Abstract
Pre-planted cultivation is widely used in tobacco seedlings in south China to avoid damage from the cold. However, the underlying mechanisms of pre-planted cultivation that can improve the cold tolerance of tobacco are still unknown. In the present study, hormone measurement was performed on seedlings with both pre-planted cultivation and floating cultivation. The results showed that pre-planted tobacco seedlings accumulated more ABA than floating ones at the same growth stage. Further exogenous application of ABA inhibitors on pre-planted seedlings decreased their freezing tolerance. However, exogenous application of ABA enhanced the cold tolerance of floating-cultivated seedlings. To explore the ABA-responsive AREB/ABF members, we conducted a genome-wide analysis of the AREB/ABF family in tobacco and identified 16 members. Duplicated NtAREB/ABF genes showed high structural conservation and contributed to gene family expansion. Amino acid sequence alignments showed high sequence similarity among the gene products, especially in the basic leucine zipper (bZIP) region, indicating that they could possess functional redundancy. Gene expression profiles revealed most of the NtAREB/ABF members showed significantly improved expression after ABA treatment by qPCR. Three NtAREB/ABFs (Ntab0169850, Ntab0398220, and Ntab0577270) were randomly selected for further analysis. Subcellular localization analysis and electrophoretic mobility shift assays (EMSAs) demonstrated that they are nuclear-localized proteins and have an affinity to ABRE in vitro. Finally, transactivation activity assays revealed that Ntab0169850 and Ntab0398220 possess transcriptional activity in yeast. Collectively, these data demonstrate why pre-planted tobacco seedlings showed enhanced cold tolerance and provide a framework for future functional analysis of the AREB/ABF subfamily in tobacco.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cold stress is an environmental factor that limits and threatens tobacco production (Hu et al. 2016). Traditional floating-cultivated tobacco is cold-sensitive. When most of the seedlings are transplanted into a field in southern China in March or April, a cold spell can cause most of the tobacco seedlings to die, which leads to high economic losses (Xu et al. 2019). The cold tolerance of seedings generated by pre-planted cultivation can be enhanced. Pre-planted cultivation indicates that floating seedlings were transferred into plastic plots before they were transferred into the field. Since pre-planted cultivated tobacco seedlings are more cold-tolerant and suffer from less damage from cold spells, their use is spreading in South China (Fang 2015). However, the underlying mechanism of how pre-cultivation can improve the cold tolerance of tobacco is still unknown.
The plant hormone abscisic acid (ABA) plays a crucial role in plants responding to various environmental cues. A significant accumulation of endogenous ABA levels, which regulate many abiotic-stress-responsive genes, is induced after exposure to abiotic stresses (Fujita et al. 2011). This establishes the ABA signaling pathway in plants. In the absence of ABA, the ABA receptors pyrabactin resistance (PYR)/pyrabactin resistance 1-like (PYL) are unable to bind to type 2C protein phosphatases (PP2Cs) and inhibit PP2Cs' activities. PP2Cs deactivate SNF1-related protein kinase 2 (SnRK2). In the presence of ABA, the binding of ABA to PYR1/PYLs inactivates PP2Cs. Their inactivation results in the accumulation of active SnRK2s, which can phosphorylate downstream transcription factors, such as ABRF-binding protein (AREB)/ABRE binding factors (ABF) transcription factors, to stimulate ABA-dependent gene responses (Raghavendra et al. 2010). One of the SnRK2s, SnRK2.6/OST1 (open stomata 1), has been demonstrated to play a critical role in plants responding to cold stress (Ding et al. 2015).
AREB/ABF transcription factors bind to the cis-acting ABA-responsive elements (ABRE; PyACGTGGC) of ABA-dependent genes to activate their expression. In some species, AREB/ABF transcription factors have been isolated and identified to regulate plants’ tolerance to abiotic stress. Arabidopsis thaliana AREB/ABF genes have been isolated with yeast one-hybrid assays using ABRE as bait (Choi et al. 2000; Uno et al. 2000). AREB1/ABF2, AREB2/ABF4, and ABF3 are highly upregulated in vegetative tissues under osmotic conditions in A. thaliana (Choi et al.2000; Uno et al. 2000; Fujita et al. 2005). Overexpression and knockout of the genes in A. thaliana confirmed that they participate in ABA-dependent osmotic stress responses (Kang et al. 2002; Kim et al. 2004; Fujita et al. 2005; Furihata et al. 2006). The AREB/ABF homolog OsbZIP23 in rice (Oryza sativa) was identified and found to enhance plant abiotic stress tolerance, including drought and high salinity conditions (Xiang et al. 2008). SlAREB1 of Solanum lycopersicum was isolated and found to confer drought and salt stress tolerance (Orellana et al. 2010). The results indicate that AREB/ABFs play essential roles in plant abiotic stress responses. However, not all AREB/ABF subfamily members in tobacco have been identified.
In addition to ABA, SA and JA also contribute to cold tolerance of plants. Numerous studies support that SA regulate cold tolerance of plants including spinach and wheat (Min et al. 2018a, b; Wang et al. 2021). Presently, SA is identified to improve cold tolerance of a Solanaceae crop potato by transcriptomics and metabolomics analyses (Chen et al. 2023). JA can regulate the cold tolerance of a Solanaceae crop tomato (Min et al. 2018a, b). Therefore, it is intriguing to know whether the hormones are also related to cold tolerance of tobacco, other a named Solanaceae crop.
