Abstract
Deoxyribonucleic acid binding with one finger (Dof) family, a kind of plant-specific transcription factor family, is involved in a variety of biological processes including signal transduction, morphogenesis, and environmental stress responses. In the present study, 46 putative Dof genes were identified from maize (Zea mays subsp. mays). The predicted ZmDof genes were non-randomly distributed within their chromosomes, and segmental duplication seemed to be the prevalent mechanism for their expansion. Most of these genes lacked introns and some only possessed one intron. Here, we classified 140 Dof proteins from Arabidopsis, rice, sorghum, and maize into seven groups by means of phylogenetic analysis. In addition, many cis-elements responding to light, endosperm specific gene expression, hormone, meristem specific expression, and stress were also found in the 1,000 bps upstream sequences of promoter regions. Selection analysis also identified some significant site-specific constraints acted on most Dof paralogs. Expression profiles based on microarray data provided insights into the functional divergence of different tissues. Results by quantitative reverse transcription polymerase chain reaction analysis indicated that some ZmDofs responded to some abiotic stresses. Taken together, our study will provide some useful information for biological evolution and functional characterization of the maize Dof genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factors are also called trans-acting factors that participate in many important cellular processes, such as signal transduction, morphogenesis, and environmental stress responses, by influencing or controlling the expression of some specific genes. Transcription factors can be bound to cis-regulatory elements of promoters and then determine the transcription rate of genes (Wasserman and Sandelin 2004). At the same time, transcription factors can also mediate protein–protein interactions in a complex network (Yanagisawa 1997). Numerous transcription factors have been identified. And they can be divided into different gene families (Yanagisawa and Schmidt 1999). Some families exist in both animals and plants, whereas others are specific for plants, such as WRKY (Eulgem et al. 2000), R2R3-MYB (Martin and Paz-Ares 1997), NAC (Olsen et al. 2005), TIFY (Vanholme et al. 2007), SBP-box (Yang et al. 2008), etc.
Deoxyribonucleic acid (DNA) binding with one finger (Dof) is a kind of plant-specific transcription factor gene family. Previous studies have shown that Dof domain proteins are active in plant processes as transcriptional activators or repressors (Yanagisawa 2004). The first Dof gene was found in maize (Yanagisawa and Izui 1993), which contained a conserved DNA binding Dof domain. This domain usually consists of 50–52 amino acid residues and a classic four Cys residues zinc-finger that can bind specifically to cis-regulatory elements containing the common core 5′-T/AAAG-3′ (Yanagisawa and Schmidt 1999). The conserved Dof domain is located at the N-terminal region, and another transcriptional regulation domain is located at the C-terminus of Dof proteins (Kushwaha et al. 2011). These regions are involved in interacting with different regulatory proteins or signals (Noguero et al. 2013), which lead to the diverse functions of Dof proteins, such as plant defense (Chen et al. 1996), seed storage protein synthesis (Vicente-Carbajosa et al. 1997; Marzábal et al. 2008), auxin response (Kisu et al. 1997; Kisu et al. 1998; Baumann et al. 1999), carbohydrate metabolism (Yanagisawa 2000), gibberellin response (Mena et al. 2002; Washio 2001), stomata guard cell specific gene regulation (Plesch et al. 2001; Negi et al. 2013), seed germination (Papi et al. 2000; Papi et al. 2002; Gualberti et al. 2002), photoperiodic control of flowering (Imaizumi et al. 2005; Imaizumi and Kay. 2006), cell cycle regulation (Skirycz et al. 2008), leaf axial patterning (Kim et al. 2010), etc.
Dof proteins are also involved in the physical interactions with other transcription factors. For example, OBP1 (a Dof protein) interacts with bZIP transcription factors OBF4 and OBF5 and then facilitates the binding of OBF to its DNA target (Zhang et al. 1995). Wheat prolamin-box binding factor, a Dof transcription factor, interacts with TaQM to activate transcription of an alpha-gliadin gene in seed development (Dong et al. 2007).
Dof gene family was studied widely in plant kingdom, from lower green unicellular alga Chlamydomonas reinhardtii and the moss Physcomitrella patens (Moreno-Risueno et al. 2007) to higher plants, such as Arabidopsis and rice (Lijavetzky et al. 2003), Brachypodium distachyon (Hernando-Amado et al. 2012), sorghum (Kushwaha et al. 2011), soybean (Guo and Qiu 2013), and tomato (Cai et al. 2013). Maize is one of the most important cultivated food plants. But only few reports described its Dof gene complement and functions. Maize Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism (Yanagisawa 2000). As an example, maize Dof1 can interact specifically with the C4 phosphoenolpyruvate carboxylase gene promoter and enhancer its promoter activity, which displays a light-regulated expression pattern matching Dof1 activity (Yanagisawa and Sheen 1998), while maize Dof2, as competitive inhibitor of Dof1, can block and repress the transactivation by means of competitive combination (Yanagisawa and Sheen 1998).
