify the occurrence of diseases and pests (Sharifi, 2021; Arantes et al., 2021; Abdulridha et al., 2019; Diao, 2020; Martins et al., 2020).
Advanced machine learning techniques have been applied to develop models, using different variables capable of associating yield with factors that influence crop growth (Bocca & Rodrigues, 2016). In conjunction with the data acquired by remote sensors, these techniques have been the focus of several studies aimed at monitoring and estimating agronomic parameters (Singhal et al., 2019; Ranđelović et al., 2020; Zha et al., 2020; Zhou et al., 2019; Sousa et al., 2021).
A number of studies describe the performance of spectral models based on machine learning algorithms in detecting categorical agricultural variables (areas under water and nutritional stress) and estimating continuous variables (agronomic yield parameters). Chemura et al., (2017) used the RF algorithm and MSI Sentinel-2 spectral data with spatial resolutions of 10 and 20 m to estimate chlorophyll indices in coffee crops. The algorithm produced significant RMSE results of 6.80 for plants in different phenological stages across the entire experimental area, using spectral bands with a spatial resolution of 10 m. Other studies used Artificial Neural Network (ANN) and linear regression to estimate plant height, based on data acquired by remote sensing platforms. Ndikumana et al., (2018) used three algorithms (multiple linear regression, support vector regression and RF) to validate the ability of Sentinel-1 multitemporal radar images to estimate plant height and dry biomass in rice plants. The RF algorithm exhibited the best performance, with R²= 0.92 and RMSE = 16%. The model performed best when applied only to plants with mature fruit, with a decline in RMSE to 5.90. In other studies, SVM, ANN and linear regression were used to estimate chlorophyll indices based on data acquired by remote sensing platforms. Sánches (2019) used hyperspectral data with the SVM algorithm to estimate chlorophyll indices in Guinea grass and recorded an average RMSE of 0.2280. Visible and red-edge spectral bands produced the most accurate chlorophyll content estimates. Oliveira et al. (2019) used algorithms based on artificial neural networks to predict the maturation of peanut pods in irrigated and dry regions. The neural networks produced significant results and performed best when the modified vegetation indices were used, with RMSE values of 0.069 and 0.088, whereas the original indices produced RMSE results of 0.090 and 0.094. To assess plant height in sugarcane plantations, Bunruang & Kaewplang (2021) tested three machine learning algorithms (generalized linear model, decision tree and support vector machine) using the reflectance values of RGB images obtained in RPA surveys and digital terrain models. The results demonstrated a correlation between measured and estimated height, with the support vector machine exhibiting the best performance (R² = 0.82 and RMSE = 0.19).
Thus, the hypothesis is that agricultural variables can be accurately estimated by spectral models based on machine learning algorithms, considering vegetation indices and spectral bands derived from RPA orbital sensors as predictor variables. In addition, given the limited literature investigating the estimation of coffee plant yield via spectral models, it remains unclear whether accurate agricultural monitoring can be applied to estimate the physiological parameters of coffee plants based on machine learning algorithms.
In order to answer these questions and test the research hypothesis, the present study aims to compare different machine learning algorithms, using input datasets compiled with remote sensing data, identify the best algorithm combinations, datasets and remote sensors, and propose a means of monitoring the agronomic physiological parameters of coffee crops submitted to nematode management treatments in an area with a history of high infestation levels, without the need for direct and destructive measurement methods. It is important to underscore that these technologies may provide a more cost-effective means of obtaining rapid, accurate and adequate information in extensive agricultural areas and allow decisions about the precise management of the nematode in a localized area.
To that end, spectral models based on machine learning algorithms were developed to monitor a nematode-infested coffee plantation submitted to 11 chemical and biological treatments. The ANN, SVM and RF algorithms were assessed and the images taken by multispectral images mounted on an RPA and orbital satellites.
Methodology
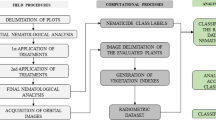
For the purposes of this study, stages were established for field analyses involving the collection of agronomic data, image acquisition and data processing, as follows: (1) definition of the study area; (2) field assessments to obtain agronomic parameters; (3) multispectral image acquisition; (4) digital image processing; (5) data mining; (6) generating predictive models for agronomic variables; (7) assessing the accuracy of the predictive models; and (8) generating spatial distribution maps for the agronomic parameters.
Figure 1 presents the flowchart for the activities carried out in the assessment stages, data processing and results analysis.
Study area
The experiment was conducted at Fazenda Juliana, a private farm in the municipality of Monte Carmelo, Minas Gerais state (MG), in the Triângulo Mineiro and Alto Paranaíba mesoregion, Brazil (-18º41’59’ latitude, -47º33’53 longitude and 826 m a.s.l.; Fig. 2). Climate in the region is tropical with a dry winter, according to the Köppen–Geiger classification system.
Biological and chemical treatments
For the management of plant-parasitic nematodes, coffee plants were treated with seven Bacillus isolates separately (B. subtilis isolates B18, B202, and B33; B. thuringiensis isolate B22; B. safensis isolate B53; B. amyloliquefaciens isolate B266). The isolates belong to the Laboratory of Microbiology and Plant Pathology of the Federal University of Uberlândia – Campus Monte Carmelo and were applied at a dose of 4 L ha− 1 and with a concentration of 1 × 109 CFU mL− 1 (colony forming unit). In addition, the plants were treated with a commercial biological product based on B. subtilis + B. licheniformis (Q; 300 g of product ha− 1); combined application of abamectin + Q (AQ; abamectin at 375 mL ha− 1); and a commercial chemical nematicide based on fluensulfone (F; 2 L ha− 1). Water was applied as a negative control (T).
Non-commercial bacterial isolates were streaked onto Petri dishes containing solid medium 523 (Kado and Heskett, 1970). The plates were kept in an incubator at 25 °C. After 2 days, 1 cm³ of the colonized medium was transferred to 250 mL conical flasks containing liquid medium 523. The flasks were shaken at 25 ± 2 °C and 150 rpm for 5 days in the dark. The bacterial suspensions were indirectly calibrated using a light spectrophotometer adjusted for optical density at 600 nm. Readings equal to 1.8 were equivalent to the concentration of 1 × 109 CFU mL− 1. The choice of this concentration was based on liquid formulations of commercial products based on Bacillus spp. All bacterial isolates and commercial products were applied on the soil surface on both sides of each plant using a backpack sprayer. A spray volume of 500 L ha− 1 was used to cover a 50-cm-wide band under the plant canopy. The organic materials on the soil surface were removed before application and were replaced after the soil treatment. The experiment was arranged in a randomized block design with 11 treatments and five replicates, with each experimental plot consisting of 28 plants, with two plants at each end used as borders.
Characterization and general conditions of the experimental area
The experimental area covers 15,113 m2 and is cultivated with the coffee species Coffea arabica L. cv. Yellow Bourbon, established in 2013, with a history of high nematode infestation levels. It is irrigated via drip irrigation, with plants spaced 0.7 m apart and 3.8 m between the rows. Nematode analyses indicated the presence of four nematode genera in the experimental area, namely Meloidogyne, Pratylenchus, Rotylenchulus and Mesocriconema. Among these, Meloidogyne and Pratylenchus are detected more frequently in coffee crops, causing substantial economic losses.