Common tobacco (Nicotiana tabacum), an economically important Solanaceae crop, is commonly used as a model system in plant biology because it can be easily transformed and has a relatively short generation time (Rabara et al. 2015). Tobacco is also an important economic crop. However, abiotic and biotic stresses have increasingly affected tobacco production. Genetic engineering approaches can help crops cope with adverse environmental conditions. Nevertheless, a lack of fundamental knowledge of the molecular mechanism underlying plant response to abiotic stress is a primary limitation slowing crop improvements using genetic engineering approaches (Ahmad et al. 2012). The draft genome sequences of two tobacco species, Nicotiana tabacum (Sierro et al. 2014) and Nicotiana benthamiana (Bombarely et al. 2012), provide a foundation to identify and functionally characterize stress-related genes.
In the present study, the accumulated ABA content has been shown to contribute to the cold tolerance of tobacco seedlings using pre-planted cultivation techniques. We also provided a comprehensive genome-wide survey of tobacco’s AREB/ABF subfamily and identified 16 AREB/ABF genes. As part of these analyses, we identified phylogenetic relationships, gene structures, and genomic locations of tobacco’s AREB/ABF subfamily. Furthermore, we determined the tissue-specific expression patterns of all the subfamily members and in response to ABA. Moreover, the DNA-binding activity of three members induced by cold conditions was determined by subcellular localization, transactivation activities, and the DNA-binding activity of three members. The data presented here provide a solid foundation for further functional characterization of each subfamily member, improving tobacco cultivation by shedding light on their roles in abiotic stress response.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of the cultivated tobacco variety K326 (N. tabacum) were germinated in MS medium and transplanted into plastic pots at 28 °C day/23 °C night with a 16-h photoperiod. For cold tolerance measurement, assays were performed according to our previous research (Fang 2015). Briefly, the six-leaf stage seedlings were transferred to a 12-h period with daytime (4 °C)/nighttime (0 °C) for 5 days. The samples were collected at 0 and 5 days after cold condition. The assays were biologically repeated for three times.
To determine the electrolyte leakage of the leaves, a previously described method with some modifications was performed (Qiu et al. 2020). In brief, 0.1 g expanded leaves after cold treatment were taken from individual plants and cut up. Then, the leaves were moved into glass tubes containing 5 mL of distilled water. Then, leaves were placed on a shaker with 160 rpm for 2 h directly at room temperature. At last, electrolyte leakage, using a FE30 Plus conductivity meter (Mettler Toledo, Ohio, USA), was determined as the ratio of conductivity measured in the water before and after the samples boiled.
For the exogenous application of ABA, 750 mM ABA was used on the floating seedlings, according to Loik and Nobel (1993) and Mang et al (2012). For the exogenous application of the ABA inhibitor nordihydroguaiaretic acid (NDGA), 100 mM NDGA was applied to the pre-cultivated seedlings using the methods of Ding et al. (2015). ABA and NDGA were applied to the seedlings 7 h before cold tolerance measurement. Roots, stems, leaves, buds, and apical buds were collected for gene expression analysis, and RNA was isolated from plants at the six-leaf stage, as previously described (Wang et al. 2015). For ABA treatment, tobacco plants at the six-leaf stage were transferred to 1/2 Murashige and Skoog (MS) solution and grown hydroponically for 1 week before treatment. After pretreatment, the plants were treated with 5 μM ABA in 1/2 MS solution. Then, roots and shoots were harvested together for immediate RNA extraction after 0, 4, 8, 12, and 24 h.
Sequence and Phylogenetic Analysis
The amino acid sequences of A. thaliana AREB/ABF subfamily members were retrieved from the Arabidopsis Information Resource (https://www.arabidopsis.org/). The tobacco AREB/ABF subfamily members were anchored using the amino acid sequences of known Arabidopsis AREB/ABF members as a query to search against the tobacco genome sequence database (https://solgenomics.net/tools/blast/?db_id=238) with the BLASTp program. An E-value of 1e-20 was applied to reduce false positives. The tobacco AREB/ABF proteins were further confirmed by the Pfam database (http://pfam.xfam.org/). The Circos software package (http://circos.ca) was used to show the location of genes on chromosomes (Zhang et al. 2018). Synteny analysis was conducted using MCScanX (Wang et al. 2012). Analysis of exon–intron organization was determined by alignment of the genomic and cDNA sequences using GSDS (http://gsds.cbi.pku.edu.cn). A phylogenetic tree of AREB/ABFs from A. thaliana and N. tabacum was constructed with MEGA X (http://www.megasoftware.net/) using the neighbor-joining algorithm, with 1000 bootstrap replications. Multiple sequence alignments of the tobacco AREB/ABF subfamily members were performed and visualized using Jalview 2.10 (http://www.jalview.org/).
