Abstract
This study aimed to evaluate the genetic diversity (GD) and association analysis for 100-kernel weight (100KW) and understand the population structure and genetic distance among Korean waxy maize inbred lines collected in South Korea. The average value of 100KW was 19.8 ± 4.7 g, with statistically significant differences between low and high groups based on the average value. GD analysis using 150 simple sequence repeat (SSR) loci showed an average of 3.94 alleles and 4.32 genotypes per locus. The average GD was 0.581, and the average polymorphic information content (PIC) was 0.522. Pairwise genetic similarity coefficients ranged from 0.229 to 0.893 with an average of 0.385. A distance-based unweighted pair group method with arithmetic mean (UPGMA) dendrogram showed all the inbred lines dividing into five subgroups at a genetic similarity of 35.1%. Group I consisted of 16 inbred lines, with nine low inbred lines and seven high inbred lines. Group II included 20 inbred lines, composed of 10 low and 10 high 100KW inbred lines. Groups III and V each had two high or low inbred lines, respectively. Group IV had only one high inbred line. The model-based population structure classified K = 2 and K = 4 based on delta K. Association analysis between 100KW and SSR markers identified five marker-trait associations (MTAs) involving four SSR markers. The information from this study may support opportunities for selecting inbred lines with desirable traits and for developing new waxy maize varieties in South Korea. This could enable maize breeders to improve crop yield and quality through marker-assisted selection (MAS).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is the most popular cereal crop and is widely cultivated for use as staple food, livestock feedstock, edible oil, and biofuel (Mackay 2009). Maize ranks second after wheat in terms of harvested area, but its production and yields have been rising globally because of increases in the harvested area of maize. World production, harvested area, and yield for maize were 1162.4 million metric tons, 202.0 million ha, and 5.8 t/ha, respectively, in 2020 (FAOSTAT 2020). Maize, a naturally cross-pollinated plant, undergoes pollination by other maize plants for over 97% of its kernels under field conditions (Brittan 2006). Maize hybrids typically exhibit greater yield and quality, such as uniform grain color and size, compared with open-pollinated varieties (OPVs) in open-field conditions owing to the effect of heterosis, which results in higher vigor and productivity (van Heerwaarden et al. 2009). Therefore, almost all commercially cultivated maize varieties are hybrids (Wagner et al. 2021). Among the different types of hybrids, such as single-cross, three-way, and double cross hybrids, single-cross hybrid refers to a hybrid maize variety produced by crossing two inbred lines, and this type is widely used in modern agriculture because of its high yield potential and uniformity compared with other types of hybrids (Duvick 2001; MacRobert et al. 2014). However, F1 seed productivity of a single-cross hybrid is lower than that of other hybrids because the female is an inbred line (MacRobert et al. 2014). To ensure effective hybrid development, it is important to possess sufficient understanding of the genetic diversity among maize inbred lines, as well as their breeding potential (Adu et al. 2021). This knowledge serves as a guide when selecting appropriate parental candidates for the desired hybrids. Consequently, development and selection of elite inbred lines are critical for the success of a hybrid breeding program. Normal maize, which is widely cultivated and utilized in food and feed worldwide, contains a starch that is approximately 25% amylose and 75% amylopectin (Nelson and Rines 1962). On the other hand, waxy maize, in which the starch is exclusively made up of amylopectin, is a distinct type of maize that is utilized in food production in China and South Korea (Devi et al. 2017). A large number of waxy maize inbred lines have been developed from various sources in South Korea. These inbred lines were either obtained from local farmers in South Korea and other countries or collected from the Genebank at the Rural Development Administration (RDA) in South Korea.
According to breeding practice, the performance of parental inbred lines is not consistent with the hybrid performance, i.e., elite hybrid varieties are not necessarily derived from elite parental inbred lines (Yu et al. 2020). Therefore, breeders and researchers for maize hybrids should evaluate a parental inbred line by its performance and potential to produce superior hybrids (Riedelsheimer et al. 2012). Information on the genetic diversity (GD) and relationships among diverse maize inbred lines has a significant impact on improvement of new cultivars because it is useful for planning crosses for hybrid and inbred line development, assigning lines to heterotic groups, and protecting plant variety (Hallauer et al. 1988; Pejic et al. 1998). Traditional methods, such as morphological data and pedigree record, had limitations in assessing GD and relationships among inbred lines because of environmental interactions and being labor intensive. Using molecular markers can overcome these limitations, and several researchers have attempted to estimate GD and relationships using various molecular markers (Melchinger et al. 1992; Russell et al. 1997; Dao et al. 2014; Wang et al. 2008). Initial studies on predicting hybrid performance using molecular markers relied on calculating the genetic distance between lines. However, correlations between GD of parents and hybrid performance were not significant in previous studies (Melchinger 1999, Jordan et al. 2003). Heterosis, also known as hybrid vigor, is a complex phenomenon that cannot be attributed to a single hypothesis. It is influenced by various factors including the genetic distance between the parents, the mode of reproduction, the specific traits being studied, and the environmental conditions in which parental lines perform well (Kaushik et al. 2018).
