Abstract
Background and aim
Consortia-based inoculants, such as the microbiome method, are advocated over individual biocontrol agents (BCAs), owing to partner reinforcement and rapid rhizospheric establishment. Herein, we screened the sole and dual suppressive effects of selected desert spurge (Euphorbia antiquorum) derived Bacillus spp. vis-à-vis Ralstonia solanacearum.
Methods
The conventional agar diffusion method was used to examine the BCAs' capacity to release antibiotics. Thereafter, the compatibility, the ability to form biofilm and the phytotoxicity of the BCAs towards tobacco leaves were screened. The Box-PCR fingerprinting technique was adopted to ascertain the endophytism of the BCAs, and the wilt suppression test followed by the specific activity of defence-related enzymes was performed.
Results
R. solanacearum was completely inhibited by the extracts from Bacillus velezensis CBv_BE1, Bacillus amyloliquefaciens CBa_BFL2, and Bacillus amyloliquefaciens CBa_RA37 at 0.625 mg/mL. The outstanding candidates could form biofilm and colonize into tomato seedlings, with a plateau at log10 CFU = 4.29/g fresh weight. The consortium from CBa_BFL2 and CBa_RA37 (CBa_BFL2/CBa_RA37) yielded vigorous plants (up to a 319% increase in biomass). Likewise, a 90% and 89% reduction in wilt incidence and severity was recorded by the CBa_BFL2-CBa_RA37 combination. The Guaiacol peroxidases (GPX), phenylalanine ammonia lyase (PAL), and superoxide dismutase (SOD) specific activity were significantly and inversely correlated with wilt severity via Pearson's test. This alludes to a greater innate defence system in tomatoes in response to bacterization.
Conclusions
Our data suggest that tomato priming with a consortium of B. amyloliquefaciens CBa_BFL2 and CBa_RA37 resulted in vigorous and healthier tomato seedlings as referred to their sole effects, with altered activities of key plant distressing enzymes. However, further studies are warranted to reveal its full potential as a pioneering alternative to agrochemicals for the control of bacterial wilt.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The inadequacy between the global food supply and ever-increasing demand is steadily imposing a shift in the current agricultural production systems. The world has faced similar concerns throughout history, which have been successfully addressed through a number of measures such as expanding agricultural lands (Gouda et al. 2018), the use of fertilizers and genetically engineered planting materials (Shrestha 2016; Gouda et al. 2018), and pest and disease mitigation through the application of chemical biocides (Eke et al. 2016; Kamkar 2016). (Eke et al. 2016; Kamkar 2016). To date, these approaches are less advocated, owing to their inherent health and environmental concerns, particularly in light of the changing climate. (Kumar et al. 2011). Besides, according to the Food and Agricultural Organization (FAO 2024), only 12.6% of the earth’s arable land is devised to crop production, of which, 25% of the land gets damaged each year due to flood and drought events, depending on geographical location. Unfortunately, this tendency is expected to become worse in the future years (The World Counts 2024). Demographers estimate that there are up to 81 million extra mouths to feed and house each year in the humanitarian sector (United Nations 2019). Thus, ensuring a tripartite trade-off between food security, environmental safety, and sustainable land management is the present unprecedented task for the world's agriculture (Gouda et al. 2018). In this regards, microbial technology has emerged as an important tool for sustainable crop production. (Ubertino et al. 2016; Xiaobo liu 2020). As a result, endophytic bacteria found in the intercellular spaces of higher plants have been isolated and tested for their functional traits, with the goal of introducing them into crop production systems. (Sheoran et al. 2016; Munjal et al. 2016; Amr et al. 2021). Known as plants primary colonizer, the former are well-springs of chemically diverse and novels compounds capable of altering host’s physiology, leading but not limited to enhanced yield and resistance toward phytopathogen (Sheoran et al. 2016; Amr et al. 2021). A wide spectrum of study endeavours have acknowledged the biocontrol potentials of endophytic bacteria, notably against rice leaf blight incited by Xanthomonas oryzae pv. oryzae, (Lingaiah and Umesha 2013), Fusarium (F. gaminearum) head blight of wheat (Ma 2024), bacteria wilt of tomato (Raza et al. 2016), peanuts (Jiang et al. 2017; Chen et al. 2019) and Phytophthora post-harvest rot of stored potato tubers (Oksana et al. 2020).
Despite the many papers demonstrating their biocontrol abilities, there has always been controversy around the effectiveness of BCAs in open fields owing to the unpredictable interactions they have with soil autochthonous microbiota (Hong and Kwon 2016). The underlined gap has led to the advocacy of consortia over individual BCAs, as they may readily thrive in hostile environment with enhanced efficiency through synergism or additivity. Henceforth, the attributes of endophytes differ drastically in function of the geographical regions from which they originate. This process commonly known as ‘habitat-adapted symbiosis’ is believed to result from the activation of distinct metabolic pathways in a face of a stressor (Ballesteros et al. 2022, Ballesteros et al. 2024). The performances of agricultural crops might therefore be enhanced by endophytes from arid and saline soils, such as those in desert and Antarctic environments (Poveda et al. 2022). Although these understudied environments are largely untapped because of their geographic isolation and inhospitable conditions, they are considered to be rich sources of endophytes that might synthesize a plethora of natural scaffolds capable of enhancing host stress tolerance (Tiwari & Bae 2022; Ballesteros et al. 2024).
This work aimed to evaluate the in vitro and in planta antagonistic properties of E. antiquorum originating endospheric Bacilli from the Sahelian region of Cameroon against the bacterial wilt of tomato.
Materials and methods
The pathogenic and biocontrol agents
The bacterial strains used, namely B. velezensis CBv_BE1, B. amyloliquefaciens CBa_BFL2 and B. amyloliquefaciens CBa_RA37, were gotten from the core collection of the Biocontrol gents Unit of the Laboratory of Phytobiochemistry and edicinal Plants Studies, of the University of Yaoundé I, Cameroon. The BCAs were previously isolated from horns and seeds of wild E. antiquorum plant from Far North Region of Cameroon (5′ 13.4988’’ N and 15° 0′ 53.3952’’E). Data pertaining to their identification (16S rRNA gene sequence) and plant growth promotion attributes were reported by Eke et al. (2019).
The pathogen, namely R. solanacearum Rs5 was isolated from tomato stem, exhibiting typical symptoms of bacterial wilt (Youmbi et al. 2023). The wilt-causing bacterium was the most pathogenic (100% incidence and 90% wilt severity) among several others upon pathogenicity test (data not shown). The bacteria (pathogenic and BCAs) were stored at -20 and -80 oC.
Antagonistic effects of BCAs against R. solanacearum
The potential of the BCAs to suppress the development of R. solanacearum was tested on Muller Hinton Agar (MHA) plates using the dual culture approach as reported by Hudzicki (2009). Single log-phase colonies of Bacillus sp. or R. solanacearum were scraped off MHA plates and subcultured overnight in Muller Hinton broth (MHB) media with stirring (28 °C, 150 rpm). MHA plates were inoculated with 100 µL of R. solanacearum RS5 culture (108 Cells/mL). After one hour, wells (5 mm diameter) were drilled in each plate and 100 µL of each Bacillus suspension were added. The plates were incubated (28 ± 2 °C, 48 h) and the diameters of clear halos (inhibition zone) formed around the wells were measured and expressed in centimetre. Each experiment was performed in triplicate and repeated twice. Control wells consisted of sterile distilled water.