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from approximately 100 mg of tissue using the SuperPure Plantpoly RNA Kit (Gene Answer, Beijing, China). RNA was treated with RNase-free DNase I (Gene Answer, Beijing, China) to remove any genomic DNA contamination. The concentration and quality of the RNA samples were evaluated with a NanoDrop 2000 instrument (Thermo Scientific, Waltham, MA, USA). Subsequently, first-strand cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Swiss). qRT-PCR analysis was performed using SYBR Green premix (2 ×) (Roche, Basel, Swiss) in a LightCycler® 96 Real-Time PCR System (Roche, Basel, Swiss) following the fast cycling protocol. The N. tabacum 26S gene was selected as an internal reference gene for normalization. The specific primer pairs for each tobacco AREB/ABF were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). All primers are listed in Table S1.
Hormone Content Measurement
Content of four types of hormones, including SA, JA, IAA, and ABA were measured. Leaf samples of 10-day-old seedlings were collected and were delivered to Metware Biotechnology Co. Ltd (Wuhan, China) (http://www.metware.cn/) to determine the endogenous hormones’ content of the samples. The quantification of the hormones was conducted using a high-performance liquid chromatography–tandem mass spectrometry (HPLC–MS/MS) system (HPLC, Agilent 1260 Infinity II LC system; MS, Agilent 6490 Triple Quad LC/MS, Agilent, Santa Clara, CA, USA). The content of SA, JA, IAA, and ABA were determined using the external standard method and is expressed as ng/g FW. Three biological replications were performed.
Subcellular Localization Analysis
The preparation and transformation of tobacco protoplasts were performed as previously described (Stigliano et al. 2013). Subcellular localization of GFP-NtAREB/ABFs was investigated 24 h after transformation by a PEG-mediated method with laser confocal fluorescence microscopy (Leica TCS‐SPE, Wetzlar, Germany) in N. benthamiana mesophyll protoplast cells. To generate the green fluorescent protein (GFP) fused target gene vectors, the full-length coding sequences (CDS) of Ntab0169850, Ntab0398220, and Ntab0577270 were amplified using tobacco cDNA as a template (refer to Table S1 for the primers used) and cloned into the pH7LIC‐N‐eGFP plasmid with Exnase II (Vazyme, Nanjing, China).
Transactivation Activity Assays in Yeast
To construct the vectors, the full-length CDSs of Ntab0169850, Ntab0398220, and Ntab0577270 were cloned into pGBKT7 (Clontech, USA), using primers listed in Table S1. Plasmids were transformed into AH109 yeast cells by the lithium acetate-mediated method (Clontech, Mountain View, CA, USA). After growth on SD/-Trp medium at 28 °C for 2 days, positive transformants were spotted in fresh, solid SD agar lacking Trp/His/Ade (SD/-Trp/-His/-Ade) to evaluate transactivation activity. A transformant harboring the empty pGBKT7 vector was used as a negative control.
Electrophoretic Mobility Shift Assays (EMSAs)
To construct the in vitro expression vectors, the CDS of Ntab0169850, Ntab0398220, and Ntab0577270 or the truncated CDS of Ntab0577270 were amplified with the specific primers listed in Table S1, and cloned in-frame in pET28a with NdeI and XhoI sites using Exnase II (Vazyme, China). N-terminal 6 × His-tagged fusion proteins were expressed in Escherichia coli Rosetta (DE3) cells (Novagen, USA) and purified using Ni Sepharose® 6 Fast Flow resin (GE Healthcare, USA) according to the manufacturer’s instructions.
For EMSAs, purified 6 × His-tagged proteins were used, and binding reactions were performed at 37 °C for 15 min with a FAM-labeled probe. For the competition assay, competitors (unlabeled ABRE or mutant ABRE) were added to reactions at 50- or 100-fold molar excess before adding the probe. Sequences of oligonucleotides were as follows: ABRE, aattccGGACACGTGGCGtaagct; mABRE, aattccGGACctacaGCCtaagct. The reaction mixtures were loaded in 1% agarose gels, subjected to electrophoresis in 1 × Tris/borate/EDTA buffer, and analyzed by an Amersham Imager 600 (GE Healthcare, USA).
Results
Seedlings with Pre-planted Cultivation Accumulated More ABA than Those with Floating Cultivation
As shown in Fig. 1a–d, pre-planted seedlings are more cold-tolerant than floating-cultivated ones after exposure to cold conditions. To avoid the effects of different vigors, agricultural traits of the seedlings with two types of cultivation were compared. As shown in Fig. 1e, there was no significant difference in height, stem diameter, leaf length, and leaf diameter between the seedlings from the two types of cultivation. The floating-cultivated seedlings show more fresh shoot weight and root weight than the pre-planted cultivated ones (Fig. 1e). This indicates that the cold tolerance of pre-planted cultivated seedlings was not determined by their vigor.
Cold resistance, agricultural traits, and hormone measurement of pre-cultivated and floating tobacco seedlings (bars = 8 cm). a Pre-planted seedlings before exposure to cold conditions. b Pre-planted seedlings after exposure to cold conditions. c Floating seedlings before exposure to cold conditions. d Floating seedlings after exposure to cold conditions; e measurement of agricultural traits of the seedlings; f hormone measurement of the seedlings. Asterisks indicate significant difference between the pre-planted seedlings and floating-cultivated ones detected by Student’s t-test, p < 0.05
As hormones play essential roles in plant abiotic stress tolerance (Zhu 2016), the hormone content of IAA, SA, ABA, and JA of the seedlings with two types of cultivation were further measured. SA, JA, and ABA were measured because the three hormones are known for their roles in cold tolerance of Solanaceae crops. The IAA was measured because the vigor of pre-planted seedlings and floating ones was apparently different in fresh root weight and fresh shoot weight. Since IAA determines the growth of plantlets and its content is affected by cold stress (Rahman 2013), whether the content of IAA in the two types of seedlings before cold treatment different or not was interesting to analyze.