In this study, we identified 46 ZmDof genes, about two and a half times more than that of the previous study (Jiang et al. 2010). And some substantial studies containing phylogenetic analysis, gene location and duplication, gene organization, cis-element analysis, and expression profiles were further performed. This research will provide useful information about Dof gene family functions in maize.
Materials and Methods
Identification of the Dof Gene Family in Maize
To identify potential members of Dof gene family in maize, we performed a database search. Arabidopsis and rice Dof sequences were retrieved (Lijavetzky et al. 2003) and were used as queries in a BLAST search against the maize B73 genome sequence (http://www.maizegdb.org) (Schnable et al. 2009). BioXM 2.6 (http://zhanglab.njau.edu.cn/) was used to analyze the physicochemical parameters of Dof proteins. CELLO v2.5 Server (http://cello.life.nctu.edu.tw; Yu et al. 2004) was used to predict the subcellular localization of each Dof protein in maize. The second structure prediction of the Dof protein was performed with CFSSP server (http://www.biogem.org/tool/chou-fasman/; Chou and Fasman 1974).
Protein Alignment and Phylogenetic Analysis
Multiple sequence alignments of full-length Dof protein sequences from Arabidopsis, rice, sorghum (Lijavetzky et al. 2003; Kushwaha et al. 2011), and maize were performed using ClustalW (http://www.genome.jp/tools/clustalw/), followed by manual comparisons and refinement. Phylogenetic analyses of Dof proteins based on amino acid sequences were carried out using the neighbor-joining (NJ) method in MEGA v5 (Tamura et al. 2011). NJ analyses were done using p-distance methods, pairwise deletion of gaps, and default assumptions that the substitution patterns among lineages and substitution rates among sites are homogeneous. Support for each node was tested with 1,000 bootstrap replicates. The distribution of amino-acid residues at the corresponding positions in domain profiles for the conserved Dof domains of ZmDofs was created using WebLogo (Crooks et al. 2004).
Chromosomal Location and Gene Structure of the ZmDof Genes
Each of the ZmDof genes was located on maize chromosomes by using their annotation information on MaizeSequence (http://www.maizesequence.org; Schnable et al. 2009). The resulting position of ZmDof genes on maize chromosome named ZmDof1–ZmDof46 was marked on the model of maize chromosome obtained from SyMAP v3.4 (Soderlund et al. 2011). Gene intron–extron structure information was also collected from genome annotations in MaizeSequence (http://www.maizesequence.org; Schnable et al. 2009).
Identification of Conserved Motif and Cis-Regulatory Element Analysis
To identify conserved motifs in Dof proteins, the deduced protein sequences containing 46 maize Dof proteins were analyzed by means of the Multiple EM for Motif Elicitation (MEME) program software version 4.4.0 (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). The maximum number of motif was set to 19, and others used the default settings. Next, 1,000 bps upstream sequences from the transcription start site (TSS) of the putative ZmDof genes were retrieved for promoter analysis. The retrieved sequences were subsequently subjected to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/ plantcare/html/; Lescot et al. 2002) for searching for cis-regulatory elements.
Inference of Duplication Time
We used ClustalW to perform the pairwise alignment of nucleotide sequences of Dof paralogs and used K-Estimator 6.0 program (Comeron 1999) to estimate the Ka and Ks values of paralogous genes. Estimates of evolutionary rates are meaningful for explaining patterns of macroevolution because Ks can be used as a proxy for time when estimating dates of segmental duplication events. We used each of gene pair to calculate Ks value and then used this value to calculate the approximate date of the duplication event (T = Ks/2λ), assuming clock-like rates (λ) of 6.5 × 10−9 synonymous/substitution site/year for maize (Gaut et al. 1996).
Site-Specific Selection Assessment and Testing
Selecton Server (http://selecton.tau.ac.il/; Stern et al. 2007), a Bayesian inference approach for evolutionary models, was used to calculate site-specific positive and purifying selection. We calculated the synonymous rate (Ks) and the non-synonymous rate (Ka), and then the values of Ka/Ks are used to estimate two types of substitutions events. In this study, three of the evolutionary models [M8 (ωs ≥ 1), M7 (beta), and M5 (gamma)] were used. Each of the models assumes different biological assumptions and enables contrasting different hypotheses through testing which model better fits the data.
