Abstract
Background and aims
Unravelling how fundamental axes of leaf and fine-root trait variation correlate and relate to nutrient availability is crucial for understanding plant distribution across edaphic gradients. While leaf traits vary consistently along soil nutrient availability gradients, the response of fine-root traits to the same gradients has yielded inconsistent results.
Methods
We studied leaf and root trait variation among 23 co-occurring plant species along a 2-million-year soil chronosequence to assess how leaf and root traits coordinate and whether this axis of trait variation relates to soil fertility.
Results
Leaf and root trait variation was primarily structured by mycorrhizal association types. However, when considering community-level traits weighted by plant species abundance, soil nutrient availability was an important driver of trait distribution. Leaves that support rapid growth on younger more fertile soils were associated with roots of larger diameter and arbuscular mycorrhizal colonization. In contrast, leaves that favor nutrient conservation on nutrient-impoverished soil were associated with greater root-hair length and phosphorus-mobilizing root exudates proxied by leaf manganese concentration. At the species level, leaf and root trait variation patterns deviated from the community-wide results, as leaves that support either rapid growth or survival were associated with a wide range of root trait syndromes.
Conclusions
Our results highlight the challenge of generalizing a specific set of root-trait values that consistently meet the requirements of leaves supporting rapid growth or survival. Hence, the leaf economic spectrum's ability to capture variation in carbon gain may not be reflected by the root economic space.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Evaluating how fundamental axes of trait variation coordinate between leaves which impact nutrient use, and fine roots, influencing nutrient acquisition, is crucial to gain deeper insights into plant functioning (Chapin 1980; Reich 2014; Weigelt et al. 2021). The alignment of these leaf and fine-root axes of variation, potentially forming a single axis (Chapin 1980; Reich 2014; Weigelt et al. 2021), might also offer valuable insights into plant distribution along resource-availability gradients (Lambers and Poorter 1992; Reich 2014). The putative association between this shared axis of variation and resource availability is particularly important, as it may mechanistically explain whole-plant strategies, especially, those ranging from supporting rapid growth under high nutrient availability to those associated with survival in nutrient-impoverished habitats (Chapin 1980; Lambers and Poorter 1992; Reich 2014). A growing number of plant species in global trait databases (Bergmann et al. 2020; Kattge et al. 2020; Wright et al. 2004) and root trait measurements such as root exudates (Wen et al. 2022) have improved our understanding of interspecific variation in leaf and root traits across biomes. However, the general lack of environmental metadata (e.g., soil fertility) in these databases leave uncertainties about leaf and root trait covariation and their coordination with soil variables (Weigelt et al. 2021). Because soil nutrient availability might impact fine-root traits that influence nutrient acquisition differently from leaf traits that influence nutrient use (Kramer-Walter et al. 2016; Weemstra et al. 2016), more studies of root and leaf trait covariation across soil nutrient availability gradients are needed.
According to nutrient economic theory, plant fitness depends on leaf trait values that support rapid growth where nutrients are abundantly available (e.g., high specific leaf area, SLA), or slow growth and conservation of acquired nutrients such as nitrogen (N) and phosphorus (P) allowing higher long-term nutrient-use efficiency under adverse conditions (e.g., high leaf tissue density, LTD; Craine 2009; Lambers and Poorter 1992). This trade-off has been reported widely in different studies on leaf traits, across and within biomes (e.g., Reich et al. 1992; Wright et al. 2004). Leaves that promote rapid growth enhance carbon (C) assimilation by minimizing both C investment and C maintenance costs associated with extended longevity but incur greater nutrient losses during leaf senescence than conservative leaves do, owing to their high nutrient investment in short-lived leaves that exhibit less-efficient nutrient remobilization. Conversely, long-lived conservative leaves enhance nutrient-use efficiency by allocating more C relative to N and P per leaf area and mass, resulting in a well-defended leaf that reduces nutrient loss, but limits the C-assimilation rate per unit leaf mass (Wright et al. 2004). This range of strategies is termed the leaf economics spectrum (LES) or referred to, in terms of return on C investment, as the ‘fast-slow’ spectrum (Wright et al. 2004).
A spectrum similar to the LES was anticipated within the root economic space (RES) to enable rapid growth or promote nutrient conservation (Reich 2014), which was later identified as the conservation gradient (i.e. root nitrogen concentration [N]-root tissue density (RTD) axis) (Bergmann et al. 2020). Analogous to the LES, this RES axis impacts root nutrient acquisition, lifespan and nutrient loss relative to C investment (Bergmann et al. 2020; Freschet et al. 2021; Tjoelker et al. 2005; Wen et al. 2022). However, since soil nutrients are less mobile than aboveground resources (e.g., CO2), plants must forage soil nutrients (Laliberté 2017). Consequently, the RES is multidimensional, including another root trait variation axis, the collaboration gradient, describing a range of nutrient-foraging strategies (Bergmann et al. 2020), enabling plants to alleviate limitations associated with low nutrient mobility. Along this gradient (i.e. diameter-specific root length (SRL) axis), for a given root mass, fine roots may forage for nutrient by increasing their length producing a root of high SRL or rely on an increased diameter to favor arbuscular mycorrhizal colonization to forage for nutrients beyond the root-depletion zone (Bergmann et al. 2020). However, root-trait variation does not appear to be clearly connected to fast growth or survival, as they are not consistently related to variation in soil nutrient availability. This creates uncertainty about how the RES coordinates with the LES (Weemstra et al. 2016), and whether a single ‘fast-slow’ spectrum (Reich 2014) is a valid concept for describing the joint variation of root and leaf traits.