To characterize nematode population distribution before and after treatment, characterization maps were created based on the laboratory test results for Meloidogyne and Pratylenchus, the genera with the highest incidence in coffee crops. The results were interpolated by inverse distance weighting (IDW), using ArcGis 10.5 software.
The characterization map shows initial nematode population distribution (before treatment application) and were generated based on the results of laboratory analyses of soil collected for the first assessment (09/23/2019) (Fig. 3). For Meloidogyne sp. (Fig. 3a), the eastern portion of the experimental area exhibited the highest concentration of juveniles/150 cm³ of soil and the western section the lowest. The western region contains areas with a population density of 200 to 600 and 600 to 800 juveniles/150 cm³ of soil, and three critical sites with 800 to 1000 juveniles/ 150 cm³ of soil. Smaller juvenile populations were observed across almost the entire western portion of the experimental area, with 0 to 200 juveniles/150cm3 of soil, and some points closer to the eastern section exhibiting densities of 200 to 600 and 600 to 800 juveniles/150 cm³ of soil. Population density for Pratylenchus sp. (Fig. 3b) is 0 to 300 individuals/150 cm³ of soil across almost the whole experimental area, with 300 to 600 individuals/150 cm³ of soil at some locations. Two critical points can be seen in the eastern section, with density populations of 1300 to 1500 individuals/ 150 cm³ of soil.
A randomized block design (RBD) was used, with each block separated by 2 rows of coffee plants and 11 nematode-management treatments/block, with 5 repetitions. The plots contained 32 coffee plants, with the 2 at the ends of each plot disregarded as border plants in order to prevent interference from treatments applied in neighboring plots. Among the plants eligible for assessment of agronomic parameters, the 3 center plants were selected for analysis, as shown in Fig. 4.
Characterization map of the experimental area. (a) Experimental area consisting of 55 plots divided into 5 blocks (BL), and distribution of the chemical and biological treatments applied during the study to manage nematodes, each plot is identified with the code of the treatments applied (B18, B202, and B33: B. subtilis isolates; B22: B. thuringiensis isolate; B53: B. safensis isolate; B266: B. amyloliquefaciens isolate; Q: B. subtilis + B. licheniformis; F: fluensulfone; AQ; abamectin + Q ; T: negative control); (b) Location of the three center plants assessed in each plot
All the plots were delimited using striped tape to indicate the beginning and end of each plot. The 3 plants selected for assessment were identified with small flags mounted on stakes placed in front of the center plant, making them easier to locate. The pair of plagiotropic branches were identified using red tape tied to the base of the branch. The experimental area, plots and plants assessed were georeferenced using coordinates obtained with a HiPer V dual-frequency global navigation satellite system (GNSS) receiver (L1/L2) by real-time kinematic (RTK) positioning.
Acquiring and measuring agronomic parameters
Sample collections to measure the agronomic parameters were conducted in 3 assessment periods. For the first assessment, initiated in the week beginning September 23, 2019, soil samples from each plot were analyzed to quantify the initial nematode population density in the experimental area, and the first application of chemical and biological treatments was carried out in the first week of October. The second application of treatments was performed in the second assessment (week beginning November 25, 2019) and in the third assessment (week beginning March 12, 2020), soil samples were collected for a second time to quantify nematode population density after treatment. Multispectral images were obtained and growth-related agronomic parameters measured in all the assessment stages.
The following agronomic physiological parameters were assessed: plant height (m), measured from the plant base to the apical bud of the orthotropic branch (m), with a 5-meter stadiametric rangefinder; number of plagiotropic branches, obtained by counting the productive branches along the orthotropic branch; number of nodes, determined by counting pairs of north and south-facing plagiotropic branches in the middle third of the plant; length of the plagiotropic branches (m), measured from the base of the orthotropic branch to the apex of the north or south-facing plagiotropic branch, with a fiberglass measuring tape; total chlorophyll index, determined in healthy leaves in the middle and upper third of the plants, using a portable chlorophyll meter (Falker CFL 1030); canopy diameter (m), measured from the multispectral images obtained in aerial surveys.
The sample data collected were measured and evaluated on the same plants for all the assessments. After collection, the data were tabulated and the arithmetic means of the 3 plants assessed per plot were considered in analyses.
Soil sampling for nematode analyses
In order to quantify the nematode population and identify the different species in the experimental area, 500 g soil samples were collected at a depth of 20 cm. Samples were taken in the middle of all the plots, from the location of the center plants. Laboratory testing was conducted in partnership with the Brazilian Laboratory of Environmental and Agricultural Analyses (LABRAS) in Monte Carmelo (MG).
Multispectral image acquisition
Concomitantly to the growth assessments, multispectral images were taken of the study area in order to estimate the agronomic parameters of coffee plants based on these images, which depict the development of the crop after treatments to manage areas infested with nematodes. In this respect, multispectral data were obtained by aerial surveys and orbital platforms.
The aerial survey images were obtained by a remotely piloted aircraft (RPA) (Phanthon 4 PRO) equipped with a native camera (Complementary Metal Oxide Semiconductor sensor) that captures blue (B) (430–460 nm), green (G) (550–570 nm) and red (R) (640 680 nm) wavelengths, with 20MP resolution.
A second multispectral camera (MAPIR Survey 3 N, non-fisheye), equipped with a sensor that captures red (R) (640–680 nm), green (G) (540–560 nm) and near-infrared (NIR) (8330 − 870 nm) wavelengths at 12MP resolution, was used in the aerial surveys. The MAPIR camera was attached to the RPA so that images could be obtained simultaneously by both sensors during the same flight. The images were taken at similar times of day.
High resolution multispectral images were obtained via orbital remote sensors from the PlanetScope-PS2 satellite constellation, taken on the same or similar dates to those of the assessments, considering the weather conditions in the region. The spatial and radiometric resolutions of the Planet satellite images are 3 m and 12 bits, respectively. The satellite constellation uses Charge Coupled Device sensors equipped with Bayer filters that capture blue (B) (455–515 nm), green (G) (500–590 nm), red (R) (590–670 nm) and near-infrared (NIR) (780–860 nm) wavelengths. The images are configured in the Universe Transverse Mercator (UTM) projection system, WGS-84 datum surface and 3B correction level, provided with an orthorectified image product and corrected to surface reflectance values.
Digital image processing
Geometric and atmospheric corrections
Drone survey images were processed by photogrammetry, where the images were imported with their respective metadata (geotags, geographic coordinates, altitudes and focal distance, referenced to SIRGAS 2000 datum). Aerotriangulation was performed automatically, following the workflows that involve photoalignment and construction of densified clouds and the digital elevation model, applying machine learning models based on the search for homologous image points and using Agisoft PhotoScan Professional software. Next, the images were georeferenced using the ENVI 5.1 program, based on the control points collected in the field with the GNSS receiver.
