Abstract
It has been reported that members of the Catharanthus roseus receptor-like kinase1-like kinase (CrRLK1L) gene family detect cell wall integrity, cell-to-cell communication, and biotic and abiotic stress. We performed a comprehensive study including the genome-wide identification, characterization, and gene expression analysis of CrRLK1Ls in apple (Malus domestica). Sixty-seven M. domestica CrRLK1Ls (MdCrRLK1Ls) were identified based on their domain structure. Molecular weight and pI ranged from 52.36–141 kDa and 5.05–8.9, respectively. They were distributed across 16 of the 18 chromosomes and classified into five phylogenetic branches. Exon-intron structural analysis indicated a wide range of exon numbers. Collinearity analysis showed that both segmental-and tandem-duplication contributed to the expansion of this family. Cis-elements in the MdCrRLK1L promoter region responded mainly to light, circadian rhythm, phytohormones, and biotic or abiotic stress. Many members exhibited tissue-specific expression patterns and differentially expressed under biotic stresses, which may contribute to the different functional roles of MdCrRLK1Ls under physiological stress and/or pathological conditions. This study provides new insights into the CrRLK1Ls in Malus spp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first receptor-like kinase (RLK) identified in plants resembled animal receptor tyrosine kinases (Walker and Zhang 1990). In the following decades, other RLKs were identified in various plant species (Shiu et al. 2004; Lehti-Shiu et al. 2009). More than 600 and 1100 RLKs have been identified in Arabidopsis thaliana and rice (Oryza sativa L.), respectively (Shiu et al. 2004). A typical RLK contains a signal sequence, a transmembrane region, and a C-terminal domain with eukaryotic protein kinase signatures (Lehti-Shiu et al. 2012). RLKs may also vary greatly in their domain organization and extracellular domain sequence identity (Lehti-Shiu et al. 2012). Plant RLKs are transmembrane proteins that can detect many extracellular and endogenous signals through their extracellular domains. They can propagate these signals via their intracellular kinase domains (Walker 1994). Therefore, RLKs play pivotal roles in plant growth, development, and biotic and abiotic stress responses (Stracke et al. 2002; Wang et al. 2008).
Catharanthus roseus receptor-like kinase1-like kinases (CrRLK1Ls) are members of a distinct RLK subfamily first discovered in C. roseus (Schulze-Muth et al. 1996). At present, 17, 16, and over 40 CrRLK1Ls have been identified in Arabidopsis thaliana, rice, and cotton (Gossypium spp.), respectively (Boisson-Dernier et al. 2011; Han et al. 2011; Nguyen et al. 2015; Niu et al. 2016). CrRLK1Ls also have a putative extracellular carbohydrate-binding malectin-like domain (Boisson-Dernier et al. 2011), the presence of which suggests that this gene family participates in cell wall surveillance. They may coordinate cell growth, cell-to-cell communication, and cell wall remodeling during both vegetative and reproductive development (Lindner et al. 2012). Some CrRLK1Ls have been reported to play key roles in cell elongation, coordination of cell wall integrity, and cell-to-cell communication between male and female gametophytes (Lindner et al. 2012). Recently, it was shown that a CrRLK1L helps powdery mildew penetrate host cells (Kessler et al. 2010). Further investigation confirmed that CrRLK1Ls were activated by a secreted peptide known as rapid alkalinization factor (RALF), which suppresses primary root cell elongation in Arabidopsis thaliana (Haruta et al. 2014). Many different RALFs have since been identified in plants. Some of them mediate the expression of CrRLK1Ls and their co-receptor brassinosteroid insensitive 1-associated kinase 1 (BAK1), which itself regulates plant immune responses (Stegmann et al. 2017). Chen et al. (2016) showed that this protein interacts with guanine nucleotide exchange factors (GEFs). GEFs regulate RhoGTPases (RAC/ROP), which influence certain stress-induced responses, such as the regulation of reactive oxygen species (ROS) production (Greeff et al. 2012). The rapid accumulation of ROS has been proposed as a key step in biotic and abiotic stress responses (Fujita et al. 2006). Therefore, this CrRLK1L may be involved in stress response regulation through GEFs. Several CrRLK1Ls are regulated in response to drought and cold stress (Nguyen et al. 2015; Wang et al. 2008).
Apple (Malus domestica Borkh; family Rosaceae) is an important fruit crop and is beneficial to the economic development and human health (Gallai et al. 2009; Kellerhals 2009). Biotic stress (such as Valsa canker, fire blight, and replant disease) and abiotic stress (for example, frost, drought, and salinity) severely impede apple tree growth (Byers and Marini 1994; Reed and Mazzola 2015; Rodrigo 2000; Wang et al. 2016; Wu et al. 2017). The completed genome sequence for the diploid “Golden Delicious” apple variety allowed us to perform a comprehensive survey of the CrRLK1L family members (Velasco et al. 2010). Although CrRLK1Ls have been investigated in several other plant species, those involved in stress responses in apple remain poorly understood.