Antibacterial potential of culture filtrates from the BCAs
The culture filtrates were obtained upon growing each Bacillus spp. (28 °C; 150 rpm) for 5 days in 100 mL sterile MHB, followed by centrifugation (10,000 rpm, 15 min) and filtration through a 0.22 µm membrane (Millipore). The inhibitory effect of resulting crude culture filtrates against R. solanacearum RS5 was evaluated using the agar dilution method (Grover and Moore 1962). In brief, filtrates were added to melted MHA medium to make final concentrations of 0.0, 6.25, 12.5, 25.0 and 50.0% (v/v), and poured in Petri dishes (55 cm Ø). After solidification, the pathogen (108 cell/mL) was inoculated (100 µL) by sprawl. Negative control plates consisted of R. solanacearum RS5 grown on extract-free MHA medium. Ciprofloxacin served as positive control. The plates (controls and tests), were sealed with parafilm tape and incubated at 28 °C for 72 h. Upon incubation, R. solanacearum survivor colonies were counted after culture on solid medium and expressed in terms of percentage of growth inhibition (RGI) with reference to negative control.
Antibacterial potential of ethyl acetate based-extracts from BCAs
Ethyl acetate extracts were prepared from previously obtained culture filtrates through liquid–liquid partitioning. The extraction solvent was then evaporated at 70 °C using a rotary evaporator (BUCHI 001). The inhibitory effect of the so-said extracts were assessed using the M38-A2 procedure with minor modifications (CLSI 2008). In 96-well microtiter plates containing 100 µL MHB (Himedia, India), 100 µL of ethyl acetate extracts were added to the first well and carefully mixed. Two-fold serial dilution was then performed to reach final extract concentrations of 10.0, 5.0, 2.50, 1.25, 0.625, 0.312, and 0.156 mg/m.
L. Then, 100 µL of R. solanacearum RS5 suspension (108 CFU/mL) were added to wells containing the test extract. Wells without inoculum served as blanks (sterility controls), whereas extract-free wells were negative controls. The plates were incubated for 48 h at 28 °C. The experiment was carried out in triplicate and repeated twice. The Minimum Inhibitory Concentration (MIC), which is the lowest inhibitor concentration that totally suppresses bacterial growth, was determined.
Biofilm formation assay
The Biofilm formation capacity of the BCAs was evaluated (O'Toole and Kolter 1998). In summary, 100 µL of each bacterial solution at 108 CFU/mL in LB culture media were seeded in flat bottomed 96-well microtiter plate. The negative control wells contained just sterile LB medium. The plates were incubated at 28 °C for 72 h, and turbidity at OD600 nm was measured using a microtiter plate reader (TECAN M200). The growth media was then withdrawn, and the plates were washed three times with sterile distilled water to eliminate any planktonic cells. The plates were stained with 150 µL of crystal violet (0.1% v/v) for 30 min. After that, they were washed three times with sterile distilled water before being resuspended in ethanol (95%). The optical density (OD) was measured at 570 nm and compared to the controls. The biofilm formation index (BFI) was developed by normalising biofilm formation with regard to bacterial growth using the following equation:
BFI stands for Biofilm Formation Index.
OD570 and OD600 are Optical Density for biofilm formed (OD570) and cell growth (OD600), respectively.
Hypersensitivity test
Prior to applying BCAs to planting material, their possible harmful impact on plant cells was tested on detached tobacco (N. tabacum) leaves (Granada and Sequeira 2011). Tobacco leaves' secondary veins were injected with 10 µL of bacterial cell solution (108 cells/mL) using a micro-syringe. The negative control leaves were injected with sterile distilled water (SDW). Both control and test leaves were incubated at room temperature (27–29 °C) for 72 h. The bioagents that caused chlorotic and/or necrotic areas surrounding the inoculation sites were deemed pathogenic and were removed from the experiment.
Seed biopriming assay
Tomato seeds (Cv. Rio Grande) were surface disinfected by immersing them in 1.5% sodium hypochlorite (NaOCl) for 3 min and then rinsing three times with SDW. The sterilised seeds were spread on Petri plates with filter papers soaked with SDW. Furthermore, 1 mL suspensions (108 cells/mL) of BCAs were applied to the Petri plates. An equal amount of SDW was applied to the negative control plates. Each Bacillus sp. was represented by four plates containing 25 seeds. The plates were incubated on a greenhouse bench, and seed germination was monitored daily. The experiment was performed three times. The germination index (GI) was calculated as stated by Bench et al. (1991).
With: n1, n2, …, n10 = Number of germinated seeds on the first (n1), second (n2) till the day x (nx) and 10, 9, …, 1 are weights given to the number of geminated seeds on the first, second and subsequent days respectively.
Assessment of endophytic performance of BCAs
Spontaneous antibiotic multi-resistant mutant generation
To facilitate the retrieval of BCAs from tomato tissues after artificial inoculation, spontaneous antibiotic resistance mutants were generated prior to seed inoculation. BCAs were seeded on Luria Berthani Agar (HIMEDIA, India) with 100 µg/mL of Ampicillin, Chloramphenicol, Ciprofloxacin, or a combination (Silvana and Maria 2016). Fifty (50) mL of 18-h (Mid log phase) BCAs suspensions were centrifuged at 8000 g for 2 min, and the cell pellets were resuspended in 5 mL of saline (0.09% NaCl). A 100µL aliquot of the solution was plated on LBA containing the aforementioned antibiotics. Distinct colonies (ground glass appearance, cream haemolytic colonies, and uneven edges) were picked and streaked onto newly prepared LBA medium containing a mixture of the three antibiotics (Sheoran et al. 2016). The antibiotic multi-resistant mutants were then kept at -20 °C in antibiotic-administered Luria Berthani Broth.
Tomato inoculation with multi-resistant mutant BCAs and gnotobiotic growth
Two (2) week-old tomato plantlets (Cv. Rio Grande) were cultivated in plastic pots (0.5 kg) using sterile garden soil. Overnight cultures of antibiotic multi-resistant mutants of each BCA were rinsed three times with saline (0.09% w/v NaCl) solution before centrifugation (8000 g, 5 min). The pellets were re-suspended in fresh saline and diluted to 0.5 × 108, 108, 2 × 108, and 4 × 108 CFU/mL. Each inoculum contained 100 mL, which was poured around the collar area of immature plantlets. The control seedlings received SDW instead. The bacterized and mock plants were cultivated under axenic conditions, with continual SDW irrigation. The endogenous bacterial population number was evaluated 21 days after inoculation (dpi).
Re-isolation and quantification of endophytic competent candidates
Tomato plantlets were carefully uprooted and surface sterilized by soaking in 2% chlorox (10 min) and 70% ethanol (1 min), then washing three times with SDW. One g of each sample was then pulverised in phosphate buffered saline (PBS) at pH 7.2. The resultant slurry was allowed to settle for 10 min before being collected and plated on LBA supplemented with the aforementioned antibiotic combination and incubated at 28 °C for 48 h. Sheoran et al. (2016) counted colonies and expressed them in terms of CFU/g of fresh weight. The experiment was carried out in triplicate.