The Planet satellite images did not require atmospheric correction because they were obtained in Surface Reflectance (SR) format. In this format, standard analytical products (radiance) are processed for top-of-atmosphere reflectance and then atmospherically corrected for surface reflectance. Surface reflectance is determined from top of atmosphere (TOA) reflectance, calculated pixel-to-pixel using search tables created from the radiative transfer code, which maps reflectance (TOA) for 2 bottom of atmosphere (BOA) reflectances for all the relevant physical condition combination ranges of satellite images from this constellation. Based on near real-time (NRT) MODIS satellite data, and if there is no overlap of water vapor and ozone data collected on the same day, a 6 S atmospheric model is selected based on the local latitude and time of the year of image acquisition, using the FLAASH atmospheric correction tool.
Radiometric normalization
In this stage, images from the first assessment served as the basis for normalizing those from the second and third assessments. The radiance of the RPA images and the surface reflectance factor of the Planet image were extracted by supervised learning for the same bright and dark targets and each image to be normalized. The values extracted were used in Eq. 1 and processed via the Band Math function in ENVI 5.1 software.
Where:
;
;
Calculating the vegetation indices
Nine vegetation indices widely applied in agricultural areas were selected as predictors for the predictive models of the agronomic parameters: NDVI, CVI, VARI, TGI, ARVI, SIPI, MPRI, SR and GNDVI (Table 1). The indices were applied to each sensor, considering its respective spectral range.
Cluster analysis
Several configurations were evaluated to select the best predictive model, varying the algorithms and input datasets with different spectral bands and vegetation indices. Cluster analysis was applied to the input datasets in order to determine the different band and index combinations for each individual sensor, using Minitab 19 software.
For this stage, the software was set to a fully connected layer with distance measured by correlation. This means that similarities between two clusters are defined by the greatest distance between any variable in one cluster and any in another, calculated by a distance matrix. Additionally, as a final partitioning criterion, the similarity level was selected using the value corresponding to three σ (sigma), that is, 67%, as stipulated by the empirical rule.
The final results of these clusters were presented in a dendrogram. Bands and vegetation indices were randomly selected for each cluster generated.
Generating predictive models
After each assessment, databases were created in table format containing the values of the agronomic variables collected in the field and the radiometric values extracted from the spectral bands and vegetation indices for all the sensors.
Based on the corrected multispectral images and points identified in the field by the GNSS receiver, shapefiles with vector polygons were created to extract the radiometric values in the region of each plant assessed in the plots, using the region of interest (ROI) tool in ENVI 5.1 software. The radiometric values from band to band were automatically extracted and calculated by the software, considering the average values of the pixels in the vectorized polygons for each plot.
This dataset was used to generate the predictive model. A total of four classification algorithms available in Weka 3.9.4 software were trained, namely; RF, ANN and SVM.
As proposed by Breiman (2001), the RF algorithm allows flexible modeling of high dimensional data, resulting in a large number of regression trees, and calculating the averages of their predictions. It uses kernels and the nearest neighbor method because it assigns weights to predictions based on the weighted average of the nearest observations. However, unlike other methods, the RF relies on data to determine which nearest neighbors will be assigned the most weight (Wager & Athey, 2018). The network settings proposed by the software were used for RF modeling: batch size (100 instances), number of iterations (100) and bag size percentage (100).
ANN neural networks consist of an input layer, an output layer and a hidden layer, referred to as units or neurons, through which the input signal travels, and use an error back-propagation algorithm. The network settings proposed by the software and applied to ANN modeling were: learning rate (0.3), momentum (0.2), and training epochs (500).
The SVM algorithm implements the SMO algorithm proposed by Platt (1999) and improved by Shevade et al., (2000), who presented an iterative algorithm, denominated sequential minimal optimization (SMO), to solve regression problems in association with a support vector machine. Unlike other learning models, the support vector machine is based on minimizing errors. Based on a dataset, this algorithm aims to create a hyperplane equidistant from the closest data of each class in order to reach a maximum margin on either side of the hyperplane, considering only the training data of each class that falls within the boundary of these margins as training data. These data are denominated support vectors. The following network settings, proposed by the software, were used for SVM algorithm modeling: batch size (100 instances) and Kernel (Polykernel).
The supplied test set was used for training. A total of 165 samples were analyzed and randomly allocated into training sets, containing 80% of the data (132 samples), and test sets, with 20% of the data (33 samples) using the supervised learning method. Data modeling was performed using the input datasets from all three assessments in a single analysis. This made it possible to create multitemporal models applied to each agronomic parameter in all the assessment periods.
It is important to underscore that, after several tests, the adjustment parameters used for all the regression algorithms were those established as standard by the Weka 3.9.4. algorithm. This is justified by the fact that adjustment values other than those proposed by the algorithm itself did not significantly improve the accuracy of the predictive models.
In order to assess the relationship between the agronomic parameters and radiometric values, three different algorithms were evaluated for the predictive model: the original spectral bands alone; vegetation indices alone; spectral band and vegetation indices combinations resulting from cluster analysis.
Metrics used to assess the models
Metrics such as the root mean square error (RMSE) and relative root-mean-squared error (RMSE%) were used to validate the quality of the predictive models and identify the best model.
The RSME and RMSE% are calculated by Eqs. 2 and 3:
where, \( {x}_{i}\) and \( {x}_{meas}\) represent the estimated and measured value; and \( n\) the number of samples.
For the trained models, only the four best and worst-performing algorithms were selected, as a function of RMSE and normalized RMSE (RMSE%), considering the error of the difference between measured and estimated variables for each agronomic parameter assessed in the experimental area.
Characterization maps of the growth rate of coffee plants
After defining the best model based on the algorithms, input datasets and sensors, interpolation images were generated using the agronomic parameter estimates for each epoch of the assessment stages, in order to create a standard grid of values to calculate plant growth rates. Inverse distance weighting (IDW) was applied to generate the interpolated images, using ArcGis 10.5 software.
The plant growth rate (GR) was calculated using the relative growth rate (RGR) equation, which shows the monthly GRs for each parameter assessed (Eq. 4). In this stage, GRs were calculated between the first (09/23/2019) and third (03/12/2020) assessments, comprising a six-month period.
where, \( {N}_{f}\) = final value; \( {N}_{i}\) = initial value; and \( t=\) time.
Results
Cluster analysis to select input dataset combinations
Based on the results presented in the dendrogram, a spectral band of vegetation index was randomly chosen for each cluster to compile an input dataset for the predictive model.
The dendrogram created for spectral bands and vegetation indices from the MAPIR camera sensor (Fig. 5) was partitioned into four clusters containing the following observations: First cluster (R/G/N); second cluster (SR); third cluster (NDVI); and fourth cluster (CVI/GNDVI). The respective observations selected for each cluster were N, SR, NDVI and CVI.
Figure 6 shows the dendrogram created after analysis of the spectral bands and vegetation indices from the RPA sensor, partitioned into two clusters with the following observations as predictive variables: first cluster (R/B/G); second cluster (MPRI/VARI/TGI). Observations G and TGI were selected for each cluster, respectively.
Analysis of the vegetation indices and original spectral bands from the Planet satellite constellation (Fig. 7) resulted in a dendrogram with two clusters, as follows: first cluster (B/G/R/SR/SIPI); second cluster (N/NDVI/ARVI/GNDVI/MPRI/VARI/TGI/CVI). Observations G and TGI were selected for each cluster, respectively.