Theoretically, rapid growth should be associated with leaf and root traits maximizing C gains, suggesting a coordination among both ‘fast’ ends of the LES and conservation gradient and fertile soil. This framework generates a single ‘fast-slow’ spectrum where variation along the diameter-SRL axis should be unrelated to a nutrient availability gradient. Meanwhile, trait values along the root [N]-RTD axis should vary with this gradient, thereby forming a ‘fast’ subspace defined by diameter-, SRL- and root [N]- and ‘slow’ RES subspace defined by diameter-, SRL- and RTD- consistently favored in fertile and infertile soils, respectively. This aligns with the fact that the majority of studies investigating leaf and root trait coordination have reported a positive root [N]—leaf [N] correlation (Weigelt et al. 2021). It is also in line with the observation that ‘fast’ leaf traits are consistently associated with more fertile soils at a global scale (Ordoñez et al. 2009), while the diameter-SRL axis exhibits contrasting responses to nutrient availability (Table S1). Among 17 studies, 10 showed increasing root diameter and decreasing SRL under higher fertility and seven showed the opposite. However, ‘fast’ fine roots with high [N] have also been associated with nutrient-impoverished soils, enabling rapid nutrient acquisition to compete with microorganisms (Freschet et al. 2017; Liu et al. 2010), Conversely, long-lived ‘slow’ fine roots with high RTD and maintenance costs should provide limited benefits in the same soils, as rhizosphere nutrients would be rapidly depleted (Laliberté et al. 2015). These findings challenge the validity of a single ‘fast-slow’ spectrum, as the ‘fast’ subspace of the RES may be linked to a ‘slow’ leaf trait syndrome in nutrient-poor soils.
A deeper-rooted limitation hindering the mechanistic association of the varying allocation of C associated with fundamental axes of root-trait variation with soil fertility and either rapid growth or survival is that the RES is incomplete (Wen et al. 2022). Belowground strategies and associated considerable C allocation involved in the acquisition of nutrients from immobile organic and inorganic resources are often overlooked, resulting in a partial comprehension of their correlation with the RES dimensions (Wen et al. 2022). Among belowground strategies, root exudation involves significant C allocation and affects nutrient availability through priming effects, inorganic nutrient mining, and among root exudates, carboxylates are particularly important as they provide access to immobile nutrients such as P in nutrient-impoverished soil (Canarini et al. 2019; Wen et al. 2022). Rapid root exudation, including release of carboxylates, has, however, been suggested to be positively associated with the endpoints of both RES dimensions, known to reflect lower C investment (high SRL: Guyonnet et al. 2018; Lv et al. 2023; Meier et al. 2020; Wang et al. 2021, high root [N] or low RTD: Sell et al. 2022; Sun et al. 2021; Wen et al. 2022; Williams et al. 2022). This complicates the task of associating variation along the RES dimensions with C gain maximization and, thereby, with fast or slow growth rates.
Similarly, C allocation toward ectomycorrhizal and ericoid symbioses which may parallel investment in AM symbiosis (Smith and Read 2008), is also often overlooked despite organic nutrients – accessible through these symbioses – constituting a substantial proportion of soil nutrients along fertility gradients (Walker and Syers 1976). Furthermore, different types of mycorrhizal symbioses have distinct impacts on root-trait values and may influence root-trait distribution within the RES (Valverde-Barrantes et al. 2017). Arbuscular mycorrhizal hosts display coarser fine roots, while ectomycorrhizal (ECM) hosts exhibit more ramified ones (Comas et al. 2014; Eissenstat et al. 2015) and ericoid mycorrhizal (ERM) hosts develop finer and denser fine roots (Read et al. 2004). Finally, as growth form (i.e. herbaceous, shrub, tree) affects a plant’s capacity to compete for light across the vertical stratification of plant communities, it may lead to different belowground C allocation (Ericsson et al. 1996) and, consequently, different trait values. Overall, this introduces uncertainty regarding the primary drivers of root trait variation and if a single ‘fast-slow’ spectrum is a valid concept for predicting covariation between leaf and root traits, where nutrient availability may represent the primary source of environmental variation.
To evaluate how fine-root and leaf trait covariation might depend on soil fertility, we conducted our study along a long-term soil chronosequence in Western Australia (Turner et al. 2018). Long-term soil chronosequences are series of adjacent soils that differ in age but are formed under similar environmental conditions from the same parent material (Walker et al. 2010). Large differences in soil age give rise to a strong natural fertility gradient and minimize plant responses to other environmental factors (e.g., climate, topography, parent material) (Walker et al. 2010). Along the Warren chronosequence, we measured common leaf and root traits of 23 of the most abundant plant species including one very common species, Agonis flexuosa (Willd.), which is one of the most abundant species in all stages of the chronosequence. We also measured foliar manganese concentrations [Mn], which can be used as a proxy for rhizosphere carboxylate concentrations and P mobilization, since Mn is also mobilized by carboxylates whereas its uptake is poorly controlled (Lambers et al. 2015a). Our objectives were to 1) determine whether soil nutrient availability has a stronger influence on root trait variation than growth form or mycorrhizal symbiosis type, and 2) determine how leaf and fine-root traits covary along a strong soil fertility gradient, with a focus on whether a single ‘fast-slow’ spectrum is a valid concept to describe this covariation and hence species distribution.
Materials and methods
Study site
Sampling took place from September to December 2016 along the Warren dune chronosequence, located in d’Entrecasteaux National Park in Western Australia. The plant communities studied are located at five stages whose soils range from unstable sand, leached calcareous sand, podzol over calcareous sand and podzol on siliceous sand (Turner et al. 2018), thereby showing only small differences in soil texture. Turner et al. (2018) posit that the ages of the dunes range from the Early Pleistocene to the Holocene as dune systems on the Western Australian coastal plain are linked to sea level changes during interglacial periods throughout the Pleistocene (Kendrick et al. 1991; Playford et al. 1976). Each major dune system in the chronosequence corresponds to a stage of fertility (Turner et al. 2018). For reference, the dune systems under study correspond to dune stages 1, 2, 3, 4 and 6 in the study by Turner et al. (2018). Within each of these stages, five 20 m × 20 m plots were systematically positioned to minimize differences in topography among plots within and across stages. As shown by Turner et al. (2018), variation in soil P and N availability corresponds to the soil formation model developed by Walker and Syers (1976). Topsoil (0–20 cm) concentrations of nutrients such as total P, total N and ‘readily-available’ P where roots were sampled, however, did not show the same patterns as the first 1 m of soil. Plant nutrient uplift likely increased soil nutrient concentration in stages 2 through 4, peaking at stage 3 (Table S2) (Turner et al. 2018). There is no significant difference in temperature and precipitation across the chronosequence over the relatively short distance (~ 10 km) from the youngest to the oldest dunes (Turner et al. 2018). The climate is Mediterranean, with hot, dry summers and mild, rainy winters, with a mean annual temperature and precipitation of 15.2 °C and 1185 mm, respectively (Turner et al. 2018).