Microarray-Based Expression Analysis
We obtained the genome-wide microarray data of maize published by Sekhon et al. (Sekhon et al. 2011) from the Gene Expression Omnibus with Accession Numbers GSE27004 in the National Center for Biotechnology Information. Expression data were gene-wise normalized and hierarchically clustered based on Pearson coefficients with average linkage in the Genesis (v 1.7.6) program (Sturn et al. 2002).
Plant Materials and Treatment
One-week-old maize seedlings (B73 genotype) were used to examine the expression patterns of ZmDof genes under salt and drought stresses. Plants were grown in a plant growth chamber at 23 ± 1 °C with a 14 h light/10 h dark photoperiod. For salt stress, the seedlings were dealt with 150 mM NaCl for 24 h. For drought treatment, the seedlings were dried between folds of tissue paper at 23 ± 1 °C for 3 h. Control seedlings were grown at 23 ± 1 °C with normal irrigation.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
Trizol total RNA extraction kit (Sangon, Shanghai, China, SK1321) was used to extract total RNA. Next, M-MLV (TakaRa, Dalian, China) was used to perform reverse transcription. Triplicate quantitative assays were performed on each cDNA dilution using SYBR Green Master Mix (TakaRa) with an ABI 7500 sequence detection system, according to the manufacturer’s protocol. The gene-specific primers were synthesized in Sangon. Eight maize Dof genes from different major branches of the phylogenetic tree were randomly selected for quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. Expression level of the maize Actin 1 (GRMZM2G126010) gene was used as the endogenous control. The relative expression level was calculated as 2−∆∆CT method (Livak and Schmittgen 2001). Their gene-specific primers are shown in Table S2.
Results and Discussion
Identification of the Dof Gene Family in Maize
The sequenced maize genome (Schnable et al. 2009) provides an opportunity to predict the complete set of non-redundant Dof genes by means of various bioinformatics tools. We used the amino acid sequences of Arabidopsis and rice Dof (Lijavetzky et al. 2003) to perform BLAST search in MaizeGDB database (www.maizegdb.org). The presence of conserved Dof domain is a common typical feature for consideration as a member of the Dof transcription factor family (Yanagisawa and Sheen 1998). To verify the reliability of our results, all of the putative Dof protein sequences were subjected to the Pfam database (Finn et al. 2014). As a result, a total of 46 Dof transcription factor genes were found in maize. To further investigate the character of Dof domains in maize, we performed their alignment analysis. The results indicated that 20 of 36 amino acids were conserved in all maize Dof domains and that a typical zinc-finger consisting of four cysteine residues has been found with the locus of 4 sites, 7 sites, 29 sites, and 32 sites, respectively (Fig. 1). Other conserved residues in maize Dof domains were Pro-5, Arg-6, Ser-9, Thr-12, Lys-13, Phe-14, Cys-15, Tyr-16, Asn-18, Asn-19, Tyr-20, Gln-24, Pro-25, Arg-26, Arg-34, and Trp-36. Compared with other plants, we also found that these amino-acid residues were also highly conserved (Lijavetzky et al. 2003; Kushwaha et al. 2011; Hernando-Amado et al. 2012; Guo and Qiu 2013). This result indicated that the unique structure of Dof proteins in maize was determined by the highly conserved character of these amino acids. The ZmDof genes encode peptides ranging from 223 to 618 amino acid residues in length with from 21.6 to 67.08 kDa (Table 1). The isoelectric point (pI) of ZmDof protein is predicted ranging from 4.49 to 10.91 (Table 1). To further analyze the Dof proteins, CELLO v2.5 Server (http://cello.life.nctu.edu.tw/) (Yu et al. 2004) was used to predict the probable protein localization. Results indicated that all maize Dof proteins possess signal sequences for targeting the nucleus (Table 1), suggesting their specific function of transcription regulation.