Box PCR fingerprinting
The BOX-PCR profiles of wild and re-isolated endophytic Bacilli were compared to determine if the BCAs effectively colonised tomato tissues (Martin et al. 1992). The amplification mixture included 100 µM of template DNA from re-isolated antibiotic-resistant and wild type strains of B. velezensis CBv_BE1, B. amyloliquefaciens CBa_BFL2, and B. amyloliquefaciens CBa_RA37, along with 2 µM BOX primer, Dntps, 10 mM Tris–HCL, 50 mM KCl, 2.5 mM MgCl2, and 1.25 µM taq DNA polymerase. The amplification was carried out with the following Thermal Cycler software: (i) a 7-min initial denaturation stage at 95 °C; (2) 30 cycles of 1 min at 94 °C, 1 min at 53 °C, and 8 min at 65 °C, followed by a 15-min final elongation step at 65 °C. The results of the PCR amplification were seen under a UV lamp and identified by electrophoresis (Sambrook and Russell 2001). Pictures were captured using the BioPrint device. A 1 KB DNA ladder was used as a baseline. To guarantee the repeatability of the patterns, amplifications were carried out in at least two different assays, and only bands that were shared by the duplicated amplifications were scored. After that, the DNA fingerprints, or band patterns, of the wild-type and antibiotic-resistant mutant BCAs were compared.
BCAs compatibility assessment
The bioagents were also assessed for their capacity to coexist in the same ecological niche. Overnight cultures of 100µL of each BCA were spread over LBA medium to make a lawn. Different bacteria were drop-inoculated (10µL) at equidistant points on the same plate, and vice versa. Plates were incubated at 28 °C for three days and checked for inhibiting halo formation (Silvana and Maria 2016). The experiment was carried out in triplicate.
Assessment of in planta synthetic induced resistance
Pre-germination and priming of tomato seeds
Multi-competent-compatible BCAs were utilised to confront R. solanacearum Rs5 in planta. Tomato seeds (var. Rio Grande) were surface sterilised and pre-germinated in plastic trays for 5 days, as reported before. The pre-germinated seeds were then immersed in a bacterial solution (108 CFU/ml) for four hours. Control seeds were steeped in an equivalent amount of SDW. The primed and non-primed seedlings were grown in plastic pots filled with sterile garden soil and watered on a regular basis.
Pathogen inoculation and experimental layout
At transplanting, 14-day-old tomato seedlings were gently uprooted and washed with SDW. The root tips were then cut with sterile scissors and immersed in Log-phase R. solanacearum Rs5 at 108 CFU/mL for 30 min (Ji et al. 2014). The diseased plantlets were placed into plastic containers. Negative control plants were dipped in the same amount of SDW. The treatments used were codified as follows: (1) Control: uninoculated seedlings (no Bacillus sp. or R. solanacearum Rs5). (2) Rs5: seedlings infected by R. solanacearum Rs5 alone, (3) BFL2_Rs5: seedlings primed by B. amyloliquefaciens CBa_BFL2 and inoculated with R. solanacearum Rs5. (4) BE1_Rs5: Seedlings primed by B. velezensis CBv_BE1 and inoculated with R. solanacearum Rs5. (5) RA37_Rs5: Seedlings treated with B. amyloliquefaciens CBa_RA37 and challenged with R. solanacearum Rs5. (6) BFL2_BE1_Rs5: seedlings were primed with B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1, and infected with R. solanacearum Rs5. (7) RA37_BE1_Rs5: seedlings dually primed with B. amyloliquefaciens CBa_RA37, B. velezensis CBv_BE1, and infected with R. solanacearum Rs5. (8) BFL2_RA37_Rs5: seedlings dually primed with B. amyloliquefaciens CBa_BFL2, B. amyloliquefaciens CBa_RA37, and infected with R. solanacearum Rs5. The pots were placed in the semi-controlled greenhouse using a Randomised Complete Block Design (RCBD) with fifteen (15) replications for each treatment.
Disease and agro-morphological parameters recording
The percentage of wilting plants (PWP) was determined for each treatment at 7, 14, 21, and 28 dpt. The wilt severity (WS) was calculated using He's (1983) 1 to 5 disease rating scale, with slight modifications: 1: no visible wilting symptom on the leaf system, 2: one wilted leaf, 3: two to three wilted leaves with no yellowing, 4: four or more wilted leaves with slight leaf chlorosis, and 5: all leaves wilted or total plant death. The WS was then determined using the following formula:
With: ni = number of plants displaying similar disease severity score; vi given disease score in relation with ni; N = Total number of examined plants per treatment; V = highest wilting severity (1 to 5). In addition, the Area Under the Disease Progress Curve (AUDPC) was calculated as reported by Simko and Hans (2011).
With: y1, y2 and yn = wilting index at day one, day two and last day of observation respectively. t1, t2, tn-1 and tn = Time of first, second to the last day of disease estimation.
Twenty-eight days post transplantation, root and shoot lengths and dried weights were recorded.
Extraction and assessment of oxidizing enzymes
The protocol described by Silvana et al. (2003) was followed in the preparation of oxidising enzyme extracts. One g fresh material (root and aerial parts) was crushed on ice using a pre-chilled mortar and pestle and suspended in 10 mL of 50 mM phosphate buffer (Ph 7.4) containing 1 mM EDTA, 1 g of polyvinylpyrrolidone (PVP), and 5% (V/V) Triton X100. The slurry was centrifuged at 10,000 g for 20 min at 4 °C, and the supernatant was used to perform the guaiacol peroxidase (GPX), superoxide dismutase (SOD), and phenylalanine ammonia lyase (PAL) tests.
Assessment of GPX-specific activity
Peroxidase activity was evaluated using Rabe and Kreeb's (1979) approach, using a reaction mixture including the supernatant and guaiacol (100 mM sodium phosphate buffer, pH 7.0 and 20 mM guaiacol). The absorbance at 470 nm increased after adding 20 µL H2O2. Enzyme activity was measured as the change in optical density per gramme of fresh weight per minute.
Assessment of superoxide dismutase specific activity
Silvana et al. (2003) assessed the activity of total superoxide dismutase by inhibiting the photochemical reduction of nitroblue tetrazolium. In a test tube, combine 200 µL of plant extract supernatant and 300 µL of buffer solution (50 mM potassium phosphate buffer, pH 7.8, and 0.1 mM EDTA) with 3.5 mL of O2-generator combination (14.3 mM methionine, 82.5 µM NBT, and 2.2 µM riboflavin). The test tubes, control (tube without supernatant), and blank (tube with supernatant but not exposed to light) were all orbitary shaken and put 30 cm apart under continual illumination from a natural light source for 10 min. Optical density was measured at 560 nm using a Tecan M200 multiplate reader (Tecan, Bangkok, Thailand) to determine the percent inhibition of NBT reduction, which corresponds to the quantity of SOD present in the enzymatic extract and is represented in SOD units per mg of protein.