Soil chemical analyses
Four soil samples, one in each quarter of the plot, were collected from the first 20 cm of the mineral soil in each plot. Samples were then combined into a composite sample, which was sieved (< 2 mm) and air-dried before conducting soil chemical analyses at the Smithsonian Tropical Research Institute (Turner et al. 2018). Nitrogen was determined by dry combustion using a Thermo Flash 1112 analyzer (Thermo Fisher Scientific, Massachusetts, United States). Total P concentration was determined by ignition at 550˚C for 1 h followed by acid extraction with 1 M H2SO4 for 16 h. Determination of exchangeable P (resin P) was carried out using anion exchange membranes (Turner and Romero 2009). Both total P and resin P were used in the analysis, since resin P represents the readily-available mobile inorganic P and underestimates the P available to some plants. Mobile organic P, and weakly sorbed inorganic and organic P, are also available to plants, although not directly (Lambers et al. 2006).
Leaf and root sampling
Roots and leaves from multiple individuals of selected plant species were sampled within each plot to obtain a composite sample per species. Twenty-three species, often occurring in multiple plots or at several stages, were sampled, resulting in a total of 111 composite samples at the species level across all plots of the chronosequence (Table S3). Among these samples, strategies (i.e. AM, ERM, ECM, AM-ECM, NM) and growth form (i.e. herbaceous, shrub, tree) had a similar representation across the chronosequence, except for ECM, which was filtered out at two stages (Tables S4-S6). Leaves directly exposed to light, ranging from five to 15 per individual, depending on their surface area, and fine roots were sampled in the first 20 cm by tracing them back to three to five individuals of the most-abundant species. These individuals, together, were sampled to represent a cumulative relative canopy coverage – estimated by the ground projection of the canopy of individual species – as close as possible to 75%, starting with the most-abundant species (Table S3).
Leaf functional traits
Leaves were photographed in the field using a Portable Imaging and Calibration Kit (PICK, Régent Instruments Inc, Quebec, Canada), Olympus TG-4 tripod cameras (Olympus corporation, Tokyo, Japan) and subsequently analyzed using WinFolia software (Régent Instruments Inc, Quebec, Canada) to determine leaf area. Leaf thickness was also measured in the field using a digital caliper (Sona Enterprise, California, United States). Following this, leaves were dried on site and re-dried in the laboratory at 60˚C for 72 h and weighed to obtain the specific leaf area and leaf tissue density. Leaf [N] was obtained using an elemental analyzer (Vario Micro Elementar, Langenselbold, Germany). Leaf [P] and [Mn] were determined after acid digestion of 2 mg of dry leaves followed by an inductively coupled plasma mass spectrometry analysis (Perkin Elmer NexION 300x, Waltham, USA).
Root functional traits
Root functional traits were measured on fine roots that were classified using a root order-based threshold. For each species, variation in color, texture, diameter and rigidity were used to distinguish fine roots, resulting typically in the first two to three root orders being sampled. Specific root length, root diameter, root branching intensity (BrInt) and root tissue density were obtained using WinRhizo Pro software (Régent Instruments Inc, Quebec, Canada). Dry mass was obtained by drying fine roots at 60 °C for 72 h to complete the measurement of SRL and RTD. Root-hair density (RHD) was measured on 10 segments of approximately 1 mm across 10 randomly sampled first-order roots and root-hair length (RHL) was measured on one randomly sampled root hair per first-order root previously sampled for RHD using images captured with a Zeiss Axio Imager 2 microscope (software: AxioVison, Jena, Germany).
Degree of mycorrhizal colonization
Fine roots were cut into sections of approximately 1 cm length and then bleached in modified syringes (Claassen and Zasoski 1992) using 10% (v/v) KOH at 90˚C. Cleared roots were acidified with 5% (v/v) acetic acid for five minutes. Subsequently, roots were stained in Shaeffer Black ink and 5% (v/v) vinegar solution for 4 min (Vierheilig et al. 1998). Afterwards, roots were kept in a lactoglycerol solution for 48 h to remove excess staining.
To derive a quantitative estimate of AM and ERM colonization intensity, 90 segments of fine roots randomly sampled from the aforementioned 1 cm sections (i.e. approximately 900 mm), were mounted on microscope slides using glycerol. Fungal structure cover (i.e. coils, arbuscules and hyphae) was estimated based on seven classes (1: 0%; 2: < 1%; 3: < 10%; 4: < 25%; 5: < 50%; 6: > 50%; 7: > 75%) within randomly sampled segments of approximately 0.15 mm along the mounted fine-root fragments until approximately 300 mm of root fragments were analyzed. The degree of colonization was obtained by summing the product of the median value of class i by the length of the segments associated with this class standardized by the total length of the analyzed segments:
Ectomycorrhizal colonization was obtained by dividing the number of colonized root tips by the total number of root tips (20 tips on average per sample).
This quantitative estimate of mycorrhizal colonization was subsequently used to assign nutrient-acquisition strategies for all composite samples. A threshold of 10% of symbiotic root colonization was used, according to the overall apparent rate of misdiagnoses for mycorrhizal structures (Brundrett 2009), except for ECM and ERM colonization, for which we were confident of the diagnosis. Accordingly, an NM strategy was assigned to composite samples with an AM colonization less than 10%, while an AM strategy was assigned to those with an AM colonization greater than 10%.