Conserved Dof domains in maize all 46 Dof proteins. The sequence logos are based on alignments of all 46 ZmDof domains. These Dof domains were used to perform multiple alignment analysis with ClustalW. The bits score means information content of each position in the sequence. Asterisks signified the conserved four cysteine (Cys) residues in the Dof domain
Phylogenetic Analyses of Dof Genes in Maize, Arabidopsis, Rice, and Sorghum
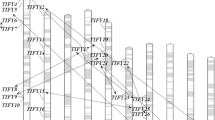
To examine the evolutionary relationship of the Dof gene family in maize, Arabidopsis, rice (Lijavetzky et al. 2003), and sorghum (Kushwaha et al. 2011), a total of 140 Dof proteins were aligned using ClustalW (Data S1), and a combined NJ tree was constructed from the alignments for the four species. We divided the 140 members into 7 groups based on the sequence similarity and phylogenetic tree topology, designated from group I to group VII (Fig. 2; Fig. S1). The largest group was Group IV containing 35 members, while the smallest group was group VI with 3 members. All these three members in group VI belong to the Dof proteins of Arabidopsis, while group III and group V are only composed of monocot Dof proteins, suggesting their specific origination between monocot and dicot. The phylogenetic analysis of four species displayed that all maize Dof genes were clustered with their sorghum or rice counterparts with high bootstrap support (Fig. S1). Furthermore, we also noted that the ZmDofs were more closely grouped with the Dofs of sorghum than those of rice, implying that some ancestor Dof genes had been existing before the divergence of maize and sorghum. It is well known that maize underwent another genome’s duplication after the divergence from sorghum (Gaut and Doebley 1997). Therefore, we should get 56 or more Dof genes in theory rather than 46 Dof genes in maize, given 28 Dof genes were identified in sorghum. A reasonable explanation of fewer Dof genes in maize was that some duplicated Dof genes were likely lost during evolution.
Unrooted neighbor-joining phylogenetic tree of Dof proteins in four plants. Dof proteins of four species A. thaliana (pink), O. sativa (green), S. bicolor (blue), and Z. mays (red) were used to construct NJ phylogenetic tree. Seven major phylogenetic groups designated from group I to group VII are indicated
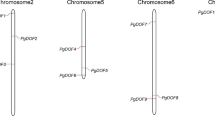
Chromosomal Location of ZmDof Genes and Genomic Duplication
To further investigate the relationship between genetic divergence within the Dof gene family and gene duplication and loss in maize genome, chromosomal location of each Dof gene was determined. As a result, the predicted 46 Dof genes in maize are located on nine of ten chromosomes. We renamed them from ZmDof01 to ZmDof46 based on their chromosome locations. Chromosome 1 has a maximum of 12 Dof genes, while chromosome 2, 4, 6, 7, and 10 only has 3 Dof genes, respectively. Maize has experienced two genome amplification events: one is an ancestral tetraploidy, another is a large-scale duplication event (Gaut and Doebley 1997). Studies have shown that gene duplication for most of gene families is due to the second large-scale duplication event in maize (Schnable et al. 2009; Chen et al. 2014). Within the identified duplication events between chromosomes 1 and 9 (Fig. 3), three Dof genes (ZmDof1, ZmDof2, and ZmDof3) are still located on the chromosome 1, while no Dof gene is found on the corresponding duplicated segment of chromosome 9. Therefore, we suggested that some Dof genes maybe lost on the maize chromosome 9 in evolution.
Chromosomal locations of ZmDof genes and genomic duplication. The 46 maize Dof genes were mapped on the maize chromosomes. Paralogous regions in the putative ancestral constituents of the maize genome are depicted using SyMAP v3.4 (Soderlund et al. 2011) and are shown on one side of each chromosome. The same colors on both sides of a chromosome region and maize chromosome color key signified the same paralogous regions in the genome
Segmental duplication, tandem duplication, and transposition events play important roles for gene family expansion (Liu et al. 2011; Cao et al. 2011). A phylogenetic tree using maize Dof protein sequences was constructed. From the phylogenetic tree, 17 pairs of paralogous Dof genes in the terminal nodes were identified and inferred from the duplication events. Within identified duplication events, 12 of 17 pairs (ZmDof04/ZmDof19 ZmDof34/ZmDof41, ZmDof08/ZmDof30, ZmDof42/ZmDof20, ZmDof15/ZmDof38 ZmDof33/ZmDof25, ZmDof32/ZmDof24, ZmDof23/ZmDof31, ZmDof28/ZmDof10 ZmDof14/ZmDof37, ZmDof40/ZmDof36, and ZmDof43/ZmDof21) are retained as duplicates in maize (Fig. 3), suggesting that segmental duplication is the main way for Dof gene expansion in maize. We also identified two pairs of tandem repeats on chromosome 1 (ZmDof11/ZmDof12) and chromosome 5 (ZmDof26/ZmDof27; Fig. 3). In addition, evolutionary dates of duplicated ZmDof genes were estimated using Ks as the proxy for time (Table 2). Two earlier observed segmental duplication events occurred in the maize ZmDof46/ZmDof13 and ZmDof07/ZmDof01 around from 70.02 to 79.10 million years ago, close to the time of origin of monocotyledons (Blanc and Wolfe 2004). We found that duplication events of the other 12 pairs of maize chromosomes had occurred during the past 7.90–25.71 million years. This period is consistent with the time when the subsequent large-scale genome duplication event is thought to have occurred in maize after separating from sorghum (Gaut and Doebley 1997). Therefore, gene duplication for most of the ZmDofs is due to the second large-scale duplication event after divergence from sorghum.