The IAA, SA, and JA content show an insignificant difference in seedlings from two types of cultivation (Fig. 1f). However, the ABA content of pre-planted cultivated seedlings exceeded that of floating cultivation (Fig. 1f). It was assumed that the higher accumulated ABA content could be related to the freezing tolerance of the pre-planted method. Therefore, an assay of exogenous application of ABA and ABA inhibitor nordihydroguaiaretic acid (NDGA) on the seedlings was performed.
ABA Contributes to the Cold Tolerance of Pre-planted Cultivated Seedlings
The ABA inhibitor was added to the pre-planted cultivated tobacco seedlings before cold tolerance measurement. Meanwhile, ABA was added to the floating seedlings before cold tolerance measurement. As shown in Fig. 2a–c, the leaves of pre-planted cultivated tobacco seedlings treated with ABA inhibitors suffered from more cold damage and showed significantly higher electrolyte leakage comparing with mock. It implied that ABA inhibitors decreased cold tolerance of pre-planted cultivated tobacco seedlings. In contrast, the leaves of floating-cultivated tobacco seedlings with ABA treatment suffered from less cold damage and showed significantly lower electrolyte leakage comparing with mock. It implied that ABA increased cold tolerance of floating-cultivated tobacco seedlings (Fig. 2d–f). Taken together, ABA was regarded to contribute to the cold tolerance of pre-planted cultivated seedlings.
Cold resistance measurement of pre-cultivated seedlings with NDGA treatment and floating tobacco seedlings with ABA treatment (bars = 2 cm). a Pre-planted seedlings with NDGA treatment before exposure to cold conditions. b Pre-planted seedlings with NGPA treatment after exposure to cold conditions. c Electrolyte leakage of pre-planted seedlings with NDGA and water treatment (mock) after exposure to cold conditions. d Floating seedlings with ABA treatment before exposure to cold conditions. e Floating seedlings with ABA treatment after exposure to cold conditions. f Electrolyte leakage of floating seedlings with ABA and water treatment (mock) after exposure to cold conditions. Bars indicate 8 cm. Asterisks indicate significant difference of between the mock and the plantlets with NDGA/ABA detected by Student’s t-test, p < 0.05
Isolation of the AREB/ABF Subfamily from Nicotiana tabacum
Since ABA signaling is transferred by AREB/ABF transcription factors, it is important to assess which AREB/ABF family members participate in the cold tolerance of seedings with pre-planted cultivation. First, the AREB/ABF subfamily members from N. tabacum were identified. Five AREB/ABF members of A. thaliana were used: ABF1, AREB1/ABF2, ABF3, AREB2/ABF4, and AREB3. We identified 16 distinct gene loci in the N. tabacum genome using a comprehensive search process. We further constructed a phylogenetic tree of the AREB/ABF subfamily to verify and elucidate the relationships between N. tabacum and A. thaliana AREB/ABF members (Fig. 3). The subfamily was divided into two small subgroups. Six putative tobacco AREB/ABFs, shown to have relatively high similarities to A. thaliana AREB3, were classified into subgroup A and contained three exons. The remaining ten putative tobacco AREB/ABF genes and A. thaliana AREB1/ABF2, ABF3, AREB2/ABF4, and ABF1 comprise subgroup B. Ntab0011370 and Ntab0577270 possess five and three exons, respectively, and the other eight putative tobacco AREB/ABFs contain four exons.
The chromosomal distributions of the sixteen putative NtAREB/ABFs were visualized using Circos software. Thirteen genes were identified on 10 of 24 chromosomes, while three genes (Ntab0225270, Ntab0370160, and Ntab0740100) were present on the scaffolds (Fig. 4). The number of AREB/ABF genes on each chromosome ranges from one to three (Fig. 4). Ntab0810900, Ntab0577270, Ntab0323520, Ntab0011370, Ntab0392500, Ntab0959150, Ntab0864800, and Ntab0169850 are localized on chromosomes 1, 4, 7, 8, 12, 16, 18, and 22, respectively. Furthermore, two genes were identified on chromosome 11 (Ntab0131720 and Ntab0766440), and three genes were present on chromosome 19 (Ntab0526530, Ntab0398220, and Ntab0634760).
Duplication analysis revealed expansion of the AREB/ABF gene family in tobacco. Synteny analysis revealed that 14 of 16 AREB/ABFs derived from expansion, including Ntab0810900, Ntab0766440, Ntab0392500, Ntab0398220, Ntab0323520, Ntab0131720, Ntab0959150, Ntab0634760, Ntab0526530, Ntab0864800, Ntab0740100, Ntab0370160, Ntab0169850, and Ntab0225270 (Fig. 4). These results indicate that segmental duplication occurred in the tobacco AREB/ABF gene family.