Determining the best algorithm for the predictive models
Table 2 show the results of analyses conducted to identify the best algorithms, input datasets and sensors, respectively, for the following parameters: total chlorophyll index; plant height (m); branch length (south-facing) (m); branch length (north-facing) (m); number of nodes; number of nodes (south-facing); number of nodes (north-facing); and canopy diameter (m).
For the total chlorophyll index, the RF algorithm performed best with the radiometric values of vegetation indices from RPA images as input data, exhibiting the lowest RMSE (4.7975) and RMSE% (9.0545). However, this same algorithm exhibited the worst performance in estimating total chlorophyll indices when the radiometric values of the spectral bands and vegetation indices selected by cluster analysis from Planet satellite images were used, producing the highest RMSE (6.1261) and RMSE%, (11.5621) values.
The best-performing algorithm for plant height (m) was SVM with the radiometric values of spectral bands and vegetation indices selected by cluster analysis of RPA images used as input data and the lowest RMSE (0.1128) and RMSE% (3.6929) values. The algorithm with the worst performance for plant height (m) was the RF, which obtained the lowest RMSE (0.1718) and RMSE% (5.6244) values and used the radiometric values of spectral bands from the MAPIR images as input data.
The most accurate branch length (south-facing) (m) estimates were obtained by the SVM algorithm, with the radiometric values of spectral bands and vegetation indices selected by cluster analysis of RPA images as input data and the lowest RMSE (0.1329) and RMSE% (15.3025) values. The least accurate predictions for this parameter were generated by the ANN algorithm, which obtained the highest RMSE (0.1691) and RMSE% (19.4707) values and used the radiometric values of spectral bands from RPA images as input data.
The most accurate branch length (north-facing) (m) estimates were obtained by the SVM algorithm, with the radiometric values of spectral bands and vegetation indices selected by cluster analysis of RPA images as input data and the lowest RMSE (0.1436) and RMSE% (16.8162) values. The RF was the worst-performing algorithm for this parameter when the radiometric values of vegetation indices from MAPIR images were used as input data, producing the highest RMSE (0.1825) and RMSE% (21.3715) values.
For number of branches, the most accurate estimate were obtained by SVM, which generated the lowest RMSE (12.2239) and RMSE% (16.5459) values used the radiometric values of spectral bands and vegetation indices selected via cluster analysis of RPA images as input data. The worst-performing algorithms for these parameters was the RF, with the radiometric values of spectral bands and vegetation indices selected via cluster analysis of Planet satellite images as input data and the lowest RMSE and RMSE% (17.0949 and 23.1391).
The most accurate predictions for number of nodes (south-facing) were obtained by the SVM algorithm, which obtained the lowest RMSE (5.1289) and RMSE% (18.5753) values the radiometric values of spectral bands and vegetation indices selected via cluster analysis of RPA images as input data, and the least accurate were those generated by the ANN, with the radiometric values of spectral bands from the MAPIR images as input data, which obtained the highest RMSE (10.5508) and RMSE% (36.8191) results.
In the same, the most accurate predictions for number of nodes (north-facing) were obtained by the SVM algorithm, which obtained the lowest RMSE (7.5937) and RMSE% (26.4997) values the radiometric values of spectral bands and vegetation indices selected via cluster analysis of RPA images as input data, and the least accurate were those generated by the ANN, with the radiometric values of spectral bands from the MAPIR images as input data, which obtained the highest RMSE (10.5508) and RMSE% (36.8191) results.
For canopy diameter (m), SVM performed best, using the radiometric values of spectral bands from the Planet multispectral images as input data and producing the lowest RMSE (0.1302) and RMSE% (7.7374) results. The worst-performing algorithm for this parameter was the RF, which generated the highest RMSE (0.1903) and RMSE% (11.3090) values with the radiometric values of spectral bands and vegetation indices selected by cluster analysis of RPA images as input data.
Characterization maps for the parameters
Figures 8, 9, 10, 11, 12, 13, 14 and 15 present the characterization maps of growth rate of coffee plants for the six-month period between the first (09/23/2019) and third assessments (03/12/2020) for total chlorophyll index; plant height (m); number of branches; branch length (south-facing) (m); branch length (north-facing) (m); number of nodes per branch (south-facing); number of nodes per branch (north-facing); and canopy diameter (m), subjected to different chemical and biological treatments aimed at reducing the population of nematodes.
Figure 8 shows the growth rate (%) for the total chlorophyll index (%). Total chlorophyll indices declined in the experimental area, exhibiting negative growth rates of 0 to -3% growth/month. Values of -3 to -4% growth/month were also recorded in isolated areas of all the plots. Positive growth rates of 0 to 3% growth/month were also recorded in all the plots, particularly in the south-facing portion of the experimental area.
Figure 9 shows the growth rate (%) for plant height (m). Plant growth was positive across the entire experimental area, varying from 0 to 1% growth/month. Values between 1 and 2% and 2 and 3% were also observed in some isolated plots, particularly those for treatments B33 and B202 (Bacillus subtilis) and treatment T (water) in blocks two and five, which exhibited the largest concentrations of these values.
Figure 10 presents the growth rate (%) for number of branches, which increased across the entire experimental area, displaying positive rates of 2 to 4% and 4 to 8% of growth/month. Negative rates (%) were also recorded, with values between 0 and − 2% growth/month throughout the area and − 2 to -4% in some plots, particularly treatments B33 (Bacillus subtilis) and B05 (Bacillus methylotrophicus) in block four and B53 (Bacillus safensis) and B33 (Bacillus subtilis) in block five, which obtained the greatest concentrations of these values.
Figure 11 depicts the growth rate (%) for number of branches (south-facing) (m), indicating the greater occurrence of positive rates for south-facing plagiotropic branches throughout the study area, varying from 0 to 4% growth/month. Some plots showed 4 to 8% growth/month, especially B202 (Bacillus subtilis) in block two and B33 and B202 (Bacillus subtilis), T (water) and B05 (Bacillus methylotrophicus) in block five, with larger concentrations of these values. Negative growth rates of 0 to -4% growth/month were found in some areas of all the plots, with the highest incidences of these values observed for B05 (Bacillus methylotrophicus) in blocks three and four.
The growth rate (%) for branch length (north-facing) (m) is presented in Fig. 12, demonstrating greater positive growth rates for north-facing plagiotropic branches across the experimental area, ranging from 0 to 3% growth/month. Growth of 3 to 6% was also observed in some points of all the plots, Negative growth was also recorded in some of the plots, ranging from 0 to -3% growth/month, with the largest concentration of these rates observed for treatments B33 (Bacillus subtilis) and B05 (Bacillus methylotrophicus) in blocks three and four and B53 (Bacillus safensis) and B266 (Bacillus amyloliquefaciens) in block five.
The growth rate (%) for number of branches is shown in Fig. 13, with south-facing nodes displaying greater positive growth of 0 to 5% and 5 to 10% per month, as well as 10 to 15% and 15 to 20% in some locations of all the plots. Negative growth rates of -5 to -10% were recorded in some plots, most notably B33 (Bacillus subtilis) and B05 (Bacillus methylotrophicus) in block four and B53 (Bacillus safensis) in block five.