Gene Structure and Motif Analysis
Gene structural diversity is the foundation of the evolution in multigene families (Zhu et al. 2011; Cao et al. 2010; Cao and Shi 2012). In order to further explore this diversity of Dof genes, we analyzed their exon-intron organization in each ZmDof gene. A detailed illustration of the exon-intron structures is shown in Fig. 4b. According to their predicted structures, 28 of the ZmDof genes lack introns whereas 15 genes contain only one intron. ZmDof22 and ZmDof27 contain two introns, and only one gene (ZmDof18) has four introns. These exon–intron structures are similar to those of Arabidopsis, rice, and other plants (Lijavetzky et al. 2003; Kushwaha et al. 2011; Hernando-Amado et al. 2012; Guo and Qiu 2013; Cai et al. 2013). The most closely-related members in the same subclade generally showed the same exon-intron structure, for instance, most of the intronless genes were clustered into Clade 2, while most members of Clade 5 contained one intron, which suggests evolutionary conservation in maize Dof gene evolution.
Phylogenetic relationships and structure analysis of maize Dof members. a The phylogenetic tree of maize Dof proteins constructed from a complete alignment of 46 ZmDof proteins using MEGA v5 by the neighbor-joining method with 1,000 bootstrap replicates. The six major clades designated 1 to 6 are indicated. b Exon–intron structures of maize Dof genes. Exons are represented by green boxes and introns by black lines. c Schematic distribution of conserved motifs. Motifs were identified by means of the MEME software. Motif 1 was labeled in red signifying the Dof motif
To further expose the diversification of maize Dof proteins, the MEME analysis (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) was also performed. We found that motif 1 (Dof motif) has been found in all of the maize proteins (Fig. 4c). Motif 2 is present in all the maize groups except for the ZmDof27, while the motif 16 only belongs to some members of Clade 2, suggesting that the motif 16 is specific to the evolution of some members in Clade 2. Motifs 4, 7, 14, and 19 are specific in Clade 1, while motifs 3, 5, 6, 8, and 9 are only found in Clade 6. We also found that the Clade 1 contains the largest number of 12 Dof members, and the smallest one is Clade 4 with only 2 Dof proteins. These similarities of motif patterns within the same clade might be related to the similar functions of the Dof proteins, while the different motifs among 5 clades might imply their functional divergence (Guo and Qiu 2013; Chen et al. 2014). Some motifs existed in maize are also found in other plants. For example, motifs 4, 5, 8, 15, and 20 in maize are presented a very similar residue composition with motifs 5, 2, 4, 3, and 24 in Arabidopsis, rice, and poplar, respectively (Yang et al. 2006). Moreover, about one-half of the motifs in Dof proteins were shared by non-Dof proteins in the three plants, indicating that motif acquisition may be one of the forces driving Dof gene diversification (Yang et al. 2006). The evolutionary diversification of Dof genes apparently increased the number of Dof-interacting molecules or proteins and consequently might contribute to the development of a complex and precise transcription and regulation mechanism.
Cis-Regulatory Elements Analysis in the Promoters of ZmDofs
Cis-regulatory element analysis was carried out by retrieving 1,000 bps upstream sequences from the TSS of 46 ZmDof genes. A large number of cis-regulatory elements were found when these promoter sequences were submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002). As a result, the cis-regulatory elements associated with five important physiological processes, such as light responsiveness, endosperm specific gene expression, hormone responsiveness, meristem specific expression, and stress responsiveness were found to be distributed among these promoters of ZmDof genes except for ZmDof02, ZmDof11, ZmDof17, ZmDof27, ZmDof33, and ZmDof40. In Table S1, we found that many light responsive elements, such as, ACE, ATC-motif, ATCT-motif, Box I, Box 4, GT1-motif, Sp1, MNF1 etc., were present in the promoter region of ZmDof genes, suggesting that they may be responding to light. Some specific cis-elements are associated with other processes such as the ABRE element for hormone responsiveness, the CCGTCC-box for meristem activation, and the CCAAT- and CGTCA-boxes for stress responsiveness, etc. (Table S1). Another four cis-elements (Skn-1 motif, GCN4-motif, O2-site and RY element) were concerned with endosperm specific gene expression (Vicente-Carbajosa et al. 1997; Mena et al. 1998). In maize, 22 of 46 promoters of the Dof genes contained at least one of these four cis-elements. Duplicated genes may have different evolutionary fates, as indicated by the divergence in their expression regulation patterns. We also investigated their cis-elements of duplicated Dof gene pairs (identified above) in maize, and found that most gene pairs did not share similar cis-elements composition in their promoters (Table S1). This indicates that substantial neofunctionalization may have occurred during the evolution of duplicated genes. The divergence of the regulation patterns between paralogs and duplicated genes may increase the adaptability of duplicated genes to environmental changes, thus conferring a possible evolutionary advantage. RY element exists in both the elements for endosperm specific gene expression and hormone responsive elements at the same time, indicating that it may have two functions and mechanisms of action in the promoter regions. These results indicated that the number and classes of cis-elements affect the responsiveness of ZmDofs to the environment and development.