Protein sequence alignment of the 16 tobacco AREB subfamily members was performed and visualized using Jalview. All five A. thaliana AREB/ABF members contained the bZIP domain, which consists of two motifs: a leucine zipper domain responsible for dimerization and a basic region involved in specific DNA target binding (Hu et al. 2016). Each bZIP domain of the 16 tobacco sequences comprises 52 amino acids (Fig. 5). Within each bZIP domain, 32 amino acids occupying 61.5% of the total region are entirely conserved (Fig. 5), indicating that the NtAREB/ABFs could recognize similar cis-elements. Four additional conserved domains are present in all of the NtAREB/ABF (Fig. 5), three of which are at the N-terminus (C-1, C-2, and C-3) and one at the C-terminus (C-4) partial protein (Choi et al. 2000; Uno et al. 2000 Finkelstein and Lynch 2000). ABA-dependent phosphorylation of multiple RXXS/T sites in the conserved regions reportedly contributes to AREB/ABFs activation (Furihata et al. 2006; Zhu et al. 2007; Fujita et al. 2011). These conserved RXXS/T sites are also distributed in the C1 to C4 domains of most tobacco AREB/ABF subfamily members (Fig. 5), indicating that these members could undergo activation regulation by altering their phosphorylation status on the RXXS/T sites.
Protein sequence analysis of tobacco AREB/ABF members. Identical and conserved residues are shaded in blue and light blue, respectively. The red panes indicate the positions of the C1 to C4 conserved domains and basic regions. Potential phosphorus residues (R-X-X-S/T) corresponding to phosphorylation sites characterized in Arabidopsis AREB1 are indicated by brown boxes. Green triangles indicate the positions of conserved Leu residues in the Leu zipper domain
The Tissue-Specific Expression Pattern of AREB/ABF Members and Their Expression Pattern in the Floating Seedlings in Response to Exogenous Application of ABA Treatment
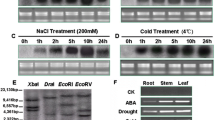
We performed qRT-PCR assays to determine the tissue-specific expression profiles of NtAREB/ABFs. Most NtAREB/ABFs showed similar tissue-specific expression patterns. Specifically, 12 NtAREB/ABFs were highly expressed in the apical bud (Fig. 6), suggesting that they could play roles in shoot growth. In contrast, Ntab0392500 and Ntab0398220 were predominantly expressed in the root tissue (Fig. 6). Tissue-specific expression patterns of NtAREB/ABFs indicated that most transcripts were abundant in both root and leaf tissue, which is consistent with known functions of ABA that broadly regulate adaptive plant responses to various stresses (Finkelstein et al. 2002; Yamaguchi-Shinozaki and Shinozaki 2006).
Tissue expression pattern analysis of NtAREB/ABF genes. The tobacco tissues, including roots, stems, leaves, buds, and apical buds, were harvested from plants with visible flowers. Nt26s (NbS00007518g0011.1) were used to normalize relative expression levels. Columns represent the mean ± standard deviation (SD, n = 3)
The expression pattern of NtAREB/ABFs in response to ABA is shown in Fig. 7. Expression of nearly all the NtAREB/ABFs increased in response to ABA except Nta0634760. It implied that most of the NtAREB/ABFs might participate in ABA signaling transferring. Seven NtAREB/ABFs genes upregulated gradually, which included, Ntab0011370, Ntab0169850, Ntab0225270, Ntab0392500, Ntab0577270, Ntab0766440, Ntab0959150. One of the NtAREB/ABFs, Ntab0398220, was induced only after 24 h. To further analyze the potential function of the NtAREB/ABFs, three randomly selected gene including Ntab0169850, Ntab0398220, and Ntab0577270 were used to further analysis.
The change in expression of the NtAREB/ABF subfamily in response to ABA treatment over time. Nt26s were used to normalize relative expression levels. Columns represent the mean ± standard deviation (SD, n = 3). For each gene, bars labeled with the same lower-case letters indicate significant differences among treatments, based on Duncan's multiple range test (p < 0.05)
Characterization of the Three AREB/ABF Genes
To identify whether the three AREB/ABF genes are functional transcription factors, subcellular localization of the three AREB/ABF members was determined in N. benthamiana mesophyll protoplasts. The results indicated that all three AREB/ABFs were specifically targeted in the nucleus (Fig. 8a).
Characterization of three NtAREB/ABF members. a Confocal images showing three AREB members localized to the nucleus. Constructs expressing GFP alone and GFP-AREB/ABF fusion proteins were transiently expressed in N. benthamiana protoplasts isolated from mesophyll cells. Images were taken at 24 hpi by CLSM. The green color indicates GFP fluorescence, and the red color depicts chlorophyll autofluorescence. Bars = 10 μm; b detection of transcriptional activation activity of three AREB/ABF members using a yeast system. The transcriptional activation capacity of these proteins was analyzed by assessing growth in the SD/-Trp-His-Ade medium. Yeast transformants carrying pGBKT7 empty vector were used as negative controls. c Electrophoretic mobility shift assays (EMSAs). Extracts of E. coli before (0 h) and after induction with 0.4 mM IPTG (16 °C for 16 h) and the proteins purified with Ni Sepharose® 6 Fast Flow resin (E) are shown on SDS-PAGE. The FAM-labeled ABRE oligonucleotide was employed in a mobility shift assay. In each assay, 1 μg of purified recombinant full-length or truncated protein was used. Triangles indicate shifted bands
A yeast-based assay was used to test the transcriptional activity of Ntab0169850, Ntab0398220, and Ntab0577270. Transformants carrying pGBK-Ntab0169850 and pGBK-Ntab0398220 normally grew on a medium lacking tryptophan, histidine, and adenine (SD-Trp-His-Ade), while transformants harboring pGBK-Ntab0577270 failed to grow, similar to the negative control (Fig. 8b). The results indicated that Ntab0169850 and Ntab0398220 have transcriptional activity.