As a result of population growth and a decrease in cultivated land, food insecurity has become a pressing issue in recent years. Consequently, improving crop yield has become the most important aspect of maize breeding programs (Li et al. 2011). Thus, there has been an increasing focus on grain yield and yield-related traits of maize. Kernel number and weight are essential components for crop yield and are also key traits for domestication and breeding (Kesavan et al. 2013). Kernel number is generally considered the main factor determining grain yield because maize accommodates environmental variation by adjusting seed number (Borrás et al. 2004; Egli 1998). Although relatively stable in response to environmental variation, kernel weight can still have an impact on grain yield (Borrás and Gambín 2010; Peltonen-Sainio et al. 2007). In South Korea, the Maize Experiment Station operated by the Kangwon Agricultural Research and Extension Services has developed a substantial number of elite inbred lines. These elite inbred lines have been used as parental lines for superior single-cross hybrid cultivars in South Korea. To ensure the long-term success of maize breeding programs in South Korea, it is necessary to conduct a genetic characterization of these inbred lines because they are rarely analyzed for GD and population structure for breeding programs using molecular markers. Therefore, this study genotyped a number of waxy maize inbred lines along with other types of maize, such as popcorn and flint maize, using simple sequence repeat (SSR) markers and evaluated 100-kernel weight (100KW) as a component of grain yield for each inbred line. The objective of this study is not only to investigate the GD and population structure using SSR markers for waxy maize inbred lines, which were used for developing superior varieties, but it is also to confirm the SSR markers related to 100KW using association analysis in waxy maize inbred lines collected from South Korea.
Materials and Methods
Plant Materials and Phenotypic Analysis
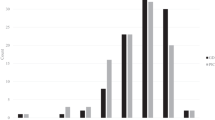
This study used 41 elite maize inbred lines that were generated and obtained from the Maize Experiment Station, Gangwon Agricultural Research and Extension Service, Hongcheon. Although almost all the inbred lines were waxy maize, there were some popcorn and flint maize inbred lines. The inbred lines are utilized as parental lines to produce various F1 hybrid varieties (Table 1). For example, HW9 and HW3 served as parental lines for Mibaek2ho, which is the most popular waxy maize variety in South Korea (Park et al. 2007). Although the series of Gangwonchal and Saekchalgyo are still unregistered, most of the maize varieties that were developed by crossing these inbred lines have been registered as cultivars in the KSVS and are being used in South Korea. Ten individuals of each inbred line were evaluated using completely randomized design with three replicates and 70 × 25 cm of planting density for 100KW of the 41 elite maize inbred lines at the College of Agriculture and Life Sciences, Kangwon National University, Chuncheon, Gangwon-do, in 2022. For association analysis, measurement of 100KW was performed using the topmost ears from each line and measured the seeds from the middle portion of each ear, and this study used three replicates for each line and an average value (Fig. 1).
DNA Extraction and SSR Amplification
Genomic DNA for the waxy maize inbred lines was obtained from young leaves using the Dellaporta et al. (1983) method with minor modifications. In the analysis of GD, population structure, and the association between markers and 100KW traits in the waxy maize inbred lines, a total of 150 SSR markers were utilized, with an average of 15 loci per each of the 10 maize chromosomes (Table 3, Supplementary Table 1). Information on the SSR markers, including their chromosome location, and sequences of forward and reverse primers were obtained from MaizeGDB (http://www.maizegdb.org/).
An amplification test for SSRs was carried out using an Ex Taq polymerase chain reaction (PCR) kit (Takara, Ohtsu, Japan). For PCR amplification of the SSR loci, a total volume of 20 µL of the product contained 20 ng of genomic DNA, 1 × Ex Taq buffer, 0.5 µM of forward and reverse primers, 0.2 mM dNTP mixture, and 1 unit of Ex Taq polymerase. The PCR protocol involved an initial denaturation step at 94 °C for 5 min, followed by a second step consisting of denaturation at 94 °C for 1 min, annealing at 65 °C for 1 min, and extension at 72 °C for 2 min. The annealing temperature was then gradually reduced in increments of 1 °C following every annealing stage until it reached a final annealing temperature of 55 °C. The second step was then repeated 36 times. After the completion of the two steps, a final third step was carried out at 72 °C for 5 min for extension.
Following the PCR reaction, DNA electrophoresis was conducted using a mini vertical electrophoresis system (MGV-202–33, CBS Scientific Company, San Diego, USA). Three µl of the PCR product was mixed with 3 µl of formamide loading dye (composed of 98% formamide, 0.02% bromophenol blue (BPB), 0.02% xylene C, and 5 mM NaOH). Two µl of the sample was loaded onto a 6% acrylamide‐bisacrylamide gel (19:1) in 0.5 × Tris–borate-EDTA (TBE) buffer and electrophoresed at 250 V for 40 ~ 60 min. The separated DNA fragments were then visualized by staining with ethidium bromide (EtBr).