Variable Selective Pressures Among Amino Acid Sites
Genes are usually affected and changed during evolution. Some genes changed slowly, while others changed fast in evolution. To further analyze the amino acid substitutions by selective pressures, we calculated Ka/Ks ratios with the Selecton Server (http://selecton.tau.ac.il) (Stern et al. 2007) to estimate the positive and negative selection of specific amino acid sites within the full-length sequences of the Dof proteins in different groups. We used three evolutionary models [M8 (ωs ≥ 1), M7 (beta), and M5 (gamma)] to perform the tests for researching the different selective pressures among the Dof proteins. M8 and M5 models predicted some positive selective sites within the Dof proteins, while M7 model was not (Table 3). We also found divergent results of the prediction of positive selective sites. This is probably due to different calculation methods between M8, M7 and M5 models. From the results, we found the Ka/Ks ratios of the sequences from different Dof groups are significantly different (p < 0.05; Table 3). Such as, higher Ka/Ks ratios existed in groups II, IV, and VI, indicating a higher evolutionary rate or site-specific selective relaxation within most members of these groups. Although the Ka/Ks values are very different among the 7 groups, all these estimated values are substantially lower than 1, indicating the Dof sequences in each group have a strong purifying selection pressure. However, some sites are predicted to undergo positive selection in evolution. For example, 16 positive selection sites are predicted in group VII with M8 model (Table 3). The detailed distribution of the positive-selection sites is showed in Fig. S2. Interestingly, none of these predicted selected sites are located the Dof domain. Further analyses indicate that four of the 16 positive selection sites in the group VII sequences are in α-helices, and two sites are located in β-turns and β-sheets, respectively. A few additional positively selected sites are distributed in other regions, suggesting that these residues might be important in maintaining the conformational stability of the proteins. These observations suggest that positive selection pressure on these residues might have changed the protein structure, thus accelerated functional divergence (Cao 2012).
The Expression Profiles of the ZmDof Genes
Expression profiling is a useful tool for understanding gene function (Chen and Cao 2014). To further understand the function of Dof genes in maize, we tested the temporal- and spatial-specific expression patterns of ZmDof genes. In this study, the gene expression levels of 60 distinct tissues signifying 11 major organ systems were identified by means of microarray data (Sekhon et al. 2011). As a result, the expression level of ZmDof genes was shown except for four genes (ZmDof02, ZmDof41, ZmDof40, and ZmDof21) for which no matching probes were present on the microarray (Fig. 5). We also found that the 42 detected transcripts produced by 42 genes are expressed with distinct levels and in different tissues, suggesting that they may be involved in various biological processes. Most members of ZmDofs displayed high expression levels in stem, root, leaf, but showed low expression levels in the embryo, endosperm and seed, implying that ZmDofs may play important roles in stem, root and leaf development (Fig. 5). Interestingly, we found that two members (ZmDof03 and ZmDof13) are different in expression profiles, and they have high expression levels in embryo, endosperm and seed, but low expression levels in other tissues. This result suggests that ZmDof03 and ZmDof13 may be involved in embryo, endosperm, and seed development. Previous study has indicated that ZmDof13 is a prolamin-box binding factor (PBF) gene, which encoding a transcriptional activator of 36 kD. PBF can regulate the temporal and spatial expression of gamma-zein gene in developing maize seeds (Marzábal et al. 2008). Therefore, PBF protein is involved in with the accumulation of gamma-zein protein in maize endosperms. The result is consistent with our research on the expression profiles of ZmDof13 (PBF) gene. From Fig. 5, we also found that ZmDof46 and ZmDof33 have high expression level in the germinating seed, implying that these genes may be involved in first 24 h germinating seed development in maize. Members in the same gene group of the phylogenetic tree usually originate from duplication of an ancestral gene and have a very similar sequence composition. Here, we also investigated the expression profiles of duplicated Dof genes in maize and found that most of duplicates exhibited divergent expression profiles (Fig. 5). It suggests that due to a relaxation of functional constraints and adaptation to new functions, duplicated genes usually exhibit divergence in expression profiles, which maybe confer an evolutionary advantage for organisms living in the changing environment (Farré and Albà 2010). Some members of Dof gene family were demonstrated to be regulated by salt and drought in Triticum aestivum (Shaw et al. 2009). However, few Dof genes in response to salt and drought treatments were reported in maize. For this purpose, we investigated the expression patterns of maize Dof genes under salt and drought treatments. Expression profiles of eight ZmDof genes responses to salt and drought treatments were identified by qRT-PCR analyses. As a result, six of eight ZmDof genes detected were upregulated during salt treatment of seedlings (Fig. 6). Among them, ZmDof22, ZmDof16, and ZmDof36 were significantly upregulated at 50 times, 25 times, and 40 times higher than the control ones, respectively. We also found that ZmDof06 was upregulated under drought treatment, implying that this gene is more likely to play critical roles in regulating drought response in maize seedlings.