Using a high-quality whole-genome sequence from a reference strain, we performed the first genome-wide identification and analyses of phylogenetic relationships, cis-elements, and expression patterns of all CrRLK1Ls in M. domestica. Our results identified candidate CrRLK1L genes for functional analysis. These data will be invaluable in breeding programs implementing genetic engineering approaches to improve the adaptation of apple trees to biotic and abiotic stresses.
Methods
Database Search and Identification of CrRLK1L Genes in Malus domestica
The M. domestica v. 3 genomic sequence was obtained from the Genome Database of Rosaceae species (GDR) (https://www.rosaceae.org/species/malus/all). The Arabidopsis thaliana protein database was obtained from the Arabidopsis information resource (TAIR: http://www.arabidopsis.org). The CrRLK1L malectin-like (PF12819) and Pkinase (PF00069) domain databases were downloaded from Pfam (http://pfam.janelia.org/; Finn et al. 2014) and used as the query to scan the apple database. All CrRLK1L protein sequences were confirmed by the SMART online database (E-value ≤ 1E-5; http://smart.embl-heidelberg.de/; Schultz et al. 1998).
Bioinformatics Analysis of CrRLK1L Genes
The physicochemical characteristics of the MdCrRLK1Ls were identified using the online ExPASyProtParam tool (http://web.expasy.org/protparam/). Multiple sequence alignments of MdCrRLK1Ls were carried out using ClustalX v. 2.0 (Larkin et al. 2007). A phylogenetic tree was constructed using MEGA v. 5.12 (www.megasoftware.net; Tamura et al. 2011) adopting the neighbor-joining method, bootstrap-replicated 1000 times. The Gene Structure Display Server (http://gsds.cbi.pku.edu.cn, Hu et al. 2014) and SMART (http://smart.embl-heidelberg.de/; Schultz et al. 1998) were used to show the exons/introns and domains. The duplication pattern for each MdCrRLK1L was analyzed as follows: (a) local BLAST was performed to search for potential homologous gene pairs (E-value < 1E-5); (b) chromosomal distribution and gene duplication data were plotted using MCScanX (Wang et al. 2012). Genes separated by five or fewer loci within 100 kb were considered tandem duplicates (Hu et al. 2010). For cis-acting element analysis, genomic DNA sequences in the promoter region (− 1500 to − 1 bp) were downloaded from GDR then scanned for in the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, Lescot et al. 2002).
Organ-Specific Expression Patterns of MdCrRLK1Ls
High throughput RNA-sequencing data (GSE42873; Celton et al. 2014) from apple (Malus × domestica Borkh) tissues (flowers, fruit, leaves, roots, stems, seedlings, and seeds) were downloaded from NCBI GEO datasets (National Center for Biotechnology Information [NCBI]). Expression data for the identified genes were extracted using gene ID. A heat map and a clustering tree were constructed using MeV 4.9.0 (open source genomic analysis software; www.tm4.org).
Expression Pattern of MdCrRLK1Ls
MdCrRLK1L expression data were retrieved from the GEO datasetGSE42873 (tissue specific; Celton et al. 2014) and the Sequence Read Archive (SRA) database SRP034726 (apple response to Valsa canker; Yin et al. 2016), downloaded from the NCBI website. Other expression data in response to apple replant disease and apple scab were obtained from supplementary data of previously published studies (Gusberti et al. 2013; Shin et al. 2016). For all data, gene IDs and names were consistent with those in the GDR database. MdCrRLK1L expression data were extracted by gene ID using Microsoft Excel software.
Plant Materials and Treatment
For the tissue-specific expression analyses, roots and leaves from a 3-year-old Malling 26 (M. 26, M. domestica) and M. baccata were collected. To investigate the gene expression of apple in response to Valsa canker, 2-year-old branches were pruned from a 12-year-old “Tian Wang No. 1” Delicious apple trees on May 15, 2016. The branches were inoculated with Valsa mali and cultured for 10 d in darkness at 28 °C. A pure conidial culture of V. mali isolate GS-03 was kindly provided by the College of Plant Protection at Gansu Agricultural University, Lanzhou, China. GS-03 culture and incubation were carried out as described by Li et al. (2015). Unincubated and incubated phloem tissues were collected and stored in liquid nitrogen at − 80 °C.