Figure 14 depicts the growth rate (%) for the number of nodes (north-facing), which was positive in most of the study area, with rates between 0 and 5% and 5 and 10% per month. A greater concentration of 10 to 15% and 15 to 20% growth rates was observed in some plots, particularly for B266 (Bacillus amyloliquefaciens) and B33 (Bacillus subtilis) in block one, treatments F (Fluensulfone) and B33 (Bacillus subtilis) in block two, AQ (Abamectin + Bacillus subtilis / Bacillus licheniformis) and B53 (Bacillus safensis) in block three, and B266 (Bacillus amyloliquefaciens) and F (Fluensulfone) in block four. Some plots also exhibited negative growth rates of 0 to -5% per month, most notably for B05 (Bacillus methylotrophicus) in block two, B22 (Bacillus thuringiensis) and F (Fluensulfone) in block three, B18 (Fluensulfone) in block four and B266 (Bacillus amyloliquefaciens) in block five.
Figure 15 presents the growth rate (%) for canopy diameter. Positive growth for this parameter was most evident in the western portion of the experimental area, ranging from 2 to 4% and 4 to 5% per month, with rates of 0 to 2% per month more frequent in the eastern section. Negative growth was only observed in two plots, namely treatments B22 and B222 in block two.
The characterization maps show the final population distribution of nematodes (after treatment) for the genera Meloidogyne and Pratylenchus (Fig. 16). The maps were generated based on the results of laboratory tests performed during the third assessment (03/12/2020). There was a decline in the concentration of juveniles/150 cm³ of soil after treatment for Meloidogyne sp. (Fig. 16a) in the eastern section, with a population density of 0 to 200 juveniles/150 cm³ of soil across the experimental area.
For Pratylenchus sp. (Fig. 16b), population density rose after treatments application, with an increase in the number of areas containing 300 to 600 individuals/150 cm³ of soil. Two points with critical concentrations, observed for this genus in the first assessment period in the eastern portion of the study area, exhibited a decline in population density, one to 0 to 300 and the other to 900 to 1300 individuals /150 cm³ of soil. Three points within the western section increased to 900 to 1300 individuals /150 cm³ of soil.
Discussion
Cluster analysis
Spectral bands and vegetation indices show great potential in estimating agricultural parameters. Vegetation indices are indicators of the dynamic biophysical properties related to yield and energy balance in plants, due to the correlation between sunlight and active photosynthetic tissue. The availability of several spectral bands makes it possible to more efficiently explore spectral characteristics in conjunction with the operating modes of sensor systems, that is, the more spectral bands available, the greater the chance of extracting information on the object analyzed (Martins et al., 2017).
For the three sensors, visible band clustering (Figs. 5, 6 and 7) into a same cluster reflects the natural high correlation between the three bands and plant characteristics sensitive to this spectral interval, such as chlorophyll a and b content, pigmentation and similar senescence for all plants. This similarity occurred because nematode infestation symptoms were initial in the experimental area, that is, phenological stage, leaf color and size were the same for healthy and infected plants. The separation of indices into different visible band clusters occurred because they are sensitive to the structural characteristics of leaves such as biomass and leaf area index (Jensen, 2009).
For the image captured by the MAPIR camera (Fig. 5), the NIR band was also clustered into the same visible spectral band cluster. This result is not significantly associated with the physiological conditions of coffee plants, but with the construction characteristics of the sensor itself, where conception of the NIR band is only possible with filters applied to the blue band, which can detect radiance at the start of the NIR spectral interval (800 nm) and is highly correlated with the other visible bands (Green et al., 2019).
Estimation performance of spectral models
The dendrograms demonstrated an input data mining option for physiological parameter prediction models, where a significant reduction in the number of predictor variables occurs, without compromising estimation accuracy, and there is less likelihood of model overadjustment. Table 2 shows that the RF algorithm model for chlorophyl estimation, obtained by the combination of the G band and TGI index derived from the RPA camera, exhibited accuracy similar to that of the most accurate model, with RF established by all the plant indices obtained by the camera.
The better chlorophyl estimation of the RPA camera demonstrates the potential of bands and indices derived from visible wavelengths in estimating photosynthesis-related agricultural variables. This condition is related to the sensitivity of the B and R bands to chlorophyl a and b (Jensen, 2009). For this case, the RF algorithm is more accurate than neural network algorithms and support vector machines because in some scenarios with few predictor variables, the RF algorithm performs better than the others. Furthermore, when data obtained from remote sensing platforms are used, the advantages of this algorithm when compared with other classifiers are its ability to deal with the substantial temporal variation of reflectance from plants, highly dynamic processing capacity, requiring only a few parameters for classification, and highly accurate classifications, among others (Zhou et al., 2019).
This premise is presented in a similar study, where Lee et al., (2020) evaluated multispectral images obtained by RPA to prevent nitrogen accumulation in the canopy of corn plants. Simple/multiple linear regression, RF and support vector regression (SVR) were selected to create the models, using individual spectral bands and vegetation index combinations as input data. The RF algorithm performed best, with an RMSE of 4.52. The authors also underscore that in order to estimate the chlorophyll and macro and micronutrients present in leaves, prediction models based on high spatial resolution images taken by an RPA camera are more accurate than their satellite counterparts. This is because of the proximity between RPA sensors and the canopy, which makes it possible to record the exact amount of energy radiated from leaves on the plant parts where there is greater nutrient concentration. Specifically for the present study, the RPA images were able to record even the energy radiated by the third basal leaves of the plants, that is, where the most mature and developed leaves are found, while orbital images were primarily limited to recording energy radiated only by the canopy.
In the mapping of specific nutrients of other crops, the same was observed by Ranđelović et al., (2020), who applied the RF algorithm to estimate soybean density using only vegetation indices from aerial RGB images captured by RPA. The results obtained showed significant model validation, with an RMSE of 7.47. Zha et al., (2020) applied machine learning algorithms and linear regression to estimate the biomass and N levels of rice plants using vegetation indices obtained from images captured by RPA. The findings demonstrated that the machine learning methods provided the most accurate estimates of plant nutrition indices when compared with linear regression, particularly RF, which exhibited the lowest RMSE (0.09).
The combination of the G band and TGI vegetation index obtained from RPA images made it possible to estimate all the agronomic parameters related to coffee plant biomass (height, branch length, number of branches and number of nodes), using the SVM algorithm. Inserting the TGI index into the models contributed significantly to estimation accuracy, since the index is highly correlated with biomass and LAI (Jensen, 2009). On the other hand, the algorithm also contributed, since the support vector machine proved to be effective in models that used multispectral images, evident in the positive characteristics of its application in remote sensing: the ability to effectively manage small training sets; efficiently dealing with highly dimensional spaces; a decision process using only a subset of training data; more efficient in terms of memory storage; and applying new kernels instead of linear boundaries (Sheykhmousa et al., 2020).
The spatial resolution of RPA images also contributed to more accurate models than those based on Planet images, due to the high spatial resolution of the images and radiometric variability of the drone images, whereas information is generalized in the orbital images, that is, the response of canopy plants predominates.