Data and Statistical Analyses
The number of genotypes and alleles, GD, PIC, and MAF for the waxy maize inbred lines was identified using PowerMarker software (Liu and Muse 2005). Genetic similarities (GS) between each pair of lines were calculated using the Dice similarity index (Dice 1945). The similarity matrix was then utilized to construct a dendrogram based on UPGMA, with the help of SAHN clustering from NTSYS-pc (Rohlf 1998). The population structure of the waxy maize inbred lines was assessed using model-based program STRUCTURE software (Pritchard and Wen 2003). This software was run five times for each simulation subgroup (K value) ranging from 1 to 10 with a burn-in of 100,000 and a run length of 100,000 in an admixture model. The ΔK value based on the degree of change for log probability by Evanno et al. (2005) was calculated using STRUCTURE HARVESTER (http://taylor0.biology.ucla.edu/structHarvester/). The subgroup was assigned using the run result with maximum likelihood among five runs of estimated numbers, with lines with membership probabilities of ≥ 0.80 assigned to subgroups and lines with less than 0.80 assigned to an admixed group (Stich et al. 2005). Association analysis was performed using TASSEL 3.0 (Bradbury et al. 2007), which was used to confirm marker-trait associations using a Q + K MLM. In this study, 100KW was considered a quantitative and qualitative trait for the Q + K MLM method. This study divided all maize inbred lines into low and high groups based on the average value. The Q + K MLM method was performed by combining the Q matrix derived from the STRUCTURE and the K matrix derived from the TASSEL at P < 0.05 (28.54). Furthermore, basic statistical analysis was performed using Microsoft Office Excel 2016. The difference between low and high groups was estimated using Student’s t-test at P < 0.01 in IBM SPSS Statistics version 21 (IBM Corp., Armonk, NY, USA).
Results
Phenotypic Analysis for 100KW in Maize Inbred Lines
Phenotypic variations for 100KW in waxy maize inbred lines are shown in Fig. 1 and Table 2. The average value of 100KW across all of the waxy maize inbred lines was 19.8 ± 4.7 g, with a range of 12.5 to 29.4 g. Based on the average value (19.8 g) for all the inbred lines in this study, the inbred lines were divided into low and high groups. The low group consisted of 21 inbred lines with an average value of 100KW of 16.5 ± 2.0 g, ranging from 12.5 to 19.4 g, while the high group contained 20 inbred lines with an average value of 23.2 ± 2.4 g, ranging from 20.4 to 29.4 g (Fig. 1, Table 2). To confirm differences in 100KW variation between the low and high groups, a t-test was performed using the phenotypic data (Table 2), and this showed statistically significant differences between the two groups (t = − 9.64, P < 0.001).
Genetic Diversity Using SSR Markers Among Maize Inbred Lines
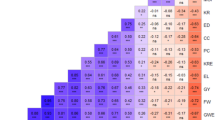
In this study, 150 SSR loci were utilized to assess the GD index, including the number of alleles, genotypes, GD, polymorphic information content (PIC), and major allele frequency (MAF) in waxy maize inbred lines (Fig. 2, Table 3, Supplementary Table 1). This study found 591 and 648 alleles and genotypes, respectively, in the waxy maize inbred lines using the 150 SSR loci. The number of alleles per locus varied between 2 and 10, with an average 3.94 alleles per locus, while the number of genotypes per locus varied from 2 to 11, with an average of 4.32 genotypes per locus. In addition, the average GD was 0.581, with a range of 0.049–0.844; and the average PIC was 0.522, with a range of 0.048–0.827. The average MAF was 0.528, with a range of 0.244–0.975 (Table 3, Supplementary Table 1). When comparing the genetic diversity index among ten chromosomes, it was found that chromosomes 2, 5, 6, and 7 showed relatively high number of both allele and genotype (Table 3). In particular, chromosome 4 exhibited the lowest average values for allele, genotype, GD, and PIC, while chromosome 7 showed the highest average number of alleles, and chromosome 2 showed the highest average number of genotypes, GD, and PIC values (Table 3).
To clearly understand genetic variation in the waxy maize inbred lines of the low and high groups for 100KW trait, this study verified the allele and genotype numbers, GD, PIC, and MAF in the low and high groups. The total number of alleles was 549 and 540 with an average of 3.66 and 3.60 in the low and high groups, respectively. The total number of genotypes was 575 and 573 with an average of 3.83 and 3.82 in the low and high groups, respectively. Furthermore, the averages of the GD, PIC, and MAF values were 0.570, 0.510, and 0.537, respectively, in the low group; and these values were 0.561, 0.501, and 0.546, respectively, for the high group (Table 4).
Genetic Similarity and Kinship Between Parental Lines for Korean Maize Varieties
Pairwise genetic similarity coefficients among the waxy maize inbred lines ranged from 0.229 to 0.893 with an average of 0.385 (Supplementary Table 2), where values close to zero indicate lack of similarity, while those close to one indicate complete similarity. The minimum similarity value was observed in combinations of GP5 and 17CS5047 (0.229), and the maximum value was observed in combinations of HCW5 and 17CS8006 (0.893). Furthermore, two combinations (HCW5 and 17CS8006, HW17 and HW18) had values above 0.800. Out of all combinations, 746 combinations had values under 0.500, while the remaining 74 combinations ranged from 0.500 to 0.893 (Supplementary Table 2).
The pairwise relative kinship coefficients among the waxy maize inbred lines ranged from 0.000 to 1.859 with an average of 0.401 (Supplementary Table 3), where values close to zero indicate lack of relationship, while those close to two indicate complete relationship. Out of all the pairs, only one combination (HF12 and HW8) had a relative kinship value of zero, and seven combinations (HF12 and KL103, GP5 and HCW2, HW3 and HW16, GP5 and 17CS5047, HW16 and 17CS8006, HF12 and 15RS8056, HW12 and 12BS5076-8) had relative kinship values close to zero (0.020 ~ 0.050). Out of all the combinations, 775 combinations varied between 0.050 and 1.000, while the remaining combinations ranged from 1.000 to 1.859 (Supplementary Table 3).