Quantitative Real-Time PCR (RT-qPCR) Assay
Total RNA was extracted with a plant RNAout kit (Tiandz Inc., Beijing, China) and reverse-transcribed to produce cDNA using a PrimeScript™ RT Reagent kit with gDNA Eraser (Takara Bio Inc., Kusatsu, Shiga, Japan). The coding sequence (CDS) of each gene was retrieved from GDR and further confirmed using the NCBI website. Primers were designed using Primer v. 3 (http://primer3.ut.ee/). Sequences of primers are listed in Supplementary Table 1. The RT-qPCR assay was conducted on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with SYBR® Premix Ex Taq™ II (Tli RNase H Plus, Takara Bio Inc., Kusatsu, Shiga, Japan). Before the RT-qPCR assay, dissociation curves were constructed to verify the specificity of each primer pair. The relative expression levels of each gene were calculated by a comparative 2-ΔΔCTmethod and normalized by the average expression level of two actin genes (Giorno et al. 2012; Bassett et al. 2014). The assay was run in triplicate.
Results
Identification of MdCrRLK1L Members
After a preliminary screen, 74 MdCrRLK1L candidates were retrieved from the apple protein database. The protein sequences ranged from 465 to 1257 amino acids (aa). Each had one or two N-terminal malectin (PF11721) or malectin-like (PF12819) domains, a transmembrane region, and a C-terminal Pkinase (PF00069) or Pkinase_Tyr (PF07714) domain. Normally, most Pkinase domain consisting of 250–300 aa is presenting RLK sequences (Hanks and Hunter 1995; Yamaguchi et al. 2001). During subsequent sequence analyses, certain candidates were excluded from the family because their domain structures differed from the others. The genes MDP0000228029, MDP0000297560, and MDP0000319483 had two Pkinase domains (one N-terminal and one C-terminal), and MDP0000322681 had two transmembrane regions. MDP0000234306, MDP0000176966, and MDP0000199384 had a C-terminal Pkinase or a Pkinase_Tyr domain shorter than 150 aa. Sixty-seven MdCrRLK1Ls were identified using Pfam and SMART (Table 1). One or two extra N- or C-terminal domains such as PDB, LRR, or Seipin (PF06775) were found in several MdCrRLK1Ls. The average molecular weight and pI of the MdCrRLK1Ls were 95.53 kDa and 6.5, respectively. The molecular weights ranged from 52.36–141 kDa and the pI ranged from 5.05–8.9.
An analysis with CELLO v .2.5(subcellular localization predictor; http://cello.life.nctu.edu.tw/) indicated that 64 MdCrRLK1Ls were localized to the plasma membrane (PM). The other three were predicted to be expressed on the chloroplast membrane (Chlo)/nuclear envelope (Nucl) (MDP0000228140), plasma membrane (PM)/Chlo (MDP0000199141), or PM/Nucl (MDP0000721825). However, the subcellular localization predictions for some of the proteins varied when different software was used. The pSORT prediction software (http://www.genscript.com/psort.html) found that 32, 23, and 6 proteins may localize to the PM, Chlo, and Nucl, respectively. A few proteins were localized to the vacuole (Vacu; MDP0000306337, MDP0000632093, and MDP0000199141), the cytosol (Cyto; MDP0000212691 and MDP0000281046) or the extracellular region (Extr; MDP0000413728).
Phylogenetic Analysis of MdCrRLK1L Family from Apple, Arabidopsis thaliana, and Rice
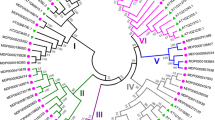
To investigate the evolutionary relationship among the CrRLK1L genes of apple, Arabidopsis thaliana, and rice, rooted phylogenetic trees were generated using 67 CrRLK1L protein sequences from apple, 17 from A. thaliana, and 16 from rice. As shown in Fig. 1, the CrRLK1L phylogram was divided into five sub-clusters. The apple MdCrRLK1L family included 23 members in cluster I, 11 in cluster II, 21 in cluster IV, and 10 in cluster V. Four Arabidopsis CrRLK1L members belonged to cluster III, 3 to cluster IV, and 10 to cluster V. Groups I and II contained only apple CrRLK1L proteins and accounted for 34 members. Thus, the CrRLK1L proteins showed a species-specific evolutionary classification.
To gain further insight into the evolution of the CrRLK1L family, the exon-intron organization of MdCrRLK1Ls was compared. As shown in Fig. 2, exon numbers varied widely (1–28). The MdCrRLK1L members in groups I and II were encoded by at least 8 exons, and several of them, such as MDP0000195070, had 28 exons. All the members of groups IV and V contained between 8and28 exons. A similar exon-intron organization was found within the same cluster.