We further performed an in vitro electrophoretic mobility shift assay (EMSA) to examine the DNA-binding affinity of Ntab0169850, Ntab0398220, and Ntab0577270. Ntab0169850 and Ntab0398220 could be successfully purified from the bacteria, while the full-length version of Ntab0577270 failed to purify. To overcome this problem, we truncated the bZIP domain of Ntab0577270 (Ntab0577270Daa 301–411) and successfully purified it. As shown in Fig. 8c, a specific shifted band was observed with all three recombinant fusion proteins. The binding was gradually abolished by adding competitor probes to the reaction mixtures. In contrast, adding an equal amount of mutant competitor probe had no effect, revealing strong binding between recombinant proteins and the ABRE element.
These results indicate that Ntab0169850 and Ntab0398220 could play important roles in response to ABA.
Discussion
Pre-planted seedlings exhibited less cold injury than those with floating cultivation (Fang 2015) (Fig. 1). Our previous research demonstrated that the cell wall in the cells of pre-planted tobacco seedlings was thicker than in seedlings produced by floating cultivation, which could contribute to the cold tolerance of pre-planted seedlings (Fang 2015). However, whether hormones participate in the cold tolerance of pre-planted seedlings is still unknown. In the present study, ABA content was higher in pre-planted tobacco seedlings than in floating-cultivated ones. Moreover, the exogenous application of ABA on the floating-cultivated seedlings can enhance cold resistance. The exogenous application of nordihydroguaiaretic acid (NDGA) on pre-planted seedlings showed reduced cold resistance. The result indicates that a higher ABA content contributes to the cold tolerance of pre-planted seedlings.
ABA contributes to cold resistance (Xin and Browse 2000). The OST1, named for its central role in cold acclimation, is a member of the SnRK2 family (Ding et al. 2015), a component of the ABA signaling pathway. However, ABA content mildly increased in Arabidopsis thaliana after exposure to cold stimulus (Ding et al. 2015), suggesting that the ABA signaling pathway, instead of ABA itself, is the critical factor in determining cold resistance. The OST1 was activated by EGR2 instead of the cold-induced increase in ABA content (Ding et al. 2015). However, our research found that the ABA content was improved in pre-planted tobacco seedlings (Fig. 1). Additionally, the exogenous application of ABA can enhance the cold resistance of cold-sensitive floating-cultivated seedlings, while the exogenous application of ABA inhibitors can reduce the cold tolerance of pre-planted seedlings (Fig. 2). This indicates that a higher ABA content contributed to the cold tolerance of pre-planted tobacco seedlings.
AREB/ABF genes play vital roles in plant stress resistance. The AREB/ABF subfamily has been identified and characterized in at least A. thaliana and Populus (Choi et al. 2000; Ji et al. 2013). The recent release of high-quality draft genomes of three main tobacco varieties provided an opportunity to evaluate the AREB/ABF family in tobacco (Sierro et al. 2014). The present study identified 16 NtAREB genes in the N. tabacum genome (Fig. 3). The identification and characterization of the gene structure and conserved motifs of NtAREB/ABF facilitates a better understanding of the evolution of the AREB/ABF gene subfamily in tobacco. The phylogenetic tree divides these subfamily members into two subgroups, and almost all members within each subgroup share a similar gene structure (Fig. 3). Protein sequence analysis of the AREB/ABFs revealed the presence of four conserved regions containing potential phosphorylation sites in addition to the highly conserved bZIP region and highly conserved bZIP regions (Fig. 5). Activation of the corresponding phosphorylation sites in Arabidopsis AREB1 plays a critical role in regulating its transcriptional activity, and overexpression of the phosphorylation-mimicking AREB1 mutant can stimulate many ABA-inducible genes independent of ABA treatment (Furihata et al. 2006).
In A. thaliana, AREB1/ABF2, AREB2/ABF4, and ABF3 all respond to ABA treatment. They are master transcription factors that cooperatively regulate the ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation (Yoshida et al. 2010). The other member of this subfamily, ABF1, which exhibits lower expression levels relative to AREB1/ABF2, AREB2/ABF4, and ABF3, was shown to be a functional homolog of the three AREB/ABFs in ABA-dependent gene expression (Fujita et al. 2005). In the present study, all ten NtAREB/ABFs are in the same phylogenetic clade of A. thaliana AREB1/ABF2, AREB2/ABF4, ABF3, and ABF1 (Fig. 3), which were significantly induced by ABA (Fig. 7). Among them, the expression of five tobacco subfamily members was dramatically induced by ABA, while five members were moderately upregulated after ABA treatment (Fig. 7). Our data suggest that these ten members could be involved in regulating ABRE-dependent ABA signaling, suggesting a more complex ABA downstream regulatory network in tobacco than in A. thaliana.