Population Structure Analysis in Korean Waxy Maize Inbred Lines
To confirm the genetic structure and relationships among the waxy maize inbred lines in South Korea, this study used a distance-based dendrogram from an unweighted pair group method with arithmetic mean (UPGMA) analysis and a model-based STRUCTURE program to subdivide the inbred lines into appropriate subgroups. A dendrogram based on distance was constructed using the 150 SSR loci (Figs. 3 and 4, Table 1). All waxy maize inbred lines were classified into five subgroups at a genetic similarity of 35.1%. Group I consisted of 16 inbred lines, with nine low inbred lines (11BS8016-7, 12S8052, 15RS8002, HW8, KL103, 12BS5076-8, 15RS8039, 17YS8003, and HW16) and seven high inbred lines (HW1, HW7, HW4, 17YS6032, HW17, HW18, and HW11). Group II consisted of 20 inbred lines, composed of 10 low inbred lines (16CLP40, 17CS8006, HW19, HCW2, 15S8021-3, 16S8068-9, 15RS8056, 16CLP23, 17CS8067, and KW7) and 10 high inbred lines (14S8025, HCW4, HW3, HCW5, 17CS5047, HCW3, HW9, HW10, HW12, and HCW1). Groups III and V had each two inbred lines: two high lines (HW15 and HF12) and low lines (GP3 and GP5), respectively. Group IV had only one high inbred line (HF22) (Fig. 3, Table 1).
Based on the STRUCTURE results, this study applied the ad hoc measure ΔK because it was difficult to separate subgroups from the LnP(D) of the data. Although the highest ΔK value was obtained for K = 2 using the 150 SSR loci in all maize inbred lines, all inbred lines were not clearly distinguished based on 100KW. By setting a membership probability of 0.8, all inbred lines were classified into three groups: Group I, Group II, and an admixed group (Fig. 4, Table 1). Group I consisted of six maize inbred lines, with three low inbred lines (16CLP40, 17CS8006, and HW19) and three high inbred lines (HCW4, HCW5, and HW19); whereas in Group II, there were 27 maize inbred lines, composed of 13 low inbred lines (11BS8016-7, 12BS5076-8, 12S8052, 15RS8002, 15RS8039, 16CLP23, 17CS8067, 17YS8003, GP3, GP5, HW16, KL103, and HW8) and 14 high inbred lines (17YS6032, HCW1, HCW3, HF22, HW1, HW10, HW11, HW12, HW15, HW17, HW18, HW4, HW7, and HW9). The admixed group contained eight maize inbred lines, with five low inbred lines (15RS8056, 15S8021-3, 16S8068-9, HCW2, and KW7) and three high inbred lines (14S8025, 17CS5047, and HF12) (Fig. 4, Table 1).
This study confirmed the population structure at K = 4 in order to obtain a more detailed classification (Fig. 4, Table 1). All maize inbred lines were divided into four main groups and an admixed group. Group I included six maize inbred lines, consisting of three low inbred lines (17CS8006, HW19, and 16CLP40) and three high inbred lines (HW3, HCW4, and HCW5). Group II comprised only two maize inbred lines, HW18 and HW17. Group III consisted of 16 maize inbred lines, with nine low inbred lines (11BS8016-7, 12S8052, 15RS8002, 17YS8003, GP3, GP5, HW16, HW8, and KL103) and seven high inbred lines (17YS6032, HF12, HF22, HW1, HW15, HW4, and HW7). Group IV was composed of three maize inbred lines, consisting of one low inbred line (17CS8067) and two high inbred lines (HCW3 and HW9). The admixed group comprised 14 maize inbred lines, with eight low inbred lines (12BS5076-8, 15RS8039, 15RS8056, 15S8021-3, 16CLP23, 16S8068-9, HCW2, and KW7) and six high inbred lines (14S8025, 17CS5047, HCW1, HW10, HW11, and HW12) (Fig. 4, Table 1).
Association Analysis Using Q + K MLM
Association analysis between a total of 150 SSR markers and the 100KW traits in the waxy maize inbred lines was performed by a population structure (Q) + kinship (K) mixed linear model (MLM). This study detected two (umc2159 and umc1612) marker-trait associations (MTAs) as qualitative character and three (umc2159, umc1716, and bnlg1246) MTAs as quantitative character, involving four SSR markers associated with 100KW at P < 0.05 (Table 5). The MTAs were located on chromosome 4 (umc1612 and umc1716), chromosome 5 (umc2159), and chromosome 6 (bnlg1246). Phenotypic variation for each marker ranged from 11.7 (bnlg1246) to 36.6% (umc2159). Among these MTAs, umc2159 showed higher phenotypic variation with overlap in qualitative and quantitative characters.