Phylogenetic relationship and gene structure analysis of MdCrRLK1Ls. a Phylogenetic tree constructed from alignments of 69 MdCrRLK1L genes using the neighbor-joining method in MEGA v. 5.1.2. b The exon/intron structure of each MdCrRLK1L gene was displayed using the Gene Structure Display Server (GSGD, http://gsds.cbi.pku.edu.cn)
Chromosomal Distribution and Duplications of MdCrRLK1Ls
To visualize the genome organization of MdCrRLK1L family members, chromosome localization maps and gene duplications were constructed using MCScanX (Fig. 3). The MdCrRLK1L proteins were widely distributed across16 of the 18 chromosomes. Nevertheless, the distribution and abundance of most members varied among chromosomes or along the same chromosome. Most members were distributed across chromosomes (Chr) 4, 5, 6, 9, 10, and 17. These carry 9, 9, 7, 6, 6, and 7 MdCrRLK1L, respectively. Additionally, six chromosomes (Chr 1, 3, 8, 11, 12, and 14) host two MdCrRLK1Ls each, and Chr 2 and 13 each have three MdCrRLK1Ls. One MdCrRLK1L was found on Chr 7 and four on Chr 16. We identified 34 gene pairs from segmental duplication and 11 gene clusters from tandem duplication (Fig. 3, Supplementary Table 2). Segmental duplication produces many homologous MdCrRLK1L genes on different chromosomes, such asMDP0000007813/MDP0000945548, and MDP0000891518/MDP0000161461. On the other hand, tandem duplication produces clusters or regions with high densities of genes, for example, MDP0000310638, MDP0000451269, and MDP0000276599 on chromosome 6. Both segmental-and tandem-duplication have expanded the number of MdCrRLK1L family members.
Cis-Elements Involved in the Transcriptional Regulation of MdCrRLK1L Genes
A 1500 bp promoter region was analyzed for each MdCrRLK1L gene. The Plant CARE database was applied, and 120 different cis-elements were identified (Table 2, Supplemental Table 4). Forty-one cis-elements were involved in light-mediated transcriptional regulation. In addition, thirteen cis-elements were responsive to phytohormones (auxin, abscisic acid (ABA), ethylene, gibberellins, methyl jasmonate (MeJA), and salicylic acid (SA)), and four to biotic stressors (elicitor, fungal elicitor) and abiotic stressors (drought, low temperature, heat, and wounding).
The distribution of cis-elements involved in the response to phytohormones and stress is shown in Table 2. Most of the cis-elements responding to phytohormones, flavonoids (such as MBSI, MBSII, and MBSIII), and stress are ubiquitous in the MdCrRLK1L promoter regions. Out of 57 MdCrRLK1L promoter regions with cis-elements responsive to MeJA, 43 contained CGTCA and TGACG motifs. Moreover, TCA, ABRE, and GARE motifs were found in the promoter regions of 40, 38, and 38 MdCrRLK1Ls, respectively. The MBS element (involved in flavonoid biosynthetic genes) and TC-rich repeats (defense and stress) were detected in 55 and 46MdCrRLK1L promoter regions, respectively. Four elicitor-induced cis-elements (ELI-box3, EIRE, AT-rich sequence, and Box-W1) were located in 5, 6, 24, and 32 MdCrRLK1L promoter regions, respectively. In addition, three abiotic stress-related cis-elements (LTR, HSE, and WUN motifs) were found in the promoter regions of 32, 42, and 4 MdCrRLK1L genes, respectively. These results indicate that the expression of many MdCrRLK1Ls in apple may be influenced by light, phytohormones, biotic stress, or abiotic stress.
Tissue-Specific Expression Patterns of MdCrRLK1L Genes
We searched for the expression pattern of each MdCrRLK1L gene in the GSE42873 dataset. It included expression profiles for seeds, seedlings, roots in vitro, healthy leaves, 1-year-old stems, flowers in full bloom, fruit parenchyma 100 d postanthesis, and postharvest fruit. For each tissue, the gene expression of two apple cultivars was analyzed. Expression profiles for 29 out of 69 MdCrRLK1Ls were identified (Fig. 4(1)). MDP0000320718 was highly expressed in fruits, whereas MDP0000263999 was significantly expressed in flowers. MDP0000493959, MDP0000450994, and MDP0000292097 exhibited low expression levels in all parts of the plants. MDP0000320718 had a higher level of expression in seeds and roots compared to other organs. MDP0000420368 was expressed mainly in the stems, leaves, and flowers. In addition, eight genes had low expression levels in the seeds but high expression levels in the seedlings, roots, stems, leaves, and flowers. In contrast, 14 MdCrRLK1Ls had high levels of expression in almost all plant organs. The tissue-specific expression profiles of seven CrRLK1Ls in M26 and M. baccata were verified with RT-qPCR. As shown in Fig. 4(2), the expression patterns of most of the CrRLK1Ls detected by RT-qPCR resembled those found by RNA-Seq. Most CrRLK1Ls had similar expression patterns between distinct cultivars. Taken together, our results suggest that the multiple potential regulatory responses of MdCrRLK1Ls may be explained by the various expression patterns in different tissues.