For the canopy diameter (m), the image from the Planet satellite sensor exhibited the best performance in estimating this parameter. For this parameter, the models constructed from orbital images are more accurate because the sensor captures the energy radiated by the canopy and to a lesser extent, the energy reflected by the lower branches and leaves (Ndikumana et al., 2018). In this respect, the models created using the RPA images tend to be less accurate due to the high spatial resolution of the images, which increases the radiometric variability of the spectral response of the canopy (Jensen, 2009).
The chlorophyll content reflects the physiological state of vegetation; it decreases in stressed plants and can therefore be used as a measurement of vegetation health (Carmona et al., 2015).This behavior can be explained by the fact that the assessments in the experimental area were conducted between the second and beginning of the fourth phenological stage of coffee, comprising part of the flowering period in September (first assessment), fruit set and rapid growth in November (second assessment), and physiological maturity and the onset of ripening in March (third assessment). Low nitrogen availability in plants that need large amounts of this nutrient can limit yield in different crops. The absence of nitrogen, important in chlorophyll synthesis and other photosynthetic processes, reduces the absorption of sunlight as an energy source and affects essential functions such as nutrient absorption (Pompelli et al., 2010). Additionally, nutrients such as nitrogen, which are directly linked to chlorophyll content in plants, play a vital role in maintaining plant vigor, particularly during the reproductive stage. Jia et al., (2021) reported that nitrogen is one of the most important elements in chlorophyll and plant enzymes, and nitrogen deficiency can decrease photosynthetic assimilation and crop yields. During the rapid growth stage of coffee berries, large quantities of carbohydrates are diverted and more than 95% of the total nitrogen absorbed is consumed, causing symptoms of nitrogen deficiency in the leaves and limiting vegetative growth (Amaral et al., 2001). During fruit set, the displacement of nutrients such as nitrogen from the leaves to the berries causes greater foliar deficiency (Salamanca-Jimenez et al., 2017).
E vidence on spatial distribution of physiological parameters.
Analysis of the maps shows the effects on the physiological parameters after treatments. For plant height (m), number of branches, branch length (north-facing), branch length (south-facing) (m), number of nodes (north-facing) (m), number of nodes (south-facing) (m) and canopy diameter (m), there was a significant increase in vegetative growth during the experiment. Two factors may explain this positive coffee plant growth under the conditions of this experiment: rainfall starting in October, which allows plants to overcome the stress naturally imposed by the phenological stages of flowering and fruit set, and the efficiency of treatments in reducing the nematode population in the area, especially those based on Bacillus spp. Bacillus spp. species are efficient antagonists against nematodes, colonizing the root system of coffee plants, thereby changing the composition of root exudates, decreasing juvenile and adult penetration of different nematode species and producing nematicide substances, that is, killing individuals of species that attack the coffee crop and producing root growth-promoting substances (Liu et al., 2013; Marin-Bruzos & Grayston, 2019; Mhatre et al., 2019). It is important to note that drip irrigation (irrigation system implemented in the experimental area of this study) only provides enough water to maintain plants turgid with a metabolism sufficient to withstand these stress phases.
Among the factors that limit the growth and production process of coffee plant, species of Meloidogyne stand out. Economic losses due to root knot nematodes vary considerably depending on the species involved and their distribution (Boisseau et al., 2009; Ferraz, 2008). Species from the genus Meloidogyne are considered sedentary parasites and spend most of their life cycle in the same place, feeding on the cells of coffee roots. Once parasitism has begun, plant metabolism changes, causing the formation of nutritive (or nurse) cells in the root galls (Muniz et al., 2008). Species from the genus Pratylenchus are classified as migratory endoparasites. These nematodes destroy plants cells during migration and feeding, damaging root tissue. The symptoms of parasitism by nematodes of these genera include reduced growth in parasitized plants, physiological imbalance and macro and/or micronutrient deficiencies, changes in agronomic characteristics and a progressive decline in yield (Inomoto et al., 2007).
The present study showed the greater efficiency of treatments in reducing the nematode populations of branches (Meloidogyne sp.) than those that damage roots (Pratylenchus sp.) One of the mechanisms by which rhizobacteria can act against nematodes consists of the production of nematocidal substances (Chitwood, 2002). These substances may exhibit specificity in relation to the target (s) and might have a nematostatic or nematocidal effect on one or more nematode species and no effect on others. For example, Pseudomonas fluorescens and Pseudomonas putida can reduce the population of Meloidogyne spp. and Radopholus similis in tomato and banana plants (Aalten et al., 1998). The production of 2,4-diacetylphloroglucinol by P. fluorescens can control Globodera rostochiensis in potato plants (Cronin et al., 1997). B. cereus and B. subtilis can reduce the population of root-knot nematodes (Meloidogyne spp.) in plant roots (Xiao et al., 2012). Several symptoms were observed in the experimental area during the first assessment, prior to chemical and biological treatment for nematodes, as shown in Fig. 17. These symptoms include leaf yellowing and dropping (Fig. 17a), dry branches (Fig. 17b) and the presence of galls on the roots of plants parasitized by Meloidogyne exigua (Fig. 17c).
In the third assessment (Fig. 18), after two treatments for nematode management, there was a notable increase in vegetative growth (Fig. 18a), plagiotropic branch emergence (Fig. 18b) and fruit growth (Fig. 18c). In this respect, measures are needed to manage plant-parasitic nematodes in order to significantly reduce populations of this pathogen. Fungi and bacteria are widely available for this purpose. As biological control agents, rhizobacteria promote vegetative growth, antibiosis, competition and systemic resistance in plants, in addition to protecting against pathogens such as nematodes, among others (Hashem et al., 2019; Tolardo et al., 2019).
Additionally, chemical products have been used to manage plant-parasitic nematodes, with significant results in infested areas. Chemical control is an effective tool when a rapid response is expected for population control and the desired damage level has not been reached; however, the use of products with low residual power is necessary to prevent health risks and environmental damage, ensuring sustainability and a better- quality product.
Conclusions
The method applied was appropriate for areas containing coffee crops with high treatment variability. Both the orbital images and those obtained by drones in aerial surveys proved to be potential tools to estimate agronomic parameters under conditions of high physiological variability. The machine learning algorithms were capable of supporting the models used to estimate the agronomic parameters. Due to the high variability of the coffee crop studied and the efficacy of the method, it is suggested that the techniques used here can be successfully applied to other regions, since the environmental and management conditions can occur in other planting areas.
The main conclusions of the article are highlighted below:
-
The results indicated that machine learning methods could significantly improve the estimation of agronomic parameters of yield in coffee crops. The SVM algorithm, G band and TGI index derived from RPA images used as predictor variables can be applied to more accurately estimate coffee plant parameters: height, branch length and number of branches and nodes. For the same sensor, the RF algorithm and MPRI, VARI and TGI indices can be used to more accurately estimate total chlorophyll.
-
Based on the four original bands of Planet orbital images, the SVM algorithm can more accurately estimate the coffee plant canopy.
-
The physiological parameter growth maps of coffee plants present the spatial distribution of agricultural yields based on treatments to manage nematode infestation.