Statistical analyses
All statistical analyses were performed in the R environment (R Core Team 2020). Principal component analysis (PCA) was performed on standardized trait values, using the rda() function from the ‘vegan’ package (Oksanen et al. 2020). The significance of the PCA axes was assessed by permuting the data and comparing the observed inertia values of the axes with those obtained by chance, while trait loading significances were assessed using the broken-stick criterion (Peres-Neto et al. 2003). A PERMANOVA analysis was conducted on standardized traits and Euclidean distance, utilizing chronosequence stage, nutrient-acquisition strategy, growth form as independent factors and their interaction to evaluate which factors explained the greatest variance within the RES and LES separately, using the adonis() function from the ‘vegan’ package (Oksanen et al. 2020). To prevent circularity, symbiotic traits were excluded from the analysis, as they were initially used to define the nutrient-acquisition strategy. To evaluate the association among standardized functional traits, Hellinger-transformed species abundance, and standardized soil fertility descriptors, a fourth-corner analysis was conducted using the fourthcorner() function from the ‘ade4’ package (Dray and Dufour 2007). Model 2 permutation was used within the fourth-corner analysis where only rows of the abundance matrix were permuted, and false discovery rate was used for multiple comparison correction. Co-inertia analysis was used to assess the degree of covariation and correlation between the LES and RES using dudi.pca() and coinertia() function from ‘ade4’ package (Dray and Dufour 2007) on unweighted and weighted traits by relative cover at the plot level (i.e. community-weighted mean (CWM); ΣPiTi). The shared inertia (RV) coefficient between two multivariate tables, derived from co-inertia analysis, is high when both structures vary along a similar axis regardless of direction, and low when they vary independently (Dray et al. 2003). To test if the observed RV coefficient significantly differed from the RV coefficient distribution under the null hypothesis, permutations using the randtest() function from the ‘ade4’ R package (Dray and Dufour 2007) were used. To assess whether community-weighted mean traits reflected trait selection, not just variation in community composition, differences in identical pairs of traits between the common species Agonis flexuosa (Willd.) and community-weighted means were assessed using linear models (lm) and generalized least squares (gls) from the ‘nlme’ package (Pinheiro et al. 2020). Generalized linear models (GLM), which provide the possibility to modify the family distribution, were not used since modeling was conducted on standardized trait values, resulting in non-positive values, thereby limiting the capacity to correct departures from normality in the residual distribution. Potential deviations from homoscedasticity were visually inspected and corrected, if necessary, by ensuring that the variance function remained constant and equal to one. Additionally, the variance function was simultaneously allowed to vary between the community-weighted mean and A. flexuosa using varIdent and form functions within the weights argument of the gls function (Zuur et al. 2009). If the residual distribution was similar between two models, selection was based on the Akaike information criterion (AIC) (Zuur et al. 2009). Post-hoc Tukey HSD tests were conducted using the ‘emmeans’ (Lenth 2020) and ‘multcomp’ (Hothorn et al. 2008) packages. Differences in strategy mean relative cover among stages were not modeled through variance analysis due to insufficient replication of strategies within specific stages. No linear analysis was performed on community-weighted mean values, since they were performed in the fourth-corner analysis. Finally, we also presented species leaf and root trait syndromes from the stages with the most contrasting nutrient availability (stage 3 and 5) to illustrate the range of strategies within community-wide trait syndromes.
Results
Root and leaf trait spectrum
A principal component analysis (PCA) performed on root traits revealed three independent axes of root trait variation. The first PCA axis, which we refer to as the root collaboration gradient, explaining 32.0% of the variance, ranged from species outsourcing nutrient acquisition through a large root diameter, high AM colonization and to a lesser extent ECM colonization to species with high SRL, BrInt, RHL and RHD values but low AM and ECM colonization intensities (Fig. 1a). The second axis of the PCA, explaining 20.4% of the variance, was more difficult to identify because root [N] was not measured and RTD contributed greatly to the first, second and third axes (Figs. 1a and S1a; Table S7). This pattern of correlation was mainly driven by ERM and ECM species, as they exhibited roots of high tissue density (Fig. S3b). Our analysis also highlighted a third significant dimension representing 15.1% of root trait variation (Figs. 1a, S1a, S2a and b). The contribution to the third dimension was also mainly driven by ECM and partly ERM colonization and their coordinating traits (Figs. 1a and S3b), since only these symbiotic traits as well as branching intensity and root tissue density were involved (Figs. 1a and S2a and b; Table S7).
Projection of species in a Mahalanobis space formed by the two first principal components. a Principal component analysis performed on root functional traits and associated PERMANOVA-derived relative sum of squares, expressed as percentage and b performed on leaf functional traits and associated PERMANOVA-derived relative sum of squares, expressed as percentage. Trait abbreviations: SLA: specific leaf area, Thick: leaf thickness, P: leaf phosphorus concentration, N: leaf nitrogen concentration, Mn: leaf manganese concentration, LTD: leaf tissue density, RTD: root tissue density, SRL: specific root length, BrInt: branching intensity, D: root diameter, AM: arbuscular mycorrhizal colonization, ECM: ectomycorrhizal colonization, ERM: ericoid colonization, RHL: root-hair length and RHD: root-hair density
The principal component analysis (PCA) conducted on leaf traits showed one major dimension, representing 42.4% of the variance, ranging from species with ‘fast’ leaves displaying high values of SLA, leaf [N] and [P] to species with ‘slow’ leaves exhibiting thick and dense leaves with high Mn concentration (Fig. 1b). Leaf tissue density contributed to an independent second dimension explaining 22.0% of the variance (Figs. 1b and S1b; Table S8). As observed in the root traits spectrum, this pattern of correlation was mainly driven by ericoid species, which had dense leaves (Figs. 1b and S3a).