Expression profiles of eight ZmDof genes in response to salt and drought treatments. qRT-PCR analyses were used to assess ZmDof transcript levels in seedlings samples after salt and drought treatment with 24 and 3 h, respectively. Control (CK) seedlings were grown with normal irrigation. Asterisk indicates a significant difference from the control (p < 0.05). Error bars indicate standard error of independent biological replicates
Conclusions
Transcriptional regulation plays an important role in the process of gene expression, and this process is determined by the number, position and interaction between different cis-elements and transcription factors. Dof gene family is a plant-specific transcription factor. Genome-wide identification, phylogenetic analyses, chromosomal location, gene structure and motif analysis, cis-elements, selective pressures, and expression profiling were performed to analyze this gene family in maize. As a result, 46 Dof genes were identified in maize. Gene structure and motif analysis revealed that the Dof motifs are highly conserved, while the other characteristics are diverse. It is notable that the Dof genes belonging to the same group or clade always display similar domain architecture, suggesting that they may have similar functions. Many cis-elements were also found in the upstream sequence of the ZmDofs, suggesting complicated regulatory relationship. We also found that site-specific selection plays an important role in Dofs multi-functionalization. Furthermore, comprehensive analysis of the expression profiles provided insights into potential function among these ZmDof genes. The different expression profiles suggest that ZmDof genes carry out different physiological functions in different tissues. Notably, ZmDof16, ZmDof22, and ZmDof36 were strongly induced by salt treatment, indicating that they may play essential roles in response to abiotic stress. These data may provide valuable and useful information for future functional investigations of this gene family.
Abbreviations
- Dof:
-
DNA binding with one finger
- NJ:
-
neighbor joining
- MEME:
-
multiple EM for motif elicitation
- TSS:
-
transcription start site
References
Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11(3):323–334
Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16(7):1667–1678
Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z (2013) Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol 55(6):552–566
Cao J (2012) The pectin lyases in Arabidopsis thaliana: evolution, selection and expression profiles. PLoS ONE 7:e46944
Cao J, Shi F (2012) Dynamics of arginase gene evolution in metazoans. J Biomol Struct Dyn 30:407–418
Cao J, Shi F, Liu X, Huang G, Zhou M (2010) Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS Lett 584:4775–4782
Cao J, Huang J, Yang Y, Hu X (2011) Analyses of the oligopeptide transporter gene family in poplar and grape. BMC Genomics 12:465
Chen Y, Cao J (2014) Comparative genomic analysis of the Sm gene family in rice and maize. Gene 539:238–249
Chen W, Chao G, Singh KB (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 10:955–966
Chen Y, Hao X, Cao J (2014) Small auxin up-regulated RNA (SAUR) gene family in maize: identification, evolution and its phylogenetic comparison with Arabidopsis, rice and sorghum. J Integr Plant Biol 56(2):133–150
Chou PY, Fasman GD (1974) Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry 13(2):211–222
Comeron JM (1999) K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763–764
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190
Dong G, Ni Z, Yao Y, Nie X, Sun Q (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63(1):73–84
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206
Farré D, Albà MM (2010) Heterogeneous patterns of gene-expression diversification in mammalian gene duplicates. Mol Biol Evol 27(2):325–335
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42(Database issue):D222–230
Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94:6809–6814
Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93:10274–10279
Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14:1253–1263
Guo Y, Qiu LJ (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8(9):e76809
Hernando-Amado S, González-Calle V, Carbonero P, Barrero-Sicilia C (2012) The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol 12:202
Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11(11):550–558
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 Fbox protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297
Jiang HY, Luo C, Jiang T, Cheng BJ, Zhu S (2010) Genome-wide analysis of Dof transcription factor family genes in maize. China J Bioinforma 8:198–201, abstract in English
Kim HS, Kim SJ, Abbasi N, Bressan RA, Yun DJ, Yoo SD, Kwon SY, Choi SB (2010) The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J 64:524–535
Kisu Y, Harada Y, Goto M, Esaka M (1997) Cloning of the pumpkin ascorbate oxidase gene and analysis of a cis-acting region involved in induction by auxin. Plant Cell Physiol 38:631–637