Assessment of PAL-specific activity
Ross and Sederoff's (1992) procedure was used to quantify PAL-specific activity. The reaction mixture of 100 µL plant extract, 500 µL 50 mM Tris HCL (pH 8.8), and 600 µL 1 mM L-phenylalanine was incubated at room temperature for 60 min. The process was stopped by adding 2N HCl. Toluene (1.5 mL) was added to the test mixture and vortexed for 30 s before centrifugation at 1000 g (CRU-5000 centrifuge ITC) for 5 min. Toluene phase containing trans-cinnamic acid was recovered and its absorbance was measured at 290 nm against a black background. Enzyme activity was measured in nanomoles of trans-cinnamic acid released per g of fresh material.
Data analysis
The data were normalised and submitted to ANOVA. Prior to analysis, results from colony counting assays were log transformed. To determine the median inhibitory concentrations (IC50) of culture filtrates against R. solanacearum Rs5, a non-linear (sigmoidal) regression model was used. The Tukey post-hoc test was used to compare mean values pairwise. Pearson's test was employed to investigate the correlations between parameters where necessary. The studies were performed using the statistical programme GraphPad Prism version 9.0. The significance thresholds were set for each test as specified by the software.
Results
Antagonistic effects of BCAs against R. solanacearum Rs5
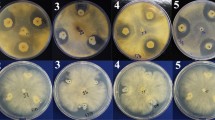
Direct confrontation was used to assess the BCAs' antagonistic impact (Fig. 1). The growth of R. solanacearum was significantly impeded by the BCAs when compared to the mock (F3, 20 = 974.1, p < 0.0001). The Turkey test revealed that B. velezensis CBv_BE1 and B. amyloliquefaciens CBa_BFL2 had significantly larger inhibition zones (27 ± 1.2 mm and 27 ± 0.6 mm, respectively) compared to B. amyloliquefaciens CBa_RA37 (23 ± 1.5 mm).
Differential antagonistic effects of B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1 and B. amyloliquefaciens CBa_RA37 against R. solanacearum Rs5. Results are presented as means ± standard deviation of 3 × 2 replicates. Bar charts bearing different alphabet letters denote significantly different effects at p ≤ 0.05, according to Turkey post-hoc test
Antibacterial potential of culture filtrates and acetate based-extracts from the BCAs.
The culture filtrates from all of the BCAs strongly (F3, 16 = 5.024, p = 0.0121) suppressed the growth of R. solanacearum Rs5, with inhibition rates ranging from 0.0 to 100%. The Pearson correlation test found a positive and substantial association (r = [0.72, 0.88]; p < 0.001) between filtrate concentration and inhibition percentages. R. solanacearum was eliminated at 5.52% (v/v), 4.65% (v/v), and 4.94% (v/v) in culture filtrates from B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1, and B. amyloliquefaciens CBa_RA37 (Fig. 2).
Non-linear representation of concentration dependent inhibitory action of culture filtrates of B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1 and B. amyloliquefaciens CBa_RA37 against R. solanacearum Rs5. Results are mean percentages of colony counts ± standard deviation of three replications (n = 3) per concentration. IC50 values were compared using the Turkey’s HSD post-hoc test. (***) indicates significantly higher LogIC50 and afferent IC50 at α = 0.001
The ethyl-acetate extracts of B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1, and B. amyloliquefaciens CBa_RA37 showed minimum inhibitory concentrations (MIC) of 0.625 ± 0.0 mg/mL, 0.625 ± 0.0 mg/mL, and 1.25 ± 0.0 mg/mL, respectively. These activity profiles revealed that B. amyloliquefaciens CBa_BFL2 and B. velezensis CBv_BE1 performed much better than B. amyloliquefaciens CBa_RA37.
Biofilm production by the BCAs
All the Bacillus spp. tested showed ability to form biofilm (Table 1) in various intensities (F3, 8 = 80.51, p < 0.0001). Significantly greater Biofilm formation index (BFI) was recorded with the BCA B. amyloliquefaciens CBa_RA37 (BFI = 1.2) compared to B. amyloliquefaciens CBa_BFL2 (BFI = 0.67) and B. velezensis CBv_BE1 (BFI = 0.77), indicating its greater suitability to resist to plant tissue environmental stresses when colonized.
Hypersensitivity and BCAs compatibility
None of the BCAs elicited a hypersensitive response to tobacco leaflets, demonstrating their safety for plant tissues. Furthermore, no hostile activity was found between the BCAs when grown in the same environment, demonstrating their capacity to coexist in the same biological area.
Seed bacterization enhances germination
Regardless of the BCAs, primed tomato seedlings resulted in 100% germination rate (GR), compared to 75% for the mock (p < 0.001). Turkey's pairwise analysis revealed considerably quicker germination and radicle development in B. velezensis CBv_BE1-primed seeds (GI = 417) compared to other BCAs and the mock treatment (Table 2, Fig. 3).
Triggered tomato seeds germination and early radicle growth as a result of seed bacterization. Sterile tomato seeds (n = 120/treatment) were inoculated with cell suspension from B. amyloliquefaciens CBa_BFL2, CBa_RA37 and B. velezensis CBv_BE1 respectively. Seed germination was recorded daily and radicles lengths were measured 10 dpi
Endophytic competence
Antibiotic-resistant mutants of the examined BCAs successfully colonized the inside of tomato tissues (Fig. 4A). The endogenous endophyte population increased with inoculum size, peaking at log10 CFU = 4.29 cells per g fresh weight 21 days after inoculation (dpi). Afterwards, the endophyte population density did not fluctuate (F2, 12 = 0.00834, p = 0.9917) regardless of the BCA's load increase, indicating a potential microbial saturation at 5 × 108 CFU/mL. Furthermore, the recovered BCAs' BOX_PCR profiles (BFL2R, BE1R, and RA37R) matched those of their wild-type counterparts (BFL2w, BE1w, and RA37w). Overall, the data clearly demonstrated the tested BCAs' endophytic competency (Fig. 4B).
Endophytic competence of BCAs by population size at 21 dpi (A) and Box-PCR fingerprint of wild and re-isolated antibiotic resistant BCAs (B) (1–2), (3–4) and (5–6): band patterns of wild (BFL2w, BE1w and RA37w) and re-isolated (BFL2R, BE1R and RA37R) B. amyloliquefaciens CBa_BFL2, B. amyloliquefaciens CBa_RA37 and B. velezensis CBv_BE1, respectively
Tomato seedling bacterization suppressed wilt incidence and severity
In pathogen-infected control, seedlings were stunted, leaves yellowed and wilted, and the plant collapsed and died. When challenged with BCAs, the proportion of wilted plants (F6, 7 = 154.6, p < 0.0001) and wilt severity (F6, 7 = 5930, p < 0.0001) decreased considerably compared to pathogen-treated seedlings alone (Fig. 5 and 6). The consortium CBa_BFL2 + CBa_RA37 was the most successful treatment, with 90% disease suppression (mean PWP = 10%) at final count (Supplementary information 1). Similarly, the same consortium (CBa_BFL2 + CBa_RA37) demonstrated greater wilt severity reduction, achieving 89% (mean severity = 11%) as compared to other treatments (Fig. 5 and 6).