(1) Expression patterns of MdCrRLK1L genes in seeds, seedlings, roots, stems, leaves, flowers, fruit 100 d postanthesis, and postharvest fruit from several cultivars. Data were retrieved from the GEO dataset GSE42873. (2) A Expression pattern of six CrRLK1Ls in the leaves and roots of the cultivars “M14,” “M49,” “GD,” and “X8877”. Data were retrieved from the GEO dataset GSE42873. B Expression patterns of the aforementioned genes from cultivars “M26” and “S” analyzed with RT-qPCR
Expression Patterns of MdCrRLK1L Genes in Response to Several Apple Diseases
To understand MdCrRLK1L transcription in response to several apple diseases, the expression profiles of apple infected with Valsa canker, replant disease, and scab were retrieved and analyzed (Fig. 5). Nineteen MdCrRLK1Ls were significantly regulated by at least one of the three apple diseases. MDP0000292097 was up-regulated by Valsa canker and replant disease. Five genes were up-regulated in ontogenetically resistant scab-free leaves but down-regulated in ontogenetically resistant leaves infected with apple scab and in “Fuji” apples infected with Valsa canker. Furthermore, two genes (MDP0000151537 and MDP0000891518) were up-regulated by scab and four (MDP0000292097, MDP0000197297, MDP0000196032, and MDP0000232699) were up-regulated by replanting disease. In contrast, these six genes were down-regulated by Valsa canker. MDP0000320718 was down-regulated by both scab and Valsa canker, and three genes (MDP0000163811, MDP0000314005, and MDP0000931970) were down-regulated by both replant disease and Valsa canker. Three genes were exclusively down-regulated by Valsa canker. The expression patterns of six MdCrRLK1Ls in “Tian Wang No. 1” infected with Valsa canker were also detected using RT-qPCR and RNA-Seq; MDP0000292097 was up-regulated, MDP0000196032, MDP0000232699, and MDP0000320718 were down-regulated. The expression of MdCrRLK1Ls was significantly influenced by the aforementioned apple diseases and most genes showed distinct expressions in response to each apple pathogen.
a Expression patterns of MdCrRLK1L genes influenced by apple scab (Gusberti et al. 2013), apple replant disease (Shin et al. 2016), and apple Valsa canker (Yin et al. 2016). A1 and A2 indicate MdCrRLK1L expression changes in old leaves (ontogenetically resistant) compared to young leaves (susceptible), and uninfected with apple scab compared to infected for 96 h, respectively. B1 and B2 represent MdCrRLK1L expression in an apple plantlet derived from a cross between a replant-tolerant Geneva (G.41) scion and a replant-susceptible Malling 26 (M.26) rootstock incubated with Pythium ultimum for 4 and 48 h, respectively. C shows the expression of MdCrRLK1Ls from “Fuji” apples infected with Valsa canker. b The expression of MdCrRLK1L genes in response to apple Valsa canker were verified by RT-qPCR
Discussion
CrRLK1Ls are receptor-like kinases common to many plants. They have been identified and analyzed in the complete genomes of A. thaliana (17 members; Shiu et al. 2004), rice (16 members; Nguyen et al. 2015), and cotton (44, 40, and 79 members from diploid G.raimondii, diploid G. arboretum, and tetraploid G. hirsutum, respectively; Niu et al. 2016). Functional investigation of CrRLK1L members in A. thaliana suggests that they function in cell wall integrity (Hématy et al. 2007), cell-to-cell communications (Escobar-Restrepo et al. 2007), and biotic or abiotic stress response (Kessler et al. 2010; Yu et al. 2012). We systematically identified MdCrRLK1Ls in the apple genome. Furthermore, we analyzed their cellular localization, chromosomal mapping, phylogenetic relationships, and expression profiles in silico. To the best of our knowledge, this is the first study to comprehensively analyze all CrRLK1Ls in the Malus genome.
Expansion of the MdCrRLK1Lgene Family in Malus
We identified 67 CrRLK1Ls in the genome of the economically important apple (Malus spp.) The total number of CrRLK1Ls in apple resembled that of cotton (40 to 79 members) (Niu et al. 2016), but was much higher than the number found in Arabidopsis (17 members) and rice (16 members) (Nguyen et al. 2015). We also confirmed many tandem and segmental duplications in MdCrRLK1Ls. In fact, the expansion of multiple gene classes resulted from genome-wide duplications (Velasco et al. 2010). Therefore, we speculated that the increase in the numbers of MdCrRLK1Ls could result from gene duplication events.