Discussion
Investigation of the characteristics for parental inbred lines is the most important for commercial success of any maize hybrid variety. Evaluating the productivity of inbred lines as parents of varieties in a production field is a means of estimating the commercial viability of maize hybrid production (Pinnisch et al. 2012). This study focused on investigating 100KW in waxy maize inbred lines. Among these waxy maize inbred lines, the lowest 100KW value was detected in GP5 (12.5 g) and GP3 (12.6 g), which are parental lines for the commercial popcorn variety Oryun2ho in Korea. In the case of popcorn, kernel size is negatively correlated with popping extension volume, indicated that a small size kernel has better popping expansion (Kaur et al. 2021). The average 100KW in the waxy maize inbred lines was 19.8 g, which is similar to the 19.6 g of 40 waxy maize inbred lines and lower than the 25.3 g of 40 flint maize inbred lines in a study by Sa et al. (2015). Moreover, this average value was lower than the 23.1 g of 27 dent maize inbred lines from a US study (Pinnisch et al. 2012). To confirm statistical differences of the four maize populations mentioned above, this study performed an analysis of variance (ANOVA) (data not shown). Although the maize population in this study included different kernel types of maize, including popcorn (GP3 and GP5) and flint maize (HF12 and HF22), it also included some duplicated inbred lines (HW3, HW4, HW7, HW8, and KW7) with the 40 waxy maize populations of Sa et al. (2015). The results showed no statistically significant difference between the two waxy maize populations (P < 0.05). However, we found statistically significant differences between the waxy population in this study and the flint population (Sa et al. 2015) and the dent population (Pinnisch et al. 2012) in the other studies (P < 0.05). Actual kernel weight may be influenced by many factors, such as market demand and environmental conditions, and waxy maize genotypes had a yield 18 ~ 27% less than that of dent and flint maize (Solaimalai et al. 2020). However, the yields of newer waxy maize hybrids are better than those of older waxy hybrids through improvement of the waxy maize inbred lines in comparison with normal maize hybrids (Darrah et al. 2019).
Assessing genetic variation is essential for understanding GD and relationships and also for the development of new cultivars in crops. This study detected an average number of 3.94 alleles per locus and an average GD and PIC of 0.581 and 0.522, respectively, in waxy maize inbred lines (Table 3, Fig. 2). In a previous study of 40 waxy and 40 flint inbred lines, which is a similar number to the total of inbred lines in this study, an average of 6.34 alleles per locus and GD and PIC values of 0.66 and 0.62, respectively, were detected in the 40 waxy inbred lines (Sa et al. 2015). Moreover, an average of 6.54 alleles per locus with GD and PIC values of 0.69 and 0.65, respectively, was confirmed in the 40 flint inbred lines (Sa et al. 2015). In another study, an average of 3.62 alleles per locus was detected in 129 maize accessions from Agriculture and Agri-Food Canada (AAFC), using 105 SSR markers, with an average PIC value of 0.68 (Reid et al. 2011). In the International Maize and Wheat Improvement Center (CIMMYT), 137 maize accessions were analyzed using 79 SSR loci, and an average of 7.2 alleles per locus was detected, with an average PIC value of 0.64 (Xia et al. 2005). Furthermore, in a study of 165 Chinese waxy maize landraces and inbred lines, an average of 5.2 alleles per locus was confirmed across 20 SSR markers, with an average PIC value of 0.7 (Zheng et al. 2013). In a comparison of the GD index of the population in this study with that of previous studies, although the allele number was found to be similar to that of another study (Reid et al. 2011), the average allele number, GD, and PIC value in this study were lower than those in previous studies (Sa et al. 2015; Xia et al. 2005; Zheng et al. 2013; Luo et al. 2020). This result indicates the presence of narrow GD in the Korean waxy maize inbred lines used in this study. Among the 150 SSR markers in this study, 13 SSR markers exhibited a high number of over 6 alleles per locus, and 79 and 56 SSR markers showed values of GD and PIC higher than 0.6 (Supplementary Table 1). These SSR markers, which have a higher allele number and GD index, have the potential to provide information for the identification and characteristics of diverse maize lines in future molecular breeding programs.
To develop new hybrid varieties with high performance, it is best to select parental lines with better phenotypic performance and a wide genetic base that can increase genetic variation (Biswas et al. 2008; Ertiro et al. 2017). To select the optimal parental combination for creating new hybrids, there are several methods including evaluation of pedigree relationships, assessment of phenotypic performance for desired traits, and analysis of genetic distances based on both phenotypic traits and molecular markers (Bertan et al. 2007). This study identified genetic distances using measures of genetic similarity and kinship among waxy maize inbred lines (Supplementary Tables 2 and 3). Among all possible combinations in this study, inbred lines with close genetic distances were selected from the same population, while inbred lines with a more distant relationship were combined with inbred lines selected from Korean landrace and unknown foreign populations. Moreover, this study confirmed the genetic similarity and kinship of 11 parental combinations for cultivars registered in the Korea Seed & Variety Service (KSVS) (http://www.seed.go.kr) (Supplementary Tables 2 and 3). The average values of similarity and kinship were 0.406 and 0.478, respectively, in the 11 parental combinations. Among these cultivars, the highest similarity and kinship values were confirmed in cultivar Oryun2ho (popcorn variety; GP5 and GP3), while the lowest values for similarity were detected in cultivar Arichal (waxy variety; HW12 and HW11) and the lowest values for kinship in Mibaekchal (waxy variety; HW4 and HW3) (Supplementary Tables 2 and 3). Previous studies have generally reported inconsistent correlations between the genetic distance of parental lines and the heterosis of progeny (Flint-Garcia et al. 2009). Similarly in this study, we also did not detect any correlations between heterosis for 100KW and genetic distance factors, such as genetic similarity and kinship (data not shown).