PERMANOVA analysis revealed that both root and leaf trait spectra were mainly structured by nutrient-acquisition strategies (RES: relative sum of squares = 25.4%; LES: relative sum of squares = 22.4%) and chronosequence stage (RES: relative sum of squares = 9.7%; LES: relative sum of squares = 25.4%), and relatively weakly by growth forms (RES: relative sum of squares = 6.4%; LES: relative sum of squares = 7.2%) (Fig. 1a and b). In both the RES and LES, all factors were significant at P < 0.0001 (Fig. 1a and b), as were their interaction terms (not presented; P < 0.05), except for the interaction between growth form and stage within the RES (not presented; 0.05 < P < 0.1). Details on the interaction terms are not shown as they do not alter the sum of squares values but rather the associated F-statistic, and, thereby, are accounted for within the p-values.
Fourth-corner analysis, which considers species abundance, revealed significant and strong associations among root and leaf traits and soil fertility variables (Fig. 2a and b). ‘Outsourcing’ root traits such as AM symbiosis (i.e. high AM colonization within abundant species) were associated with exchangeable inorganic P (resin P) concentration (P = 0.052), soil total concentration ([P]) (P = 0.031) and total [N] (P = 0.0027). High root diameter was also associated with high soil total [N] (P = 0.0027) and to a lesser extent with high resin P (P = 0.064). Root traits associated with a ‘do-it-yourself’ strategy (i.e. high SRL, BrInt, and RHD) were weakly associated (0.05 < P < 0.1) with high soil total [P], total [N] and to resin [P]. High root-hair length was the only morphological trait displaying a significant association with high soil total [P] (P = 0.044) (Fig. 2a). The only trait that relates to the conservation gradient, the RTD, although not significantly associated with soil descriptors (P > 0.05), displayed a trend in which a high RTD was associated with higher resin [P]. Leaf traits exhibiting a ‘fast’ strategy were all associated with high soil nutrient concentrations, as SLA was positively associated with soil total [P] (P = 0.0018), total [N] (P = 0.0006) and resin [P] (P = 0.0031); leaf [P] was associated with high soil total [P] (P = 0.004), total [N] (P = 0.036), and to a lesser extent resin [P] (P = 0.061). High leaf [N] was associated with high soil total [N] (P = 0.0046) and total [P] (P = 0.0018). Finally, leaf thickness, which is representative of a ‘slow’ strategy, was associated with low soil total [P] (P = 0.0006) and total [N] (P = 0.0006) and resin [P] (P = 0.013), and high leaf [Mn] was associated with soil total [P] (P = 0.0027) and not with other soil descriptors (P > 0.05), and LTD was not significantly associated with any soil descriptors (P > 0.05) (Fig. 2b).
Fourth-corner analysis produced on the vegetation cover (Hellinger transformation), roots a and leaves b functional traits (standardized) within a cumulative relative cover of 75% and soil descriptors (normalized). The stars indicate the level of significance: (.) = P < 0.1, (*) = P < 0.05, (**) = P < 0.01 and (***) = P < 0.001. Trait abbreviations: SLA: specific leaf area, Thick: leaf thickness, P: leaf phosphorus concentration, N: leaf nitrogen concentration, Mn: leaf manganese concentration, LTD: leaf tissue density, RTD: root tissue density, SRL: specific root length, BrInt: branching intensity, D: root diameter, AM: arbuscular mycorrhizal colonization, ECM: ectomycorrhizal colonization, ERM: ericoid colonization, RHL: root-hair length and RHD: root-hair density. Soil descriptor abbreviation: resinP: exchangeable inorganic phosphorus, TotalN: total soil nitrogen concentration and TotalP: total soil phosphorus concentration
Community-weighted mean trait values and abundance of associated nutrient-acquisition strategies showed that stage-specific trait syndromes comprised multiple mycorrhizal strategies (Fig. 3a-e). However, irrespective of strategies, abundant species converged to thicker roots in soils displaying the highest nutrient concentration, with the exception of Lepidosperma gladiatum (Labill.) (Cyperaceae), a less common NM species (Fig. S4). They also converged to longer root hairs or high SRL in nutrient-poor soils, with the exception of Patersonia occidentalis (R.Br.), a less common AM species (Fig. S5) and Leucopogon obovatus subsp. revolutus (R.Br.), a less common ERM species (Fig. S5). Intraspecific variation in nutrient-acquisition strategy was substantial. Among the 15 species occurring in multiple plots, five of them exhibited different mycorrhizal status on at least two occasions (Table S3). This variation was further highlighted by the fact that each community comprised species displaying distinct root trait syndromes, as evidenced in this study by the stages with the most pronounced contrast in nutrient availability (Figs S4; S5). Likewise, Agonis flexuosa, a common species occurring across all stages, differed in its mycorrhizal nutrient-acquisition strategies across stages (i.e. NM and ECM in stage 1, AM-ECM and ECM in stage 2, AM-ECM in stages 3 and 4 and AM-ECM and ECM in stage 5) (Table S3). Soil fertility-induced intraspecific variation was further evidenced by the fact that two of the most abundant species (i.e. Agonis flexuosa and Bossiaea linophylla (R.Br.)) at the stage displaying the highest soil [N] and [P], although ECM at the youngest stage, converged to an AM-ECM strategy, as strictly ECM strategy was filtered out (Table S3; Fig. 3e). Finally, the impact of soil fertility on trait value variation was evidenced by the fact that the stage-specific leaf and root trait syndromes of common species Agonis flexuosa aligned with those of the entire plant community (Fig. 3b and d).