Complex Evolutionary Relationships of MdCrRLK1Ls
Phylogenetic analysis revealed that most of the MdCrRLK1Ls were closely related to those from Arabidopsis and rice. The MdCrRLK1Ls in clusters IV and V were common to Malus, rice, and Arabidopsis, suggesting that those in the same subgroup may be derived from a common ancestor. In clusters I and II, however, the MdCrRLK1L members were only present in Malus, suggesting that they evolved to form independent branches. These results further confirm that the MdCrRLK1L members in clusters I and II are distinct from the CrRLK1Ls in Arabidopsis and rice. For intronless genes, duplication events may cause rapid evolution (Lecharny et al. 2003). Analysis of chromosomal distribution showed that many MdCrRLK1Ls in the same cluster were located in close proximity to each other (Fig. 3). The exon numbers of CrRLK1Ls ranged from 1 to 28 in Malus, 1–7 in cotton, and 1–2 in Arabidopsis (Niu et al. 2016), indicating that the CrRLK1L gene structure is more complex in Malus than in cotton or Arabidopsis. Therefore, MdCrRLK1Ls may have had a highly complex evolutionary process.
Potential Regulatory Response of MdCrRLK1Ls
Our results showed that 29 out of 67 MdCrRLK1Ls were expressed in at least one tissue. Previous studies have shown that CrRLK1L proteins have distinct functions in different tissues. Examples include pollen tube reception (Escobar-Restrepo et al. 2007), circadian rhythm (Nguyen et al. 2015), and fungal invasion (Kessler et al. 2010). Furthermore, RALF, a family of secreted peptides, has been found to regulate CrRLK1L expression and function and its function in both cell elongation and plant immune responses (Haruta et al. 2014; Stegmann et al. 2017). These studies, therefore, suggest that CrRLK1Ls have multiple functions in cell growth and response to stress.
Cis-element analysis showed that 41 out of 120 cis-elements were modulated by light, and one was modulated by circadian rhythm (Table 2). Similar regulatory patterns of CrRLK1Ls were found in Arabidopsis and rice (Nguyen et al. 2015). Multiple cis-elements were also detected that respond to phytohormones such as auxin, ABA, ethylene, gibberellins, MeJA, and SA. Most of these elements are located in the MdCrRLK1L promoter region. For example, the TGA-element, ERE, GARE-motif, ABRE, TCA-element, CGTCA-motif, and TGACG-motif were detected in the promoter regions of 23, 20, 32, 38, 40, 43, and 43 MdCrRLK1Ls, respectively. Important roles of CrRLK1Ls in Arabidopsis cell elongation, growth, and development have been verified (Guo et al. 2009; Huck et al. 2003; Kanaoka and Torii 2010; Lindner et al. 2012; Nibau and Cheung 2011; Nissen et al. 2016). THESEUS1, a plasma-membrane-bound receptor-like kinase of the CrRLK1L family, is found in elongating cells and mediates their growth (Hématy et al. 2007). Therefore, regulation of circadian rhythm and modulation of growth and development may be conservative functions of CrRLK1Ls in plants.
Biotic stress (such as diseases) and abiotic stress (such as poor weather conditions) are major concerns in agriculture because they cause significant yield losses in many crops. To date, many stress-regulated cis-elements have been identified. These include TC-rich repeats (defense and stress), Box-W1 (fungal elicitor), LTR (low-temperatures), and HSE (heat stress). Analysis of MdCrRLK1L expression levels revealed that 19 out of 67 members were influenced by at least one of the three apple diseases (scab, replant disease, and Valsa canker). The expression patterns of several genes were verified by RT-qPCR (Figs. 4 and 5). Previous reports investigating the functions of CrRLK1Ls in other plant systems confirm that they participate in the regulation of biotic stress. For example, Flagellinsensing2 (FLS2), Ef-Tu receptor (EFR), and chitin elicitor receptor kinase 1 (CERK1) are important plasma membrane-localized pattern recognition receptors (PRR) activating pattern-triggered immunity (PTI) (Zipfel 2008). Arabidopsis malectin-like/LRR-RLK IOS1 plays an important role in bacterial resistance. It regulates FLS2-, EFR-, and CERK1-mediated signaling pathways and contributes to pathogen resistance (Yeh et al. 2016). An LRR/malectin RLK mediates non-host resistance in barley and resistance to non-adapted powdery mildew fungi in wheat barley (Rajaraman et al. 2016). These results suggest that apple CrRLK1Ls may be actively involved in disease resistance. The identification of cis-elements responding to low-temperature and heat stress in MdCrRLK1Ls might be indicative of their roles in abiotic stress tolerance and requires further investigation.