Determining the population structure in a set of cultivars is very important for marker-assisted selection (MAS) and association analysis (Wang et al. 2008; Flint-Garcia et al. 2005). Our study used two methods to evaluate population structure: a distance-based phylogenetic method and a model-based clustering method (Figs. 3 and 4, Table 1). This study divided the waxy maize inbred lines into five groups by the distance-based method (Fig. 3, Table 1). At a genetic similarity of 37.3%, Group I was divided into three subgroups: Group I-1, consisting of nine inbred lines; Group I-2, consisting of the five inbred lines; and Group I-3, consisting of the two inbred lines. Similarly, Group II was also divided into two subgroups, at a genetic similarity of 39.5%: Group II-1, consisting of 11 inbred lines, and Group II-2, consisting of nine inbred lines. Additionally, since the model-based method included 65% of the lines (27 inbred lines) in Group II at K = 2, these maize inbred lines were further subdivided at K = 4, which had the next highest ΔK value (Fig. 4). Although the number of maize inbred lines in the admixed group increased at K = 4 compared with K = 2, the maize inbred lines assigned to the main group were subdivided into four subgroups instead of two groups (Fig. 4). In the results, the maize inbred lines of G1 at K = 2 were consistent with G1 at K = 4, and the inbred lines of G2 at K = 2 were subdivided into G2, G3, G4, and admixed group at K = 4. Although G3 and the admixed group at K = 4 with the model-based method still showed inconsistencies with the distance-based method, both methods were in good agreement with the patterns of population structure identified by the K = 4 of the model-based method and the subgroup of the distance-based method (Table 1). The relationship between lines within the same group at K = 4 has been confirmed based on pedigree data. HW3 was located on G1 with five different inbred lines, and these five inbred lines were near-isogenic lines generated from HW3. Moreover, HW17 and HW18 inbred lines were included in G2, which were selected same population made by a foreign collection. Although pedigree of inbred lines in G3 was complexed or unknown, this group contained inbred lines generated by Korean landrace. Maize inbred lines, HW9 was located on G4 with two different inbred lines (HCW3 and 17CS8067), and these two inbred lines were generated from HW9.
Compared with traditional quantitative trait locus (QTL) mapping, in recent years, association analysis has become a more powerful tool for detecting molecular markers associated with specific morphological traits (Liu and Qin 2021). However, association analysis suffers from the problem of false positives (Type-I errors), which can result in invalid associations due to confounding effects from population structure and kinship (Zhang et al. 2010). In this study, a Q + K MLM, using both the Q and K matrices, identified five MTAs associated with 100KW including four significant SSR markers, umc1716, umc1612, umc2159, and bnlg1246 (Table 5). Among these SSR markers associated with 100KW, SSR marker umc1612 at GRMZM2G012391 (cyp37), which encodes cytochrome P450 37, was located on the middle of the long arm of chromosome 4 (bin 4.08). The cytochrome P450 enzyme family is also involved in various physiological processes, including the regulation of seed size and development in Gramineae crops (Li and Wei 2020). Although umc2159 has been detected in both qualitative and quantitative characters for 100KW, it has not been fully characterized. However, as this marker was identified as a flanking marker for a QTL that conditions kernel weight on kernel length in maize (Zhang et al. 2014), further analysis is needed to better understand this locus. SSR marker, umc1716 at GRMZM2G119393, which encodes DNA-directed RNA polymerase II polypeptide, was located on the distal tip of the long arm of chromosome 4 (bin 4.11) (Haag et al. 2014). Moreover, bnlg1246 was mapped different four chromosomes on 3, 5, 6, and 8 (http://www.maizeGDB.org). Furthermore, bnlg1246 has been identified as a flanking marker for QTLs related to aboveground biomass dry weight traits on chromosome 5 in a previous study (Chen et al. 2008).
Data Availability
Data is contained within the article.
References
Adu GB, Badu-Apraku B, Akromah R (2021) Strategies for selecting early maturing maize inbred lines for hybrid production under low soil nitrogen and striga infestation. Agronomy 11:1309. https://doi.org/10.3390/agronomy11071309
Bertan I, Carvalho FIF, Oliveira AC (2007) Parental selection strategies in plant breeding programs. J Crop Sci Biotechnol 10:211–222
Biswas MK, Mondal MAA, Hossain M, Islam R (2008) Utilization of genetic diversity and its association with heterosis for progeny selection in potato breeding programs. Science 6:882–887
Borrás L, Gambín BL (2010) Trait dissection of maize kernel weight: towards integrating hierarchical scales using a plant growth approach. Field Crop Res 118:1–12. https://doi.org/10.1016/j.fcr.2010.04.010