Biological control effect of singly and binary inoculated BCAs on bacterial wilt incidence and severity in tomato seedlings. Standing plants were counted four weeks after R. solanacearum Rs5 infection and expressed in percentages with reference to the total number of assayed plants (n = 15). Data are means of 15 replicates ± SD. Control: Uninoculated seedlings (Neither Bacillus sp nor R. solanacearum Rs5); Rs5: Seedlings infected with R. solanacearum Rs5 alone; BFL2_Rs5: Seedlings primed by B. amyloliquefaciens CBa_BFL2 and infected with R. solanacearum Rs5; BE1_Rs5: Seedlings primed by B. velezensis CBv_BE1 and infected R. solanacearum Rs5; RA37_Rs5: Seedlings primed with B. amyloliquefaciens CBa_RA37, challenged with R. solanacearum Rs5; BFL2_BE1_Rs5: Seedlings dually primed with B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1 and infected with R. solanacearum Rs5; RA37_BE1_Rs5: Seedlings dually primed with B. amyloliquefaciens CBa_RA37, Bacillus velezensis CBv_BE1 and infected with R. solanacearum Rs5; BFL2_RA37_Rs5: Seedlings dually primed with B. amyloliquefaciens CBa_BFL2, B. amyloliquefaciens CBa_RA37, and infected with R. solanacearum Rs5. ANOVA of disease severity at day 1 (F6, 7 = 1.745, p = 0.2413), day 7 (F6, 7 = 48,455, p < 0.0001), day 14 (F6, 7 = 120.1, p < 0.0001), day 21 (F6, 7 = 884.8, p < 0.0001) day 28 (F6, 7 = 5930, p < 0.0001). ANOVA of disease incidence at day 1 (F6, 7 = 0.7019, p = 0.6589), day 7 (F6, 7 = 0.8462, p = 0.5778), day 14 (F6, 7 = 615.6, p < 0.0001), day 21 (F6, 7 = 518.6, p < 0.0001) and day 28 (F6, 7 = 154.6, p < 0.0001). Distance between mean values was measured by the Turkey’s multiple range test at P < 0.001
Biological control of bacterial wilt and growth performance in tomato seedlings as influenced by single and binary inoculation of BCAs. (A) Tomato seedlings dually inoculated with B. amyloliquefaciens CBa_RA37 and B. velezensis CBv_BE1 and challenged with R. solanacearum Rs5. (B) Uninoculated seedlings (control) neither by the Bacillus sp. nor R. solanacearum Rs5. (C) R. solanacearum Rs5 infected plants without any antagonist. (D) Plantlets co-inoculated with B. amyloliquefaciens CBa_RA37 and CBa_BFL2 and infected with R. solanacearum Rs5. (E) Plantlets primed with B. amyloliquefaciens CBa_RA37 alone and grown on R. solanacearum Rs5 infested soil. (F) Plantlets primed with B. velezensis CBv_BE1 alone and grown on R. solanacearum Rs5 infested soil. (G) Plantlets dually-primed with B. amyloliquefaciens CBa_BFL2 and B. velezensis CBv_BE1 and challenged with R. solanacearum Rs5. (H) Plantlets primed with B. amyloliquefaciens CBa_BFL2 and grown on R. solanacearum Rs5 infested substrate. The values presented in Fig. 5I. were taken on a weekly basis and were generated from fifteen replicates at each recording date
Area under the disease progress curve (AUDPC)
The trapezoidal integration approach was used to determine the summary of disease intensity across the study period (see Table 3). The results showed that the AUDPC varied from 14 to 556.5. In comparison to Rs5-treated control plants (AUDPC = 556.5), binary inoculation of seedlings with CBa_BFL2 + CBa_RA37 provided the greatest cumulative disease suppression throughout the study period (AUDPC = 14).
Effect of bacterial priming on agro-morphological parameters
The overall agro-morphological parameters changed dramatically as a result of pathogen infection. Shoot and root dry weights decreased by—66% and—86%, respectively, as compared to stress-free (uninfected and unprimed control) plants. In contrast, seedling bacterization resulted in dramatically increased seedling vigour, ranging from 157 to 392% compared to the mock. The consortium priming with CBa_BFL2 + CBa_RA37 was very successful, resulting in 229%, 392%, 157%, and 319% increases in shoot length, root length, and root and shoot dry weights, respectively (Table 3).
Bacterial priming triggered specific activities of some defence-related enzymes
The specific activity of selected defence-related enzymes synthesized by primed tomato plants and challenged with R. solanacearum Rs5 is shown (Fig. 7). R. solanacearum Rs5 infection significantly increased the synthesis of GPX (F7, 16 = 27.18, p < 0.0001), SOD (F7, 16 = 17.23, p < 0.0001) and PAL (F7, 16 = 37.03, p < 0.000). Meanwhile, significantly (p < 0.001) lowered activity of the overall enzymes was noticed upon bacterization with BCAs when compared to the pathogen control plants (Rs5). No statistical variation was found in the responses of single and consortium of BCAs be it for SOD (F7, 16 = 1.885, p = 0.176), GPX (F7, 16 = 2.008, p = 0.1557) or PAL (F7, 16 = 1.720, p = 0.2105). Whereas, the Pearson model depicted positive and significant correlations between the specific activities of GPX (r = 0.85; p = 0.006), PAL (r = 0.77; p = 0.02) and SOD (r = 0.72; p = 0.05) and wilt severity (Table 4). Hence, the higher the disease severity, the greater the activities of GPX, SOD and PAL enzymes.
Activities of GPX, SOD and PAL in tomato plants affected by R. solanacearum Rs5 and primed by BCAs. Control (uninoculated seedlings); Rs5 (R. solanacearum Rs5 infected plantlets without antagonist); CBa_RA37 + Rs5 (plantlets primed with B. amyloliquefaciens CBa_RA37 alone and grown on R. solanacearum Rs5 infested soil); CBa_BFL2 + Rs5 (plantlets primed with B. velezensis CBv_BE1 alone and grown on R. solanacearum Rs5 infested soil); CBv_BE1 + Rs5 (plantlets primed with B. velezensis CBv_BE1 alone and grown on R. solanacearum Rs5 infested soil); CBa_RA37 + CBv_BE1 + Rs5 (seedlings primed with both B. amyloliquefaciens CBa_RA37, B. velezensis CBv_BE1 and challenged with R. solanacearum Rs5); CBa_BFL2 + CBv_BE1 + Rs5 (plantlets co-inoculated with B. amyloliquefaciens CBa_BFL2, B. velezensis CBv_BE1 and challenged with R. solanacearum Rs5). CBa_BFL2 + CBa_RA37 + Rs5 (plantlets co-inoculated with B. amyloliquefaciens CBa_BFL2, CBa_RA37 and challenged with R. solanacearum Rs5). Mean values bearing different alphabet are significantly different (P < 0.01) as given by the Turkey’s HSD Post Hoc test
Discussion
The interplay of the components at the three vertices of the "disease triangle" including virulent pathogen, vulnerable host, and environmental conditions has been identified as determining factors underpinning phytopathogen's destructive effects. Disrupting this balance by acting directly or indirectly on any of the components of the disease triangle is the foundation of plant disease management techniques (Wang et al. 2018). Agriculture has long been and continues to be heavily reliant on chemical inputs to fulfil this role, but their lethal effects have prompted a search for novel and dependable technologies for higher quantity and quality food provision while protecting human health (Alori and Fawole 2017; Alori and Babalola 2018). Microbial inoculants are increasingly being used for this purpose because their inherent traits, such as site colonization and nutrient competition, plant growth promotion (PGP), antibiotic production, cell lysis effects, and induced systemic resistance (ISR), can influence the plant-environment-pathogen system to support plant health and productivity. We found that B. velezensis CBv_BE1, B. amyloliquefaciens CBa_BFL2, and CBa_RA37 derived from desert triangular spurge had promising biocontrol potential both in vitro and in planta. The former demonstrated exceptional antibacterial activity with their respective culture filtrates and ethyl acetate-based extracts, reducing pathogenic R. solanacearum growth by up to 100% at lower concentrations (0.625 mg/mL). Bacillus species have been shown to inhibit a wide range of agriculturally important pathogens (Karimi et al. 2016) and bacteria, including R. solanacearum, which causes bacterial wilt in peanuts (Karimi et al. 2016), tobacco (Tahir et al. 2017), and tomatoes (Almoneafy et al. 2014). Our findings give further support for antibiosis as a biocontrol mechanism provided by Bacilli. Chen et al. (2019) found 32 gene clusters involved in the biosynthesis of antimicrobial compounds in Bacillus velezensis LDO2 when sensing the plant pathogenic Aspergillus flavus, including 7 well-known antimicrobials such as bacilysin, surfactin, fengycin, and polyketides (bacillaene, butirosin, macrolactin).