Conclusions
We performed genome-wide identification, bioinformatics analysis, and gene expression profiling of CrRLK1Ls in Malus. Sixty-seven members of this gene family were identified and divided into five phylogenetic branches. An increase in the number of MdCrRLK1Ls might have occurred as a result of gene duplication events. The analysis of cis-elements and expression pattern profiling revealed that MdCrRLK1Ls may participate in circadian rhythms, growth and development, and biotic and abiotic stress responses. Expressions of several members such as MDP0000292097, MDP0000196032, MDP0000232699, and MDP0000320718 were significantly influenced by multiple diseases.
Abbreviations
- RLK:
-
receptor-like kinase
- CrRLK1L :
-
Catharanthus roseus receptor-like kinase1-like kinase
- MdCrRLK1L :
-
Malus domestica CrRLK1L
- GDR:
-
Genome database of Rosaceae species
- GEF:
-
guanine nucleotide exchange factor
- RAC/ROP:
-
GEF-regulated Rho GTPase
- ROS:
-
reactive oxygen species
- RT-qPCR:
-
quantitative real-time PCR
- FLS2:
-
Flagellin sensing2
- EFR:
-
Ef-Tu receptor
- CERK1:
-
chitin elicitor receptor kinase 1
- PRR:
-
pattern recognition receptor
- PTI:
-
pattern-triggered immunity
References
Bassett CL, Baldo AM, Moore JT, Jenkins RM, Soffe DS, Wisniewski ME, Norelli JL, Farrell RE (2014) Genes responding to water deficit in apple (Malus× domestica Borkh.) roots. BMC Plant Biol 14:1–12
Boisson-Dernier A, Kessler SA, Grossniklaus U (2011) The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot 62:1581–1591
Byers RE, Marini RP (1994) Influence of blossom and fruit thinning on peach flower bud tolerance to an early spring freeze. HortSci 29:146–148
Celton JM, Gaillard S, Bruneau M, Pelletier S, Aubourg S, Martin-Magniette ML, Navarro L, Laurens F, Renou JP (2014) Widespread anti-sense transcription in apple is correlated with siRNA production and indicates a large potential for transcriptional and/or post-transcriptional control. New Phytol 203:287–299
Chen J, Yu F, Liu Y, Du C, Li X, Zhu S, Wang X, Rodriguez PL, Liu D, Chen L, Luan S (2016) FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc Natl Acad Sci U S A 113:E5519–E5527
Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317:656–660
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Gallai N, Salles JM, Settele J, Vaissière BE (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68:810–821
Giorno F, Guerriero G, Baric S, Mariani C (2012) Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13(1):639
Greeff CC, Roux MM, Mundy JJ, Petersen MM (2012) Receptor-like kinase complexes in plant innate immunity. Front Plant Sci 3:209
Guo H, Ye H, Li L, Yin Y (2009) A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal Behav 4:784–786
Gusberti M, Gessler C, Broggini GA (2013) RNA-Seq analysis reveals candidate genes for ontogenic resistance in Malus-Venturia pathosystem. PLoS One 8:e78457
Han YF, Yang Q, Zhang SW, Sun DY, Sun Y (2011) Receptor like kinase CrRLK1-L subfamily: novel motifs in extracellular domain and biological functions in plants. Prog Biochem Biophys 38:891–899
Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase super family: kinase (catalytic) domain structure and classification. FASEB J 9:576–596
Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343:408–411
Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Höfte H (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17:922–931
Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol 10:145
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Huck N, Moore JM, Federer M, Grossniklaus U (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130:2149–2159
Kanaoka MM, Torii KU (2010) FERONIA as an upstream receptor kinase for polar cell growth in plants. Proc Natl Acad Sci U S A 107:17461–17462
Kellerhals M (2009) Introduction to apple (Malus× domestica). In: Genetics and genomics of Rosaceae. Springer, New York, pp 73–84
Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330:968–971
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M (2003) Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genom 3:111–116
Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150:12–26
Lehti-Shiu MD, Zou C, Shiu SH (2012) Origin, diversity, expansion history, and functional evolution of the plant receptor-like kinase/pelle family. In: Receptor-Like Kinases in Plants. Springer, Berlin Heidelberg, pp 1–22
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li Z, Yin Z, Fan Y, Xu M, Kang Z, Huang L (2015) Candidate effector proteins of the necrotrophic apple canker pathogen valsa mali can suppress bax-induced PCD. Front Plant Sci 6:579
Lindner H, Muller LM, Boisson-Dernier A, Grossniklaus U (2012) CrRLK1L receptor-like kinases: not just another brick in the wall. Curr Opin Plant Biol 15:659–669
Nguyen QN, Lee YS, Cho LH, Jeong HJ, An G, Jung KH (2015) Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 241:603–613