Borrás L, Slafer GA, Otegui ME (2004) Seed dry weight response to source-sink manipulations in wheat, maize and soybean: a quantitative reappraisal. Field Crop Res 86:131–146. https://doi.org/10.1016/j.fcr.2003.08.002
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brittan K (2006) Methods to enable the coexistence of diverse corn production systems. University of California Cooperative Extension Agricultural Biotechnology in California Series Publication 8192. https://doi.org/10.3733/ucanr.8192
Chen J, Xu L, Cai Y, Xu J (2008) QTL mapping of phosphorus efficiency and relative biologic characteristics in maize (Zea mays L.) at two sites. Plant Soil 313:251–266. https://doi.org/10.1007/s11104-008-9698-x
Dao A, Sanou J, Mitchell SE, Gracen V, Danquah EY (2014) Genetic diversity among INERA maize inbred lines with single nucleotide polymorphism (SNP) markers and their relationship with CIMMYT, IITA, and temperate lines. BMC Genet 15:127. https://doi.org/10.1186/s12863-014-0127-2
Darrah LL, McMullen MD, Zuber MS. Breeding, genetics and seed corn production. In Corn, 3rd ed, Serna-Saldivar, S.O., Ed, AACC International Press: Oxford, UK. 2019. https://doi.org/10.1016/B978-0-12-811971-6.00002-4
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21. https://doi.org/10.1007/BF02712670
Devi EL, C9Hossain F, Muthusamy V, Chhabra R, Zunjare RU, Baveja A, Jaiswal SK, Goswami R, Dosad S (2017) Microsatellite marker-based characterization of waxy maize inbreds for their utilization in hybrid breeding. 3 Biotech 7:316. https://doi.org/10.1007/s13205-017-0946-8
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Duvick DN (2001) Biotechnology in the 1930s: the development of hybrid maize. Nat Rev Genet 2:69–74. https://doi.org/10.1038/35047587
Egli DB (1998) Seed biology and the yield of grain crops. CAB International, Wallingford. https://doi.org/10.1023/A:1006182810327
Ertiro BT, Semagn K, Das B, Olsen M, Labuschagne M, Worku M, Wegary D, Azmach G, Ogugo V, Keno T, Abebe B, Chibsa T, Menkir A (2017) Genetic variation and population structure of maize inbred lines adapted to the mid-altitude sub-humid maize agro-ecology of Ethiopia using single nucleotide polymorphic (SNP) markers. BMC Genom 18:777. https://doi.org/10.1186/s12864-017-4173-9
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
FAOSTAT (2020) Statistical Database. Food and Agriculture Organization of the United Nations, Rome
Flint-Garcia SA, Buckler ES, Tiffin P, Ersoz E, Springer NM (2009) Heterosis is prevalent for multiple traits in diverse maize germplasm. Plos One 4: e7433. https://doi.org/10.1371/journal.pone.0007433
Flint-Garcia SA, Thuillet AC, Yu JM, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44:1054–1064. https://doi.org/10.1111/j.1365-313X.2005.02591.x
Haag JR, Brower-Toland B, Krieger EK, Sidorenko L, Nicora CD, Norbeck AD, Irsigler A, LaRue H, Brzeski J, McGinnis K, Ivashuta S, Pasa-Tolic L, Chandler VL, Pikaard CS (2014) Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep 9:378–390. https://doi.org/10.1016/j.celrep.2014.08.067
Hallauer AR, Russell WA, Lamkey KR (1988) Corn breeding. In: Corn and corn improvement (Sprague GF and Dudley JW, eds). 3rd edn. Agron, Monogr. 18, Madison. 463–564
Jordan DR, Tao Y, Godwin ID, Henzell RG, Cooper M, McIntyre CL (2003) Prediction of hybrid performance in grain sorghum using RFLP markers. Theor Appl Genet 106:559–567. https://doi.org/10.1007/s00122-002-1144-5
Kaur S, Rakshit S, Choudhary M, Das AK, Kumar RR (2021) Meta-analysis of QTLs associated with popping traits in maize (Zea mays L.). Plos One 16: e0256389. https://doi.org/10.1371/journal.pone.0256389
Kaushik P, Plazas M, Prohens J, Vilanova S, Gramazio P (2018) Diallel genetic analysis for multiple traits in eggplant and assessment of genetic distances for predicting hybrids performance. Plos One 13:e0199943. https://doi.org/10.1371/journal.pone.0199943
Kesavan M, Song JT, Seo HS (2013) Seed size: a priority trait in cereal crops. Physiol Plant 147:113–120. https://doi.org/10.1111/j.1399-3054.2012.01664.x
Li JZ, Zhang ZW, Li YL, Wang QL, Zhou YG (2011) QTL consistency and meta-analysis for grain yield components in three generations in maize. Theor Appl Genet 122:771–782. https://doi.org/10.1007/s00122-010-1485-4
Li Y, Wei K (2020) Comparative functional genomics analysis of cytochrome P450 gene superfamily in wheat and maize. BMC Plant Biol 20:93. https://doi.org/10.1186/s12870-020-2288-7
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu S, Qin F (2021) Genetic dissection of maize drought tolerance for trait improvement. Mol Breeding 41:8. https://doi.org/10.1007/s11032-020-01194-w
Luo M, Shi Y, Yang Y, Zhao Y, Zhang Y, Shi Y, Kong M, Li C, Feng Z, Fan Y, Xu L, Xi S, Lu B, Zhao J (2020) Sequence polymorphism of the waxy gene in waxy maize accessions and characterization of a new waxy allele. Sci Rep 10:15851. https://doi.org/10.1038/s41598-020-72764-3
Mackay TFC (2009) A-maize-ing diversity. Science 325:688–689. https://doi.org/10.1126/science.1178420
MacRobert JF, Setimela P, Gethi J, Regasa MW (2014) Maize hybrid seed production manual. CIMMYT, Mexico, DF
Melchinger AE (1999) Genetic diversity and heterosis, in The genetics and exploitation of heterosis in crops, J G Coors and S Pandey, Eds., ASA, Madison, Wis, USA. https://doi.org/10.2134/1999.geneticsandexploitation.c10