Other processes by which BCAs respond with protection and increase plant growth include a triggered metabolic shift and reprogramming of the host transcriptome that ends in the activation of the innate immune system. Colonization of the host's interior tissues is necessary first (Bais et al. 2004; Liu et al. 2017). In this context, recent omics studies have highlighted critical features that allow endophytes to colonize and translocate in host internal tissues, such as chemotaxis, motility, plant cell-wall disintegration, and biofilm formation (Sheibani-Tezerji et al. 2015).
Our results corroborate these findings where the tested BCAs produced and released polymers degrading enzymes (data not shown) and formed biofilm under in vitro conditions. It is worth noting that biofilm plays an important role in bacterial adhesion to plant outer tissues, which initiates colonisation. Interestingly, perfect similitude was observed in the Box-PCR profiles of antibiotic resistant mutants recovered from tomato tissues grown in gnotobiotic conditions and the wild strains, irrefutably proving the endophytism of the studied BCAs. In fact, Straub et al. (2013) detected a large number of copies of genes encoding cell-wall degrading enzymes in the metagenome of rice endophytic bacteria, probably to break plant cell walls and translocate in the apoplast. Upon colonization, researchers have agreed on the involvement of a finely balanced molecular regulatory network mediated by hormone-controlled signalling pathways, for instance, the jasmonic and salicylic acids (JA, SA) regulation pathways. The cross-talk between these pathways modulate the plant endogenous endophytes population to a tolerable level. Hein et al. (2008) and Sheoran et al. (2016) suggested that Pseudomonas putida PpBP25 may partly repress and stimulate 74 and 131 genes, respectively, including defence and salicylic acid (SA) signalling components, allowing for colonisation and establishment in Arabidopsis thaliana. Thereafter, the upregulation of the transcription factor WRKY regulating the cross-talk between JA and SA pathways might have re-activated the previously repressed Jasmonate signalling pathway, leading to the restriction of supplementary endophyte entry and subsequently an induced systemic resistance to future pathogens ingress (Sheoran et al. 2016). Here, we recorded a relatively constant endophyte population density (approximately log10 CFU = 4.29/g fresh plant tissues) despite the increase in BCAs inoculum load, irrespective of the bacterial strain.
When compared to pathogen-infected and unprimed seedlings, all of the investigated bioagents substantially inhibited the bacterial wilt, resulting in an enhanced defensive response (Rs5). The most effective consortium of B. amyloliquefaciens CBa_RA37 and B. amyloliquefaciens CBa_BFL2 (CBa_BFL2 + CBa_RA37) resulted in a 10% illness incidence and 11% severity. Our data are in agreement with previous findings advocating the application of consortium-based microbial inoculants for sustainable disease control rather than individual BCA (Sarma et al. 2014; Mauricio et al. 2016; Eke et al. 2016). This technology is being touted for its reliability and improved efficacy and consistency under diverse soils and environmental conditions (Sarma et al. 2014). In this study, we further dissected the induced systemic resistance (ISR) at the subcellular level through some biochemical indicators such as the oxidative burst, and the triggered phenylpropanoid pathway. The Pearson correlation model showed positive and significant correlations between the specific activities of GPX (r = 0.85; p = 0.006), PAL (r = 0.77; p = 0.02) and SOD (r = 0.72; p = 0.05) and bacteria wilt severity, indicating an enhanced expression of these genes and their role in disease suppression. It’s indeed interesting that polyphenol production is a sounding signal in reaction to developing systemic resistance. When the pathogen's epitopes, or conserved pathogen associated molecular patterns (PAMPs) are detected by plant pattern recognition receptors (PRRs), a set of adaptive measures emerge, including enhanced antioxidant activity, host cell membrane thickening by lignification to block pathogen entry, and the synthesis of protective agents against pathogen invasion (Jetiyanon 2007; Al-askar and Rashad 2010; Abdel-Fattah et al. 2011). Although the antioxidant enzymes activities were discovered enhanced upon bacterization, with regard to unprimed and uninfected control, significantly higher activities were recorded in pathogen infected and unprimed plants compared to primmed seedlings. It is widely acknowledged that the generation of reactive oxygen species (ROS) by hosts in response to pathogen detection signals the plant's metabolic pathways to redirect their attempts towards safeguarding plant machinery and limiting damage through pathogen confinement (necrotrophic pathogen). On the contrary, accumulating evidence suggests that susceptible hosts infected with biotrophic pathogens like R. solanacearum not only produce antioxidant enzymes but also activate them more rapidly. This results in decreased levels of reactive oxygen species (ROS) within the cells and expedites the proliferation of the phytopathogen (Ramzan et al. 2021). The observed decline in the specific activities of the aforementioned enzymes following inoculation with endophytes may be attributed to the downregulation of antioxidant genes, which reestablishes oxidative stress caused by the accumulation of ROS in order to impede the spread of R. solanacearum (Narayan et al. 2017). An alternative rationale posits that endophytes have the ability to diminish reactive oxygen species (ROS) within plant cells by enhancing scavenging through non-enzymatic mechanisms, including redox glutathione and ascorbate production, phenolic and terpenoids, flavonoids, glutathione, ascorbate, tocopherol tannins, carotenoids, hydroxyanthraquinones, and carotenoids (Narayan et al. 2017).