References
Aalten, P. M., Vitour, D., Blanvillain, D., Gowen, S. R., & Sutra, L. (1998). Effect of rhizosphere fluorescent Pseudomonas strains on plant-parasitic nematodes Radopholus similis and Meloidogyne spp. Letters Applied Microbiology, 27(6), 357–361. https://doi.org/10.1046/j.1472-765X.1998.00440.x
Abdulridha, J., Ehsani, R., Abd-Elrahman, A., & Ampatzidis, Y. (2019). A remote sensing technique for detecting laurel wilt disease in avocado in presence of other biotic and abiotic stresses. Computers and Electronics in Agriculture, 156, 549–557. https://doi.org/10.1016/j.compag.2018.12.018
Alves, H. M. R., Volpato, M. M. L., Vieira, T. G. C., Maciel, D. A., Gonçalves, T. G., & Dantas, M. F. (2016). Characterization and spectral monitoring of coffee lands in Brazil. The International Archives of the Photogrammetry, 8. https://doi.org/10.5194/isprsarchives-XLI-B8-801-2016. Remote Sensing and Spatial Information Sciences. XLI-B
Amaral, J. A. T., Matta, F. M. da, & Rena, A. B. (2001). Effects of fruiting on the growth of Arabica coffee trees as related to carbohydrate and nitrogen status and to nitrate reductase activity. Revista Brasileira de Fisiologia Vegetal, 13(1), 66–74. https://doi.org/10.1590/S0103-31312001000100008
Arantes, B. H. T., Moraes, V. H., Geraldine, A. M., Alves, T. M., Albert, A. M., Silva, G. J. da, & Castoldi, G. (2021). Spectral detection of nematodes in soybean at flowering growth stage using unmanned aerial vehicles. Ciência Rural, 51(5). https://doi.org/10.1590/0103-8478cr20200283
Bocca, F. F., & Rodrigues, L. H. A. (2016). The effect of tuning, feature engineering, and feature selection in data mining applied to rainfed sugarcane yield modeling. Computers and Electronics in Agriculture, 128, 67–76. https://doi.org/10.1016/j.compag.2016.08.015
Boisseau, M., Aribi, J., de Sousa, F. R., Carneiro, R. M. D. G., & Anthony, F. (2009). Resistance to Meloidogyne paranaensis in wild Coffea Arabica. Tropical Plant Pathology, 34 (1). https://doi.org/10.1590/S1982-56762009000100006
Breiman, L. (2001). Random Forests. Machine Learning Springer, 45, 5–32. https://doi.org/10.1023/A:1010933404324
Bunruang, P., & Kaewplang, S. (2021). Evaluation of Sugarcane Plant Height using UAV Remote Sensing. Engineering Access, 7(2), 98–102. https://doi.org/10.14456/mijet.2021.15
Campos, V. P., & Silva, J. R. (2008). Management of Meloidogyne spp. in Coffee Plantations. In R. M. Souza (Ed.), Plant-Parasitic Nematodes of Coffee (pp. 165–190). Dordrecht: Springer. https://doi.org/10.1007/978-1-4020-8720-2_8
Carmona, F., Rivas, R., & Fonnegra, D. C. (2015). Vegetation Index to estimate chlorophyll content from multispectral remote sensing data. European Journal of Remote Sensing, 48(1), 319–326. https://doi.org/10.5721/EuJRS20154818
Chemura, A., Mutanga, O., & Odindi, J. (2017). Empirical Modeling of Leaf Chlorophyll Content in Coffee (Coffea Arabica) Plantations with Sentinel-2 MSI Data: Effects of Spectral Settings, Spatial Resolution, and Crop Canopy Cover. IEEE Journal of selected topics in applied earth observations and remote sensing, 10(12), 5541–5550. https://doi.org/10.1109/JSTARS.2017.2750325
Chitwood, D. J. (2002). Phytochemical based strategies for nematode control. Annual Review of Phytopathology, 40, 221–249. https://doi.org/10.1146/annurev.phyto.40.032602.130045
Cronin, D., Dowling, D. N., Dunne, C., Fenton, A., Moenne-Loccoz, Y., & O’gara, F. (1997). Role of 2,4-Diacetylphloroglucinol in the Interactions of the Biocontrol Pseudomonad Strain F113 with the Potato Cyst Nematode Globodera rostochiensis. ASM Journals - Applied and Environmental Microbiology, 63(4), https://doi.org/10.1128/aem.63.4.1357-1361.1997
Diao, C. (2020). Remote sensing phenological monitoring framework to characterize corn and soybean physiological growing stages. Remote Sensing of Environment, 248. https://doi.org/10.1016/j.rse.2020.111960
Ferraz, L. C. (2008). Brazil. In R. M. Souza (Ed.), Plant-Parasitic Nematodes of Coffee (pp. 225–248). Dordrecht: Springer. https://doi.org/10.1007/978-1-4020-8720-2_12
Green, D. R., Hagon, J. J., Gómez, C., & Gregory, B. J. (2019). Using Low-Cost UAVs for Environmental Monitoring, Mapping, and Modelling: examples from the coastal zone. Coastal Management - Elsevier, 21, 465–501. https://doi.org/10.1016/b978-0-12-810473-6.00022-4
Hashem, A., Tabassum, B., & AbdAllah, E. F. (2019). Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi Journal of Biological Sciences, 26(6), 1291–1297. https://doi.org/10.1016/j.sjbs.2019.05.004
Jensen, J. R. (2009). Remote Sensing of the Environment: An Earth Resource Perspective (2nd Edition). Pearson Education India. New Delhi
Jia, M., Colombo, R., Rossini, M., Celesti, M., Zhu, J., Cogliati, Cheng, T. … Yao, X. (2021). Estimation of leaf nitrogen content and photosynthetic nitrogen use efficiency in wheat using sun-induced chlorophyll fluorescence at the leaf and canopy scales. European Journal of Agronomy, 122. https://doi.org/10.1016/j.eja.2020.126192
Lee, H., Wang, J., & Leblon, B. (2020). Using Linear Regression, RFs, and Support Vector Machine with Unmanned Aerial Vehicle Multispectral Images to Predict Canopy Nitrogen Weight in Corn. Remote Sensing, 12(13), 2071. https://doi.org/10.3390/rs12132071
Liu, Z., Budiharjo, A., Wang, P., Shi, H., Fang, J., Borriss, R. … Huang, X. (2013). The highly modified microcin peptide plantazolicin is associated with nematicidal activity of Bacillus amyloliquefaciens FZB42. Applied genetics and molecular biotechnology. 97,10081–10090. https://doi.org/10.1007/s00253-013-5247-5
Marin-Bruzos, M., & Grayston, S. (2019). Biological Control of Nematodes by Plant Growth Promoting Rhizobacteria: Secondary Metabolites Involved and Potential Applications. In H. B. Singh, C. Keswani, M. S. Reddy, E. Sansinenea, & C. García-Estrada (Eds.), Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms (pp. 253–264). Singapore: Springer. https://doi.org/10.1007/978-981-13-5862-3_13
Martins, G. D., Galo, M. L. B. T., & Vieira, B. S. (2017). Detecting and mapping root-knot nematode infection in coffee crop using remote sensing measurements. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 10(12), 5395–5403. https://doi.org/10.1109/JSTARS.2017.2737618