Community-weighted mean (CWM) stage-average trait values, Agonis flexuosa stage-average trait values and overall strategies mean relative cover (%) across chronosequence stages 1 to 5. a CWM leaf traits; b A. flexuosa leaf traits; c CWM root traits; d A. flexuosa root traits; and e overall mean relative cover of mycorrhizal types across the chronosequence. No linear analyses were performed on CWM, since these analyses were performed in the fourth-corner analysis. Stars in a to d indicate Tukey’s honest significant difference (HSD) groupings (P ≤ 0.05) for pairs of identical root and leaf traits between the CWM and A. flexuosa. Error bars in e represent standard deviation. Trait abbreviations: SLA: specific leaf area, Thick: leaf thickness, P: leaf phosphorus concentration, N: leaf nitrogen concentration, Mn: leaf manganese concentration, LTD: leaf tissue density, RTD: root tissue density, SRL: specific root length, BrInt: branching intensity, D: root diameter, AM: arbuscular mycorrhizal colonization, ECM: ectomycorrhizal colonization, ERM: ericoid colonization, RHL: root-hair length and RHD: root-hair density. Strategy abbreviations: AM: arbuscular mycorrhizal, ECM: ectomycorrhizal, AM-ECM: dual arbuscular mycorrhizal and ectomycorrhizal, ERM: ericoid mycorrhizal and NM: non-mycorrhizal
Leaf and root traits coordination
Co-inertia analysis showed that leaf and root traits were weakly coordinated for abundance-unweighted traits (RV = 0.29; P < 0.001) and strongly coordinated for abundance-weighted traits (RV = 0.96; P < 0.008) (Fig. 4a and b). Similar to results of the fourth-corner analysis, co-inertia analysis performed on abundance-weighted traits revealed that ‘fast’ species traits within the LES covaried with ‘outsourcing’ species traits within the RES. Arbuscular mycorrhizal colonization, the pair ECM colonization and BrInt, the pair ERM colonization and RTD, root diameter, SLA, leaf [P] and [N] were superimposed in leaf and root traits covariance spectrums. ‘Slow’ species traits within the LES covaried with ‘do-it-yourself’ species traits, as leaf thickness, RHD and RHL were also superimposed along the axis explaining more of the covariance (93.44%), while leaf [Mn] loaded onto both co-inertia axes (Fig. 4a and b).
Leaf economic spectrum and root economic spectrum co-inertia. RV coefficient is high when both structures vary along a similar axis, regardless of direction, and low when they vary independently. For unweighted root and leaf traits a and for leaf and root traits weighted by relative cover. b. Green is associated with foliar traits and brown with root traits. Trait abbreviations: SLA: specific leaf area, Thick: leaf thickness, P: leaf phosphorus concentration, N: leaf nitrogen concentration, Mn: leaf manganese concentration, LTD: leaf tissue density, RTD: root tissue density, SRL: specific root length, BrInt: branching intensity, D: root diameter, AM: arbuscular mycorrhizal colonization, ECM: ectomycorrhizal colonization, ERM: ericoid colonization, RHL: root-hair length and RHD: root-hair density
Discussion
Mycorrhizal strategies structured the LES and RES; nevertheless, root and leaf traits were coordinated among each other across a very strong soil P gradient once relative abundance was accounted for. Accounting for species abundance may have diluted the signal of rarer species that may acquire less abundant forms of nutrients, as observed in arctic plant communities for N (McKane et al. 2002) or tropical forests for P (Guilbeault-Mayers et al. 2020). Our results suggest that not only plant communities, but also common species whose leaves possess a ‘fast’ trait syndrome, meet their nutrient demands by coordinating with roots that exhibit a ‘collaborative’ trait syndrome in richer soils. On the other hand, in nutrient-impoverished soils, the ‘slow’ leaf trait syndrome was associated with a ‘do-it-yourself’ root trait syndrome, which was characterized by longer root hairs and high leaf [Mn], pointing toward rhizosphere P mobilization through carboxylate exudation (Lambers et al. 2015a; Lambers 2022). Together, our results indicate that root traits defining the collaboration gradient responded to soil P availability and these traits were closely correlated with the aboveground ‘fast-slow’ leaf trait spectrum, which responded in the same manner. However, the species-level signal revealed that multiple nutrient-acquisition root-trait syndromes might meet leaf demands along the LES, suggesting that a single ‘fast-slow’ spectrum might be an invalid concept to describe root and leaf trait coordination.
Variation in root and leaf traits
Our PERMANOVA analysis results indicate that mycorrhizal nutrient-acquisition strategies, when used as a grouping factor, explained a larger proportion of the variance within the RES than soil fertility did, as also found by Valverde-Barrantes et al. (2017). Mycorrhizal association types may have played a significant role in explaining the observed variance, as a third significant dimension within the RES, contrasting ECM and partly ERM colonization, was observed. The contrasting investment in mycorrhizal association types might provide plants along this axis an increasing capacity to access immobile and organic nutrients through contrasting investment in ERM and ECM symbioses (Ward et al. 2022). This suggests that the two dimensions of root-trait variation identified by Bergmann et al. (2020) adequately capture readily-available inorganic nutrient-acquisition strategies but may not encompass strategies needed to access organic nutrient forms that are not readily available to fine roots or AM fungi. Meanwhile, nutrient-acquisition strategies explained slightly less variance than soil fertility within the LES which provides evidence that leaf-trait values, irrespective of mycorrhizal association type, converged toward specific trait syndromes at all positions along the fertility gradient (Bernard-Verdier et al. 2012). We also found that growth forms explained a relatively small amount of variance within the RES and the LES, suggesting that a similar ecological trade-off structured variance, irrespective of growth form, for both LES (Wright et al. 2004) and RES (Bergmann et al. 2020). Overall, trait distribution within the LES and RES was primarily influenced by nutrient-acquisition strategies defined by mycorrhizal colonization threshold. Modification of nutrient-acquisition strategies in several common species occurring at more than one stage (Table S3) along the chronosequence and the strong association between soil descriptors, species traits and abundance, however, supports the contention that trait distribution reflected variation induced by soil fertility.