Melchinger AE, Boppenmmaier J, Dhillon BS, Pollmer WG, Herrmann RG (1992) Genetic diversity for RFLPs in European maize inbreds: II. Relation to performance of hybrids within versus between heterotic groups for forage traits. Theor Appl Genet 84:672–681. https://doi.org/10.1007/BF00224167
Nelson OE, Rines HW (1962) The enzymatic deficiency in the waxy mutant of maize. Biochem Biophys Res Commun 9:297–300. https://doi.org/10.1016/0006-291X(62)90043-8
Park KJ, Park JY, Ryu SH, Goh BD, Seo JS, Min HK, Jung TW, Huh CS, Ryu IM (2007) A new waxy corn hybrid cultivar, “Mibaek 2” with good eating quality and lodging resistance. Korean J Breed 39:108–109
Pejic I, Marsan PA, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLP. Theor Appl Genet 97:1248–1255. https://doi.org/10.1007/s001220051017
Peltonen-Sainio P, Kangas A, Salo Y, Jauhiainen L (2007) Grain number dominates grain weight in temperate cereal yield determination: evidence based on 30 years of multi-location trials. Field Crop Res 100:179–188. https://doi.org/10.1016/j.fcr.2006.07.002
Pinnisch R, Mowers R, Trumpy H, Walejko R, Bush D (2012) Evaluation of maize (Zea mays L) inbred lines for yield component traits and kernel morphology. Maydica 57:1–5
Pritchard JK, Wen W (2003) Documentation for STRUCTURE software: version 2
Reid LM, Xiang K, Zhu X, Baum BR, Molnar SJ (2011) Genetic diversity analysis of 119 Canadian maize inbred lines based on pedigree and simple sequence repeat markers. Can J Plant Sci 91:651–661. https://doi.org/10.4141/cjps10198
Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet 44:217–220. https://doi.org/10.1038/ng.1033
Rohlf FJ (1998) NTSYS-pc: Numerical taxonomy and multivariate analysis system. Version: 2.02. Exeter Software, Setauket, New York
Russell RJ, Fuller JD, Macaulay M, Hatz BG, Jahoor A, Powell W, Waugh R (1997) Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theor Appl Genet 95:714–722. https://doi.org/10.1007/s001220050617
Sa KJ, Park JY, Choi SH, Kim BW, Park KJ, Lee JK (2015) Genetic diversity, population structure, and association mapping of agronomic traits in waxy and normal maize inbred lines. Genet Mol Res 14:7502–7518. https://doi.org/10.4238/2015.July.3.26
Solaimalai A, Anantharaju P, Irulandi S, Theradimani M (2020) Maize crop: improvement, production, protection and post harvest technology (1st ed.). CRC Press. https://doi.org/10.1201/9781003090182
Stich B, Melchinger AE, Frisch M, Maurer HP, Hecknberger M, Reif JC (2005) Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor Appl Genet 111:723–730
van Heerwaarden J, Hellin J, Visser R, van Eeuwijk F (2009) Estimating maize genetic erosion in modernized smallholder agriculture. Theor Appl Genet 119:875–888. https://doi.org/10.1007/s00122-009-1096-0
Wagner MR, Tang C, Salvato F, Clouse K.M, Bartlett A, Vintila S, Phillips L, Sermons S, Hoffmann M, Balint-Kurti PJ, Kleiner M (2021) Microbe-dependent heterosis in maize. Proc Natl Acad Sci USA 118:e2021965118. https://doi.org/10.1073/pnas.2021965118
Wang R, Yu Y, Zhao J, Shi Y, Song Y, Wang T, Li Y (2008) Population structure and linkage disequilibrium of a mini core set of maize inbred lines in China. Theor Appl Genet 117:1141–1153. https://doi.org/10.1007/s00122-008-0852-x
Xia XC, Reif JC, Melchinger AE, Frisch M, Hoisington DA, Beck D, Pixley K, Warburton ML (2005) Genetic diversity among CIMMYT maize inbred lines investigated with SSR markers: II. Subtropical, tropical midaltitude, and highland maize inbred lines and their relationships with elite U.S. and European maize. Crop Sci 45:2573–2582. https://doi.org/10.2135/cropsci2005.0246
Yu K, Wang H, Liu X, Xu C, Li Z, Xu X, Liu J, Wang Z, Xu Y (2020) Large-scale analysis of combining ability and heterosis for development of hybrid maize breeding strategies using diverse germplasm resources. Front Plant Sci 11:660. https://doi.org/10.3389/fpls.2020.00660
Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JK, Buckler ES (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360. https://doi.org/10.1038/ng.546
Zhang Z, Liu Z, Hu Y, Li W, Fu Z, Ding D, Li H, Qiao M, Tang J (2014) QTL analysis of kernel-related traits in maize using an immortalized F2 population. Plos One 9:e89645. https://doi.org/10.1371/journal.pone.0089645
Zheng H, Wang H, Yang H, Wu J,Shi B, Cai R, Xu Y, Wu A, Luo L (2013) Genetic diversity and molecular evolution of Chinese waxy maize germplasm. Plos One 8:e66606. https://doi.org/10.1371/journal.pone.0066606
Funding
This study was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2021R1A6A1A03044242), Republic of Korea.
Author information
Authors and Affiliations
Contributions
K.J.S., H.P., and S.J.J. performed the experiment and analyzed the data. K.J.S and J.K.L. wrote the manuscript and designed the experiments. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sa, K.J., Park, H., Jang, S.J. et al. Genetic Variation and Association Analysis of Elite Waxy Maize Inbred Lines in South Korea. Plant Mol Biol Rep 42, 97–110 (2024). https://doi.org/10.1007/s11105-023-01405-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-023-01405-6