Three members (Ntab0169850, Ntab0398220, and Ntab0577270) were selected for further functional analysis. Subcellular localization and in vitro DNA-binding assays confirmed that they all targeted the nucleus and bound to ABRE (Fig. 8), a highly conserved cis-element in ABA/stress-inducible promoters (Fujita et al. 2013). These results are consistent with previous studies showing that A. thaliana AREB1, AREB2, and ABF3 are localized in nuclei in various tissues (Yoshida et al. 2010). Furthermore, A. thaliana ABF1 and ABF3 bind ABRE in vitro (Choi et al. 2000). Transactivation analysis of the three tobacco NtAREB/ABF members in yeast indicated that, despite sharing high sequence similarity, Ntab0169850 and Ntab0398220 possessed transactivation activity, while Ntab0577270 did not. A recent study reported that OsbZIP46, a member of the AREB/ABF bZIP subfamily in rice, had no transactivation activity in yeast. Further investigation indicated an intrinsic D domain that repressed OsbZIP46 activity (Tang et al. 2012). In the future, it is important to determine if a similar domain is present in Ntab0577270 that is involved in regulating its activity.
The homolog of Ntab0169850 in Arabidopsis is AtABF4 (abscisic acid-responsive element binding factor 4), while the homolog of Ntab0398220 in Arabidopsis is AtABF3. AtABF3 and AtABF4 have largely overlapping functions. Both of them are master transcription factors that cooperatively regulate the downstream ABRE-dependent gene expression of ABA signaling under drought stress (Yoshida et al. 2010). In addition to drought tolerance, both AtABF3 and AtABF4 are also salty-responsive. The salt treatment can induce the expression of AtABF3 and repress the expression of AtABF4 (Fernando et al. 2018). Moreover, both AtABF3 and AtABF4 regulate salinity tolerance of Arabidopsis (Du et al. 2023). Although both AtABF3 and AtABF4 regulate drought and salinity tolerance in Arabidopsis, whether AtABF3 and AtABF4 can regulate cold tolerance are still unknown. It was speculated that AtABF3 and AtABF4 probably regulate cold tolerance. It would be interesting to decipher whether the homologs of AtABF3 and AtABF4 in tobacco, Ntab0169850 and Ntab0398220 can also regulate cold tolerance. Their transgenic lines are under the way and we would decipher them function in cold tolerance in our further research.
Conclusions
In this study, we found that a higher ABA content contributes to enhanced cold tolerance of pre-cultivated seedlings. A comprehensive analysis of the AREB/ABF subfamily in tobacco was conducted, and detailed information on its members was provided. Sixteen putative AREB/ABF genes were identified in tobacco; these duplicated NtAREB/ABF genes showed high structural conservation and contributed to gene family expansion. The high amino acid sequence similarity among the gene products, especially in the basic leucine zipper (bZIP) region, indicated that they could possess overlapping roles. Gene expression profiles showed that three NtAREB/ABFs were dramatically upregulated in response to ABA. Moreover, three NtAREB/ABFs (Ntab0169850, Ntab0398220, and Ntab0577270) were confirmed to be nuclear-localized proteins and bind to ABRE in vitro. Ntab0169850 and Ntab0398220 were demonstrated to possess transcriptional activity in a yeast system, indicating that they could act as functional transcription factors to regulate ABA-responsive gene expression in tobacco plants. These results provide practical information for future efforts to characterize specific gene functions of this subfamily.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Ahmad P, Ashraf M, Younis M, Hu X, Kumar A, Akram NA, Al-Qurainy F (2012) Role of transgenic plants in agriculture and biopharming. Biotechnol Adv 30(3):524–540
Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant-Microbe Interact 25(12):1523–1530
Chen L, Zhou FY, Chen Y, Fan YQ, Zhang KK, Liu Q, Tu W, Jiang FJ, Li GC, Zhao HB, Song BT (2023) Salicylic acid improves the constitutive freezing tolerance of potato as revealed by transcriptomics and metabolomics analyses. Int J Mol Sci 24(1):609–629
Choi HI, Hong JH, Ha JO, Kang KSY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275(3):1723–1730
Ding Y, Hui L, Zhang X et al (2015) OST1 Kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32(3):278–289
Du J, Zhu X, He K, Kui M, Zhang J, Han X, Fu Q, Jiang Y, Hu Y (2023) CONSTANS interacts with and antagonizes ABF transcription factors during salt stress under long-day conditions. Plant Physiol
Fang T (2015) Study on flue-cured tobacco two section type floating system. J Anhui Agric Sci 43(15):47–48
Fernando VCD, Al Khateeb W, Belmonte MF, Schroeder DF (2018) Role of Arabidopsis ABF1/3/4 during det1 germination in salt and osmotic stress conditions. Plant Mol Biol 97(1):149–163
Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(suppl 1):S15–S45
Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12(4):599–609
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17(12):3470–3488
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124(4):509–525
Fujita Y, Yoshida T, Yamaguchi-Shinozaki K (2013) Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant 147(1):15–27
Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103(6):1988–1993
Hu R, Zhu X, Xiang S, Zhan Y, Zhu M, Yin H, Zhou Q, Zhu L, Zhang X, Liu Z (2016) Comparative transcriptome analysis revealed the genotype specific cold response mechanism in tobacco. Biochem Biophys Res Commun 469:535–541
Ji L, Wang J, Ye M, Li Y, Guo B, Chen Z, Li H, An X (2013) Identification and characterization of the Populus AREB/ABF subfamily. J Integr Plant Biol 55(2):177–186
Kang J-y, Choi H-i, Im M-y, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14(2):343–357
Kim JB, Kang JY, Kim SY (2004) Over-expression of a transcription factor regulating ABA-responsive gene expression confers multiple stress tolerance. Plant Biotechnol J 2(5):459–466
Loik ME, Nobel PS (1993) Exogenous abscisic acid mimics cold acclimation for cacti differing in freezing tolerance. Plant Physiol 103(3):871–876
Mang HG, Qian W, Zhu Y, Qian J, Kang HG, Klessig DF, Hua J (2012) Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24(3):1271–1284
Min D, Li F, Zhang X, Cui X, Shu P, Dong L, Ren C (2018a) SlMYC2 involved in methyl jasmonate-induced tomato fruit chilling tolerance. J Agric Food Chem 66(12):3110–3117
Min K, ShowmanL PA, Arora R (2018b) Salicylic acid-induced freezing tolerance in spinach (Spinacia oleracea L.) leaves explored through metabolite profiling. Environ Exp Bot 156:214–227
Orellana S, Yanez M, Espinoza A, Verdugo I, Gonzalez E, RUIZ-LARA S, Casaretto J (2010) The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ 33(12):2191–2208
Qiu S, Zhang J, He J, Sha W, Li M, Zhao Y, Zhai Y (2020) Overexpression of GmGolS2-1, a soybean galactinol synthase gene, enhances transgenic tobacco drought tolerance. Plant Cell, Tissue Organ Cult 143:507–516
Rabara RC, Tripathi P, Reese RN, Rushton DL, Alexander D, Timko MP, Shen QJ, Rushton PJ (2015) Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genomics 16(1):484
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15(7):395–401
Rahman A (2013) Auxin: a regulator of cold stress response. Physiol Plant 147(1):28–35
Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5:3833
Stigliano E, Faraco M, Neuhaus J-M, Montefusco A, Dalessandro G, Piro G, Gi DS (2013) Two glycosylated vacuolar GFPs are new markers for ER-to-vacuole sorting. Plant Physiol Biochem 73:337–343
Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158(4):1755–1768
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97(21):11632–11637
Wang W, Wang X, Huang M, Cal J, Zhou Q, Dai T, Jiang D (2021) Alleviation of field low-temperature stress in winter wheat by exogenous application of salicylic acid. J Plant Growth Regul 40:811–823
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T, Jin H, Marler B, Guo H (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49–e49
Wang Z, Wei P, Wu M, Xu Y, Li F, Luo Z, Zhang J, Chen A, Xie X, Cao P (2015) Analysis of the sucrose synthase gene family in tobacco: structure, phylogeny, and expression patterns. Planta 242(1):153–166
Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148(4):1938–1952
Xin Z, Browse J (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23(9):893–902
Xu J, Chen Q, Liu P, Jia W, Chen Z, Xu Z (2019) Integration of mRNA and miRNA analysis reveals the molecular mechanism underlying salt and alkali stress tolerance in tobacco. Intl J Mol Sci 20:2391–2408
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61(4):672–685
Zhang H, Jin J, Jin L, Li Z, Xu G, Wang R, Zhang J, Zhai N, Chen Q, Liu P, Chen X, Zheng Q, Zhou H (2018) Identification and analysis of the chloride channel gene family members in tobacco (Nicotiana tabacum). Gene 676:56–64
Zhu JK (2016) Abiotic Stress Signaling and Responses in Plants. Cell 167(2):313–324
Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, Xu YH, Zhang XY, Zhang DP (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19(10):3019–3036
Funding
This research was funded by the Chenzhou Company of Hunan Tobacco Company (CZYC2019JS03), Key Scientific and Technological Project of Henan Province (192102110003), Grants of Zhengzhou Tobacco Research Institute (902014CA0440, 902017CA0200). The funding body has no roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
QC and MF designed and performed experiments, analyzed data, and wrote draft manuscript. PL, ZL, QZ, HZ, JL, and PC analyzed data and revised the manuscript. QC conceived and supervised the project and critically revised the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
Pre-planted cultivation is widely used in tobacco seedlings to avoid cold damage. In the present study, higher content ABA was proven to contribute to cold tolerance of pre-planted cultivation by content measurement, exogenous application of ABA and its inhibitors. Furthermore, the ABA-responsive AREB/ABF family members were identified and analyzed.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Q., Liu, P., Li, Z. et al. Accumulated Endogenous Abscisic Acid Contributes to the Cold Tolerance of Pre-planted Cultivated Tobacco. Plant Mol Biol Rep 42, 151–164 (2024). https://doi.org/10.1007/s11105-023-01412-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-023-01412-7