Kisu Y, Ono T, Shimofurutani N, Suzuki M, Esaka M (1998) Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol 39:1054–1064
Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D (2011) Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep 38:5037–5053
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3:17
Liu Y, Jiang H, Chen W, Qian Y, Ma Q, Cheng B, Zhu S (2011) Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul 63:225–234
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25(4):402–408
Martin C, Paz-Ares J (1997) MYB transcription factors in plants. Trends Genet 13(2):67–73
Marzábal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD (2008) The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol Biol 67(5):441–54
Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P (1998) An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J 16(1):53–62
Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130:111–119
Moreno-Risueno MA, Martínez M, Vicente-Carbajosa J, Carbonero P (2007) The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics 277(4):379–390
Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, Hashimoto-Sugimoto M, Schroeder JI, Nishitani K, Yanagisawa S, Iba K (2013) A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr Biol 23:479–484
Noguero M, Atif RM, Ochatt S, Thompson RD (2013) The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci 209:32–45
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10(2):79–87
Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2000) Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev 14:28–33
Papi M, Sabatini S, Altamura MM, Hennig L, Schafer E, Costantino P, Vittorioso P (2002) Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol 128:411–417
Plesch G, Ehrhardt T, Mueller-Roeber B (2001) Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 28:455–464
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66(4):553–563
Shaw LM, McIntyre CL, Gresshoff PM, Xue GP (2009) Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomic 9(4):485–498
Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor MI, Lohmann JU, De Veylder L, Witt I, Mueller-Roeber B (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 56:779–792
Soderlund C, Bomhoff M, Nelson WM (2011) SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res 39:e68
Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T (2007) Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res 35(Web Server issue):W506–511
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18:207–208
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12(6):239–44
Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94(14):7685–7690
Washio K (2001) Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim Biophys Acta 1520:54–62
Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5:276–287
Yanagisawa S (1997) Dof DNA binding domains of plant transcription factors contribute to multiple protein–protein interactions. Eur J Biochem 250(2):403–410
Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21:281–288
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45(4):386–91
Yanagisawa S, Izui K (1993) Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem 268(21):16028–16036
Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17(2):209–214
Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10:75–89
Yang X, Tuskan GA, Cheng MZ (2006) Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142(3):820–830
Yang Z, Wang X, Gu S, Hu Z, Xu H, Xu C (2008) Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407(1–2):1–11
Yu CS, Lin CJ, Hwang JK (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13:1402–1406
Zhang B, Chen W, Foley RC, Buttner M, Singh KB (1995) Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 7:2241–2252
Zhu X, Ma H, Chen Z (2011) Phylogenetics and evolution of Su(var)3-9 SET genes in land plants: rapid diversification in structure and function. BMC Evol Biol 11:63
Acknowledgments
This project is supported by grants from the National Science Foundation of China (No. 31100923), the National Science Foundation of Jiangsu Province (BK2011467), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Jiangsu University “Youth Backbone Teacher Training Project” (2012–2016) to JC.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Data S1
Alignment data used in this study. (TXT 129 kb)
Fig. S1
Neighbor-joining phylogenetic tree of Dof proteins in Arabidopsis, rice, sorghum, and maize. (PDF 61 kb)
Fig. S2
Distribution of the predicted positive selection sites on the secondary structure of Dof protein in group VII. The predicted positive selection sites are indicated with arrows and red bold letters. (PDF 56 kb)
Table S1
Distribution of five class’s cis-elements in the promoter regions of the 46 ZmDof genes. (XLS 39 kb)
Table S2
Primers used in this study. (DOC 31 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Cao, J. Comparative Analysis of Dof Transcription Factor Family in Maize. Plant Mol Biol Rep 33, 1245–1258 (2015). https://doi.org/10.1007/s11105-014-0835-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0835-9