Overall, though substantial applied and basic work are still required, the findings herein presented motivate further vulgarization of the use of microbes to enhance crop production. However, biocontrol candidates are usually poorly effective when tested in field conditions. In order to mitigate this limitation, this study investigated Bacillus species as they are thought to bear particular biocontrol traits and survivorship in harsh environments, offering an outstanding avenue for its concrete applicability. We demonstrated that, in addition to being non-toxic toward tobacco leaflets and perfectly compatible, the consortium B. amyloliquefaciens CBa_RA37 and B. amyloliquefaciens CBa_BFL2 could substantially promote tomato growth and suppression of bacterial wilt induced by R. solanacearum more than any other treatment under our experimental conditions. Ultimately, we recorded an altered oxidative burst and phenylpropanoid pathway in response to the BCAs inoculation. The findings gained provide further proof that the application of bioagents in binary form performs better than solo application in controlling plant diseases. However, detailed information of their mechanisms of action is essential since it may reveal whether the observed outputs are the result of the addition of individual performances or of microbe-microbe or microbial-plant interactions.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and can be made available when needed.
References
Abdel-Fattah G, El-Haddad E, Hafez E, Rashad Y (2011) Induction of defense responses in common bean plants by Arbuscular mycorrhizal fungi. Microbiol Res 166(4):268–281
Agarwal M, Dheeman S, Dubey R, Kumar P, Maheshwari D, Bajpai V (2017) Differential antagonistic responses of Bacillus pumilus MSUA3 against Rhizoctonia solani and Fusarium oxysporum causing fungal diseases in Fagopyrum esculentum Moench. Microbiol Res 205:40–47
Al-Askar A, Rashad Y (2010) Arbuscular mycorrhizal fungi; biocontrol agant against common bean fusarium root rot disease. Plant Pathol J 9(1):31–38
Almoneafy A, Kaleem U, Guan-Lin X (2014) Tomato plant growth promotion and antibacterial related-mechanisms of four rhyzobacterial Bacillus strains against Ralstonia solanacearum. Symbios 63:59–70
Alori E, Babalola O (2018) Microbial inoculants for improving crop quality and human health in Africa. Front Microbiol 9:2213. https://doi.org/10.3389/fmicb.2018.02213
Alori E, Fawole O (2017) Impact of chemical inputs on arbuscular mycorrhiza spores in soil: response of AM Spores to fertilizer and herbicides. Alban J Agric Sci 16:10–13
Amr F, Ahmed M, Abaraa E, Ehab F, Mohammed G, Ehab A, Saad AAE (2021) Plant growth-promoting endophytic bacteria community inhabiting the leaves of Pulicaria incisa (Lam) DC inherent to arid regions. Plant 10:76–82
Bais H, Park S, Weir T, Callaway R, Vivanco J (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9:26–32
Ballesteros GI, Acuña-Rodríguez IS, Barrera A, Gunde P, Newsham KK, Molina-Montenegro MA (2022) Seed fungal endophytes promote the establishment of invasive Poa annua in maritime Antarctica. Plant Ecol Divers 15:199–212. https://doi.org/10.1080/17550874.2022.2145579
Ballesteros GI, Newsham KK, Acuña-Rodríguez IS, Atala C, Torres-Díaz C, Molina-Montenegro MA (2024) Extreme environments as sources of fungal endophytesmitigating climate change impacts on crops inMediterranean-type ecosystems. Plants People Planet 6:148–161
Bench A, Fenner M, Edwards P (1991) Change in germinability, ABA content and ABA embryonic sensitivity in developing seeds of Sorghum bicolor (L.) Moench induced by water stress during grain filling. New Phytol 118:339–347
Chen SH, Heng J, Wang D, Bian K (2019) Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol Res 218:41–48
Eke P, Nana WL, Toghueo KRM, Tsouh FPV, Jesus M, Fekam BF (2016) Integrated Assessment of Phytostimulation and Biocontrol Potential of Endophytic Trichoderma spp Against Common Bean (Phaseolus vulgaris L.) Root. Int J Pure Appl Biosci 4:50–68
Eke P, Aundy K, Kuleshwar PS, Wakam NL, Neelam S, Mushineni A, Asharani P, Fabrice FB (2019) Corrigendum to Endophytic bacteria of desert cactus (Euphorbia trigonas Mill) confer drought tolerance and induce growth promotion in tomato (Solanum lycopersicum L.). Microbiol Res 228:126302
Figueiredo L, Marcela C, Pereira S, Sorgi C, de Bezerra S, Paulo N (2017) Successful and failed mini-inplants: microbiological evaluation and quantification of bacterial endotoxin. J Appl Oral Sc 4:151–156
Gouda S, Kerry R, Das G, Paramithiosis S, Shin H, Kumar P (2018) Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol Res 206:131–140
Granada G, Sequeira L (2011) A hypersensitive reaction induced in tobacco leaves by a compatible (Race1) isolate of Pseudomonas solanacearum. J Phytopathol 65:731–734
Grover R, Moore S (1962) Taxometric studies of fungicides against brown rot organisim sclerotinia fructicola and S. laxa. Phytopathol 52:876–880
He (1983) Characteristics of Strains of Pseudomonas solanacearum from China. Plant Dis 67:1357–1361
Hein J, Wolfe G, Blee K (2008) Comparison of rhizosphere bacterial communities in Arabidopsis thaliana mutants for systemic acquired resistance. Microb Ecol 55 (2): 333e343.
Hong C, Kwon J (2016) Biocontrol activity of Paenibacillus polymyxa AC-1 against Pseudomonas syringae and its interaction with Arabidopsis thaliana. Microbiol Res 185:13–21
Hudzicki J (2009) Kirby-Bauer disk diffusion susceptibility test protocol. American Society for Microbiology, Washington, DC, USA
Jetiyanon K (2007) Defensive-related enzyme response in plants treated with a mixture of Bacillus strains (IN937a and IN937b) against different pathogens. Biol Cont 42(2):178–185
Ji S, Sun M, Zheng X, Sun L, Chen D, Sun Q (2014) Cell-surface localization of Pellino antagonizes Toll mediated innate immune signalling by controlling MyD88 turnover in Drosophila. Nat Commun 5:3458
Jiang G, Zhong W, Jin X, Chen H, Yong Z, Xiaoman S, Macho P, Wei D, Liao B (2017) Bacterial wilt in China: history current status, and future perspectives. Front Plant Sci 8:1549
Kamkar B (2016) Sustainable development principles for agricultural activities. Adv Plant Agric Res 3:163–164
Karimi E, Naser S, Masoud S, Bagher M (2016) Bacillus amyloliquefaciens SB14 from rhizosphere alleviates rhizoctonia damping-off disease on sugar beet. Microbiol Res 192:221–230
Kim K, Lee Y, Kim C, Kang J (2009) Isolation and characterization of surfactin produced by Bacillus polyfermenticus KJS-2. Arch Pharm Res 32:711–715
Kumar S, Pathak A, Mangal M (2011) Trends of fatal poisoning in Saurashtra region of Gujarat. J Indian Academ Forens Med 33:197–199
Laurent S, Celeste Q, Marc O, Nuri C, Andrea B, Marco A, Molina M, Frederic F, Claudio R (2020) Induced systemic resistance by a plant growth-promoting rhizobacterium impacts development and feeding behaviour of aphids. Insect 11:234