Martins, J. A., Fiorio, P. R., Barros, P. P. da, Demattê, S., Molin, J. A. M., Cantarella, J. P., H., & Neale, C. M. U. (2020). Potential use of hyperspectral data to monitor sugarcane nitrogen status. Acta Scientiarum. Agronomy, 43. https://doi.org/10.4025/actasciagron.v43i1.47632
Mhatre, P. H., Karthik, C., Kadirvelu, K., Divya, K. L., Venkatasalam, E. P., Srinivasan, S. … Shanmuganathan, R. (2019). Plant growth promoting rhizobacteria (PGPR): A potential alternative tool for nematodes bio-control. Biocatalysis and agricultural biotechnology, 17, 119–128. https://doi.org/10.1016/j.bcab.2018.11.009
Muniz, M. F., Carneiro, R., Almeida, M. R., Campos, V. P., & Castagnone-Sereno, P. (2008). Diversity of Meloidogyne exigua (Tylenchida: Meloidogynidae) populations from coffee and rubber tree. Nematology, 10(6), 897–910. https://doi.org/10.1163/156854108786161418. & Cunha e Castro, J. M. da
Ndikumana, E., Minh, D. H. T., Nguyen, H. T. D., Baghdadi, D. C., Hossard, L., & Moussawi, I. (2018). Estimation of Rice Height and Biomass Using Multitemporal SAR Sentinel-1 for Camargue, Southern France. Remote Sensing, 10(9), 1394. https://doi.org/10.3390/rs10091394
Oliveira, M. F., Santos, A. F. de, Lacerda, L. N., Silva, R. P. da, & Vellidis, G. (2019). Estimation of peanut maturation using remote sensing and artificial neural networks. Annals of the XVI Meeting of Peanut Culture, 3. https://doi.org/10.17648/amendoim-2019-107243
Platt, J. C. (1999). Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods. Advances In Large Margin Classifiers, 10, 61–74. http://citeseer.ist.psu.edu/viewdoc/summary?doi=10.1.1.41.1639
Pompelli, M. F., Martins, S. C. V., Antunes, W. C., Chaves, A. R. M., & Matta, F. M. (2010). da. Photosynthesis and photoprotection in coffee leaves is affected by nitrogen and light availabilities in winter conditions. Journal of Plant Physiology, 167(13) 1052–1060. https://doi.org/10.1016/j.jplph.2010.03.001
Ranđelović, P., Đorđević, V., Milić, S., Balešević-Tubić, S., Petrović, K., Miladinović, J., & Đukić, V. (2020). Prediction of soybean plant density using a machine learning model and vegetation indices extracted from RGB images taken with a UAV. Agronomy, 10, 1108. https://doi.org/10.3390/agronomy10081108
Salamanca-Jimenez, A., Doane, T. A., & Horwath, W. R. (2017). Coffee response to nitrogen and soil water content during the early growth stage. Journal of Plant Nutrition and Soil Science, 180(5), 614–623. https://doi.org/10.1002/jpln.201600601
Sharifi, A. (2021). Yield prediction with machine learning algorithms and satellite images. Journal of the Science of Food and Agriculture, 101, 891–896. https://doi.org/10.1002/jsfa.10696
Shevade, S. K., Keerthi, S. S., Bhattacharyya, C., & Murthy, K. R. K. (2000). Improvements to the SMO Algorithm for SVM Regression. IEEE Transactions On Neural Networks, 11(5), https://doi.org/10.1.1.146.375&rep=rep1&type=pdf
Sheykhmousa, M., Mahdianpari, M., Ghanbari, H., Mohammadimanesh, F., Ghamisi, P., & Homayouni, S. (2020). Support Vector Machine Versus RF for Remote Sensing Image Classification: A Meta-Analysis and Systematic Review. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 13, 6308–6325. https://doi.org/10.1109/JSTARS.2020.3026724
Singhal, G., Bansod, B., Mathew, L., Goswami, J., Choudhury, B. U., & Raju, P. L. N. (2019). Chlorophyll estimation using multi-spectral unmanned aerial system based on machine learning techniques. Remote Sensing Applications: Society and Environment, 15. https://doi.org/10.1016/j.rsase.2019.100235
Sousa, I. C., Nascimento, M., Silva, G. N., Nascimento, A. C. C., Cruz, C. D., Silva, F. F. … Caixeta, E. T. (2021). Genomic prediction of leaf rust resistance to Arabica coffee using machine learning algorithms. Scientia Agricola, 78(4), https://doi.org/10.1590/1678-992x-2020-0021
Tolardo, A. L., Alves, G. C. S., Silva, G. F., da, Pereira, W. J., Silva, D. Z. da, & Silva, S. A. S. (2019). Biological control: isolated and in mixtures and genetic control of Meloidogyne exigua in coffee. Coffee Science, 14(2), 147–156. https://doi.org/10.25186/cs.v14i2
Villain, L., Sarah, J. L., Hernández, A., Bertrand, B., Anthony, F., Lashermes, P. … Carneiro, R. M. D. G. (2013). Diversity of root-knot nematodes parasiting coffee in Central America. Nematropica, 43, 194–206. https://journals.flvc.org/nematropica/article/view/82708
Wager, S., & Athey, S. (2018). Estimation and Inference of Heterogeneous Treatment Effects using Random Forests. Journal of the American Statistical Association, 113, 1228–1242. https://doi.org/10.1080/01621459.2017.1319839
Xiao, T., Tan, S., Shen, Q., & Ran, W. (2012). Bacillus cereus X5 suppresses root-knot nematode of tomato by colonizing in roots and soil. African Journal of Microbiology Research, 6(10), 2321–2327. https://doi.org/10.5897/AJMR11.1244
Zerbato, C., Rosalen, D. L., Furlani, C. E. A., Deghaid, J., & Voltarelli, M. A. (2016). Agronomic characteristics associated with the normalized difference vegetation index (NDVI) in the peanut crop. Australian Journal of Crop Science, 10(5), 758–764. https://doi.org/10.21475/ajcs.2016.10.05.p7167
Zha, H., Miao, Y., Wang, T., LI, Y., Zhang, J., Sun, W. … Kusnierek, K. (2020). Improving Unmanned Aerial Vehicle Remote Sensing-Based Rice Nitrogen Nutrition Index Prediction with Machine Learning. Remote Sensing, 12, 215. https://doi.org/10.3390/rs12020215
Zhou, C., Ye, H., Xu, Z., Hu, J., Shi, X., Hua, S. … Yang, G. (2019). Estimating Maize-Leaf Coverage in Field Conditions by Applying a Machine Learning Algorithm to UAV Remote Sensing Images. Applied Sciences, 9, 2389. https://doi.org/10.3390/app9112389
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pereira, F.V., Martins, G.D., Vieira, B.S. et al. Multispectral images for monitoring the physiological parameters of coffee plants under different treatments against nematodes. Precision Agric 23, 2312–2344 (2022). https://doi.org/10.1007/s11119-022-09922-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-022-09922-2