Root and leaf trait coordination along a strong nutrient-availability gradient
Multivariate associations among soil-fertility descriptors, species abundance and species traits revealed that all leaf traits, with the exception of LTD, were significantly associated with soil characteristics. Leaf traits linked with the ‘fast’ trait syndrome (i.e. high SLA, leaf [N] and [P]) were associated with more fertile soils and ‘slow’ trait values, represented by thick leaves were associated with nutrient-poor soils, as observed across and within biomes (Jager et al. 2015; Ordoñez et al. 2009). By contrast, among all root traits structuring the RES dimensions, only root traits associated with the ‘outsourcing’ trait syndrome (i.e. high AM colonization/root diameter) were significantly associated with high soil total [N], total [P] and exchangeable [P], while high values of root-hair length combined with carboxylate-releasing P-mobilizing traits, proxied by high leaf [Mn] (Lambers et al. 2015a; Lambers 2022), were associated with extremely low soil total [P]. Although our results are not consistent with the trade-off underlying the collaboration gradient (i.e. diameter-SRL axis), they support a more comprehensive trade-off between root-hair length and mycorrhizal symbioses that might effectively support rapid growth and survival at both ends of a nutrient-availability gradient. Long root hairs may enhance nutrient acquisition by expanding the root-depletion zone – including zones where neighboring plants' carboxylate exudates might increase P availability – while matching the high leaf nutrient-use efficiency that supports survival in P-impoverished soil, given their resource-efficient nature (Lynch and Ho 2005). Meanwhile, AM hyphae entail a lower carbon construction cost (Fitter 1991), a higher P-acquisition rate per unit of length compared with fine roots (Jakobsen et al. 1992) and allow nutrient acquisition beyond the root-depletion nutrient-impoverished zone. AM colonization might, therefore, be favorable to growth in less infertile soil (Treseder and Allen 2002), provided that the potential increase in photosynthesis associated with greater nutrient translocation to leaves (Eissenstat et al. 1993) offsets the higher respiration cost of colonized fine roots (Jakobsen and Rosendahl 1990). The latter is supported by our study, as more abundant species displaying large-diameter and high AM colonization also exhibited high SLA, leaf [N], and [P], suggesting high mass-based photosynthetic activities (Wright et al. 2004). Overall, our results show that a tendency of high values of SRL, branching intensity, and root-hair density – indicating an increase in nutrient foraging per unit length or volume – may be beneficial when nutrient availability is low. Nevertheless, our study suggests a coordination between the LES and the collaboration gradient to support rapid growth in the most fertile soil and survival in severely nutrient-impoverished soil.
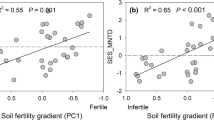
Co-inertia results confirm that the LES was strongly coordinated with the collaboration gradient – such that ‘outsourcing’ and ‘do-it-yourself’ trait syndromes were positively associated with ‘fast’ and ‘slow’ leaf trait syndromes, respectively – when species abundance was accounted for. This aligns with the single ‘fast-slow’ spectrum, as an association between a trait syndrome along the diameter-SRL axis and specific soil nutrient concentration does not invalidate that this axis might be unrelated to variation in soil nutrient availability. Instead, it suggests that any trait syndrome along the diameter-SRL axis cannot be consistently associated with a given soil nutrient availability, as proposed by Guilbeault-Mayers and Laliberté (2024). However, since the community-level RTD values show a tendency for species to be positioned within the ‘slow’ RES subspace in the most fertile soil and in the ‘fast’ subspace in severely nutrient-impoverished soil, our results are not in line with a single ‘fast-slow’ spectrum (Ordoñez et al. 2009; Reich 2014; Weigelt et al. 2021). Our results, however, align with Freschet et al.'s (2017), Liu et al.'s (2010), and Wen et al.'s (2022) proposition, which associates either rapid nutrient uptake for effective competition with microorganisms or rapid root exudation in nutrient-impoverished soil with the ‘fast’ end of the conservation gradient (i.e. low RTD and high root [N]) which is generally required for these functions. The high RTD tendency observed at the most-fertile soil (stage 3), however, was attributable to only two abundant species, Banksia grandis (Willd.) and Leucopogon obovatus subsp. revolutus. All other species within this stage, including two abundant AM-ECM species (Agonis flexuosa and Bossiaea linophylla), exhibited lower RTD values than the chronosequence mean (Fig. S4), thereby supporting a single ‘fast-slow’ spectrum. Likewise, our community-level results support that the ‘fast’ end of the conservation gradient was associated with severely P-impoverished soils (stage 5). Overall, species exhibited lower RTD values at stage 5, where soils are severely P-impoverished. However, a significant variation in RTD was observed at the species level. Notably, the common species A. flexuosa exhibited high RTD values in severely P-impoverished soils (Fig. S5). These latter two observations again upheld the validity of both the single ‘fast-slow’ spectrum and a decoupling between ‘fast’ end of the LES, the ‘fast’ RES subspace and nutrient availability, depending on the scale of where the observations were made. Overall, this high local variation challenges the validity of a single ‘fast-slow’ spectrum at the whole plant scale.