Nibau C, Cheung A (2011) New insights into the functional roles of CrRLKs in the control of plant cell growth and development. Plant Signal Behav 6:655–659
Nissen KS, Willats WG, Malinovsky FG (2016) Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci 21:516–527
Niu E, Cai C, Zheng Y, Shang X, Fang L, Guo W (2016) Genome-wide analysis of CrRLK1L gene family in Gossypium and identification of candidate CrRLK1L genes related to fiber development. Mol Gen Genomics 291:1137–1154
Rajaraman J, Douchkov D, Hensel G, Stefanato FL, Gordon A, Ereful N, Caldararu OF, Petrescu A, Kumlehn J, Boyd LA, Schweizer P (2016) An LRR/malectin receptor-like kinase mediates resistance to non-adapted and adapted powdery mildew fungi in barley and wheat. Front Plant Sci 7:1836
Reed A, Mazzola M (2015) Characterization of apple replant disease-associated microbial communities over multiple growth periods using next-generation sequencing. Phytopathology 105:S4
Rodrigo J (2000) Spring frosts in deciduous fruit trees-morphological damage and flower hardiness. Sci Hortic 85:155–173
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95:5857–5864
Schulze-Muth P, Irmler S, Schröder G, Schröder J (1996) Novel type of receptor-like protein kinase from a higher plant (Catharanthusroseus) cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto- and substrate phosphorylation. J Biol Chem 271:26684–26689
Shin S, Zheng P, Fazio G, Mazzola M, Main D, Zhu Y (2016) Transcriptome changes specifically associated with apple (Malus domestica) root defense response during Pythium ultimum infection. Physiol Mol Plant Pathol 94:16–26
Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234
Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355:287–289
Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959–962
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Ri AD, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel C, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Peer YV, Salamin F (2010) The genome of the domesticated apple (Malus [times] domestica Borkh.). Nat Genet 42:833–839
Walker JC (1994) Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol 26:1599–1609
Walker JC, Zhang R (1990) Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 345:743–746
Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu C, Woods-Tör A, Zipfel C, de Wit PJ, Jones JD, Tör M, Thomma BP (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol 147:503–517
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wang S, Hu T, Wang Y, Luo Y, Michailides TJ, Cao K (2016) New understanding on infection processes of Valsa canker of apple in China. Eur J Plant Pathol 146:531–540
Wu R, Wang Y, Wu T, Xu X, Han Z (2017) MdMYB4, an R2R3-type MYB transcription factor, plays a crucial role in cold and salt stress in apple. J Am Soc Hortic Sci 142:209–216
Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J (2001) Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell 7:1047–1057
Yeh YH, Panzeri D, Kadota Y, Huang YC, Huang PY, Tao CN, Roux M, Chien HC, Chin TC, Chu PW, Zipfel C, Zimmerli L (2016) The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28:1701–1721
Yin Z, Ke X, Kang Z, Huang L (2016) Apple resistance responses against valsa mali, revealed by transcriptomics analyses. Physiol Mol Plant 93:85–92
Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li X, Lu C, Li H, Hou C, Li L, Buchanan BB, Chen L, Cheung AY, Li D, Luan S (2012) FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci U S A 109:14693–14698
Zipfel C (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20:10–16
Funding
This work was supported by the horticultural innovation fund of Gansu Agricultural University (GAU-XKJS-2018-217), the National Natural Science Foundation of China (31860549; 31501728), the Science and Technology Major Project of Gansu Province (18ZD2NA006), and the Fund for higher education of Gansu province (2018B-034).
Author information
Authors and Affiliations
Contributions
ZC, CB, and ZW conceived and designed the study; ZC, MZ, CM, MJ, and AZ performed the experiments, analyzed the data, and wrote the manuscript. All the authors agreed on the content of this manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Data Archiving Statement
All identified amino acid and/or CDS of MdCrRLK1L gene sequences were deposited into the GDR (https://www.rosaceae.org/species/malus/all) and/or NCBI database (http://www.ncbi.nlm.nih.gov/). The accession numbers are listed in Table 1.
Electronic supplementary material
Supplementary Table 1
(DOCX 13 kb)
Supplementary Table 2
(DOCX 13 kb)
Supplementary Table 3
(DOCX 14 kb)
Supplementary Table 4
(DOCX 24 kb)
Supplementary Table 5
(DOCX 24 kb)
Supplementary Table 6
(XLSX 15 kb)
Rights and permissions
About this article
Cite this article
Zuo, C., Zhang, W., Ma, Z. et al. Genome-Wide Identification and Expression Analysis of the CrRLK1L Gene Family in Apple (Malus domestica). Plant Mol Biol Rep 36, 844–857 (2018). https://doi.org/10.1007/s11105-018-1125-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-018-1125-8