Lingaiah S, Umesha S (2013) Pseudomonas fluorescens inhibits the xanthomonas oryzae pv. Oryzae: the bacterial leaf blight pathogen in rice. Can J Plant Prot 1:147–153
Liu H, Carvalhais L, Crawford M, Singh E, Dennis P, Pieterse C, Schenk P (2017) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8:2552. https://doi.org/10.3389/fmicb.2017.02552
Ma J, Gao C, Lin M, Sun Z, Zhao Y, Li X, Zhao T, Xu X, Sun W (2024) Control of fusarium head blight of wheat with Bacillus velezensis E2 and potential mechanisms of action. J Fungi 10(6):390. https://doi.org/10.3390/jof10060390
Martin B, Humbert O, Camara M, Guenzi E, Walker J, Michell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R (1992) A highly conserd repeated DNA element located in the chromosome of Streptococcus pneumonia. Nucleic Acids Res 20:3479–3483
Mauricio S, Lopez M, Humberto S, Oscar M, Erick Z (2016) Combined application of microbial consortium and humic substances to improve the growth performance of blueberry seedlings. J Soil Sc Plant Nutrit 16:4–12
Munjal V, Nadakkakath AV, Sheoran N, Kundu A, Venugopal V, Subaharan K, Rajamma S, Eapen SJ, Kumar A (2016) Genotyping and identification of broad spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium BP17. Biol Control 92:66–76
Narayan OP, Verma N, Singh AK, Oelmüller R, Kumar M, Prasad D, Kapoor R, Dua M, Kumar AJ (2017) Antioxidant enzymes in chickpea colonized by Piriformospora indica participate in defense against the pathogen Botrytis cinerea. Sci Rep 7:13553. https://doi.org/10.1038/s41598-017-12944-w
Oksana L, Andrey B, Aysylu S, Irina S, Liudmila P, Recep P (2020) Effects of endophytic Bacillus subtilis and salisilic acid on postharvest diseases (Phytophtora infestans, Fusarium oxysporum) Development in stored potato tubers. Plants 9(1):76–86
O’Toole G, Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
Poveda J, Baptista P, Sacristán S, Velasco P (2022) Beneficial effects of fungal endophytes in major agricultural crops. Front in Plant Sci 13:1061112. https://doi.org/10.3389/fpls.2022.1061112
Rabe R, Kreeb K (1979) Enzyme activities and chlorophyll and protein content in plants as indicators of air pollution. Environ Pollut 19:119–137
Ramzan M, Sana S, Javaid N, Shah AA, Ejaz S, Malik WN, Yasin NA, Alamri S, Siddiqui MH, Datta R, Fahad S, Tahir N, Mubeen S, Ahmed N, Ali MA, El Sabagh A, Danish S (2021) Mitigation of bacterial spot disease induced biotic stress in Capsicum annuum L. cultivars via antioxidant enzymes and isoforms. Sci Rep3 11(1):9445. https://doi.org/10.1038/s41598-021-88797-1
Raza W, Ling N, Yang L, Huang Q, Shen Q (2016) Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci Lett 4:40–43
Ross W, Sederoff R (1992) Phenylalanine ammonia lyase from loblolly pine purification of the enzyme and isolation of complementary DNA clones. J Plant Physiol 98:380–386
Sambrook J, Russell DW (2001) Molecular cloning: A Laboratory manual, 3rd edn. Cold spring habor Laboratory Press, New York, p 999
Sarma K, Yadav K, Sing S, Singh B (2014) Microbial consortium-mediated plant defence against phytopathogens: Readdressing for enhancing efficacy. Soil Biol Biochem 87:25–33
Sheibani-Tezerji R, Rattei T, Sessitsch A, Trognitz F, Mitter B (2015) Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 6:e0062115. https://doi.org/10.1128/mBio.00621-15
Sheoran N, Munjal V, Kumar A, Agisha V, Eapen S (2016) Pseudomonas putida BP25 alters root phenotype and triggers salicylic acid signaling as a feedback loop in regulating endophytic colonization in Arabidopsis thaliana. Physiol Mol Plant Pathol 93:99–111
Shrestha J (2016) A review on sustainable agriculture intensification in Nepal. Int J Bus Soc Sci Res 4:152–156
Silvana B, Susana G, Marria P, Maria L (2003) Behavior of antioxidant defense system in the adaptive reponse to salt stress in Helianthus annusL. cells. J Plant Growth Regul 49:81–88
Simko I, Hans P (2011) The area under the disease progress stairs: calculation, advantage and application. Phytopatol 102:381–389
Straub T, Zabel A, Gilfillan G, Feller C, Becker P (2013) Different chromatin interfaces on the Drosophila dosage compensation complex revealed by high-shear ChIPseq. Genome Res 23(3):473–485
Tahir H, Gu Q, Wu H, Niu Y, Huo R, Gao X (2017) Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci Rep 7:40481. https://doi.org/10.1038/srep40481
Tiwari P, Bae H (2022) Endophytic fungi: key insights, emerging prospects, and challenges in natural product drug discovery. Microorganisms 10(2):360. https://doi.org/10.3390/microorganisms10020360
Ubertino S, Mundler P, Tamini L (2016) The adoption of sustainable management Practices by Mexican coffee producers. Sustain Agric Res 5:1–12
Youmbi DY, Eke P, Kouokap LRK, Nya VD, Ghomsi TG, Nana WL, Fekam BF (2023) Endophytic bacteria from Euphorbia antiquorum L. protect Solanum lycopersicum L. against bacterial wilt caused by Ralstonia solanacearum. Egypt J Biol Pest Control 32:77. https://doi.org/10.1186/s41938-022-00575-x
CLSI M27-A3 (2008) Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard CLSI Wayne, PA
FAO (Food and Agricultural Organisation) (2024) Sustainable Food and Agriculture. Retrieved April 13, 2024, from https://www.fao.org/sustainability/en/
Liu X (2020) Microbial technology for the sustainable development of energy and environment. Biotechnol Rep 1865–4403. https://doi.org/10.1016/j.btre.2020.e00486
Silvana D, Maria D (2016) Wheat seeds habour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol Res 186–187
The World Counts (2024) Impact through Awareness. Retrieved April 13, 2024, from https://www.theworldcounts.com/challenges/forests-and-deserts/global-land-degradation
United Nations (2019) The 2019 Revision of world population prospects. https://population.un.org/wpp/. Accessed on 10/28/2020
Wang X, Zhao D, Shen L, Jing C, Zhang C (2018) Application and mechanisms of Bacillus subtilis in biological control of plant disease. Pest integrated management key laboratory of China Tobacco, Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, Springer Nature Singapore Pte Ltd. pp 27
Acknowledgements
This work was supported by the NAM (S&T) Research Training Fellowship for developing countries to Dr. Pierre Eke and the Seeding Labs’ Instrumental Access Grant (SL2012-2) to Prof. Fabrice Boyom. The authors are thankful to the Indian Agricultural Research Institute (IARI) through the Bacteriology Laboratory headed by Prof. Aundy Kumar for bacterial characterization. The authors are also thankfully acknowledging the assistance of Dr. Patrick Valere Tsouh Fokou for reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Hans Lambers.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eke, P., Yimta Youmbi, D., Kuleshwar, P.S. et al. Synergistic effects of Bacillus amyloliquefaciens strains CBa_BFL2 and CBa_RA37 from the desert triangular spurge on the bacteria wilt disease in tomato. Plant Soil (2024). https://doi.org/10.1007/s11104-024-06878-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11104-024-06878-2