Challenging a single ‘fast-slow’ spectrum at the whole plant scale
The lack of convergence toward ‘fast’ or ‘slow’ root trait values among species calls into question the validity of a single ‘fast-slow’ spectrum (Reich 2014). Stated differently, questioning its validity does not imply that a ‘fast-slow’ spectrum is not shared between leaf and root functioning, but the ‘fast’ or ‘slow’ ends of these spectra do not consistently align. That is, permanently categorizing fine roots as ‘slow’ or ‘fast’ solely based on individual trait values (Dallstream et al. 2022), or even trait syndromes may not be valid, contrasting with leaf functioning, for which such categorization is widely supported (Ordoñez et al. 2009; Reich et al. 1997; Wright et al. 2004). However, a ‘fast’ leaf in terms of nutrient use requires effective acquisition and translocation of belowground resources (Reich 2014) and this could be met using a diversity of root strategies and at different costs (Raven et al. 2018). That is, a ‘slower’ root strategy in a nutrient-richer soil, entailing, for example, faster metabolism and higher maintenance respiration costs, could be labeled as ‘fast’. This would hold true if that root strategy enabled the acquisition and translocation of adequate nutrients to meet the demands of a ‘fast’ leaf, and if the net C cost associated with this strategy allowed for sufficient C allocation to growth. For instance, within the stage where soil [N] and [P] were higher (stage 3), we observed both at the species level (i.e. five out of seven species) and at the community level ‘fast’ leaf trait syndromes. Nutrient acquisition to meet the demand of this leaf strategy, however, was achieved via high ERM colonization in L. revolutus, high AM colonization in Hibbertia cuneiformis (Labill.) and Hibbertia grossulariifolia (Salisb.), high AM-ECM colonization in A. flexuosa and long root hairs with low mycorrhizal colonization intensity in B. linophylla (Fig. S4). Furthermore, the latter two species have been recognized as carboxylate-dependent, as their nutrient uptake is partially reliant on P mobilization through carboxylate exudation by neighboring plants (Abrahão et al. 2018; Huang et al. 2017). Conversely, within the same stage, ‘slow’ leaf traits were displayed by B. grandis and Lepidosperma gladiatum, a Proteaceae and Cyperaceae, respectively. This might be accounted for by the higher overall cost associated with their carboxylate-releasing P-acquisition traits which involves rapid root turnover (Lambers et al. 2015b) and rapid respiratory metabolism (Funayama-Noguchi et al. 2021; Shane et al. 2004).
Overall, our results indicate that nutrient-acquisition strategies beyond the ‘fast’ RES subspace and even extending outside the RES plane as defined by Bergmann et al. (2020) into an axis opposing ERM and ECM colonization, as observed in the present study, might be labeled as ‘fast’. From this perspective, the resulting community’s abundance profile, which represents a species’ relative abundance, may arise from the selection of the most-effective strategies during plant community assembly. This selection process, as suggested by Raven et al. (2018), might be influenced by competition for the same form of nutrient, resource partitioning (Turner 2008), and facilitation of P acquisition, where one strategy may involve the mobilization of a specific form of nutrient that becomes available to plants that exhibit another strategy (Lambers et al. 2018, 2022). This suggests that leaf and root traits may exhibit local coordination within the single ‘fast-slow’ spectrum framework. Considering that the ‘fast’ or ‘slow’ ends of both the leaf and root ‘fast-slow’ spectrum do not consistently align, however, it is unlikely that a specific coordination pattern among leaf and root trait values will be observed consistently. This highlights the potentially limited capacity of the single ‘fast-slow’ spectrum to describe and predict species distribution both at local scales and across biomes.
Conclusions
Our study provides new insights into factors explaining variance within the RES. Nutrient-acquisition strategies, defined by mycorrhizal colonization threshold, explained more variance within the RES than soil fertility. This was most likely, in part, due to a third dimension observed within the RES defined by ERM and ECM colonization, reflecting P mining from sources that are not readily accessible. When species abundance was taken into account, however, we found a strong association among root trait values and soil fertility. In P-depleted and less-infertile soils, beneficial root strategies depend, respectively, on root hairs that extend the root’s influence zone or AM symbiosis that allows foraging beyond it, and an axis formed by these opposing strategies aligned with the LES. The outsourcing trait syndrome was coordinated with the ‘fast’ leaf syndrome in less infertile soils, and root hair length was coordinated with the ‘slow’ leaf trait syndrome in P-depleted soils, respectively. We also found that the community-weighted mean RTD values showed a trend to be positioned within the ‘slow’ RES subspace in less-infertile soils and the ‘fast’ subspace in nutrient-depleted soil. Root traits commonly categorized as ‘fast’ or ‘slow’, however, coexisted at local sites among species irrespective of soil nutrient concentration. Therefore, our results challenge the validity of a single ‘fast-slow’ spectrum at the whole-plant scale as a framework to comprehend and predict species distribution along nutrient-availability gradients.
Given that root [N] was not measured, more studies including root exudation, leaf and root trait measurements along strong soil nutrient-availability gradients are needed to test if there is a single ‘fast-slow’ plant economic spectrum in which high root [N] might be consistently labeled as a ‘fast’ trait. Despite this, overall, the present study suggests that an axis of fundamental trait variation is shared and explains leaf and root functioning (Chapin 1980; Reich 2014). However, our results did not substantiate a single ‘fast-slow’ spectrum, in which leaf and root trait values labeled as ‘fast’ or ‘slow’ are consistently associated (Reich 2014). A single ‘fast-slow’ spectrum is, therefore, likely an invalid concept for understanding differences in relative growth rates across species occurring at both ends of a nutrient-availability gradient, and hence, species distribution.
Data availability
The data that support the findings of this study are openly available in ‘figshare’ at https://doi.org/10.6084/m9.figshare.6025748, reference no. https://doi.org/10.6084/m9.figshare.25838536.
Abbreviations
- AM:
-
Arbuscular mycorrhizal
- BrInt:
-
Branching intensity
- C:
-
Carbon
- D:
-
Diameter
- ECM:
-
Ectomycorrhizal
- ERM:
-
Ericoid mycorrhizal
- LTD:
-
Leaf tissue density
- Mn:
-
Manganese
- N:
-
Nitrogen
- NM:
-
Non-mycorrhizal
- P:
-
Phosphorus
- RHD:
-
Root hair density
- RHL:
-
Root hair length
- RTD:
-
Root tissue density
- SLA:
-
Specific leaf area
- SRL:
-
Specific root length
- Thick:
-
Thickness
References
Abrahão A, Ryan MH, Laliberté E, Oliveira RS, Lambers H (2018) Phosphorus- and nitrogen-acquisition strategies in two Bossiaea species (Fabaceae) along retrogressive soil chronosequences in south-western Australia. Physiol Plant 163:323–343. https://doi.org/10.1111/ppl.12704
Bergmann J, Weigelt A, van der Plas F, Laughlin DC, Kuyper TW, Guerrero-Ramirez N, Valverde-Barrantes OJ, Bruelheide H, Freschet GT, Iversen CM et al (2020) The fungal collaboration gradient dominates the root economics space in plants. Sci Adv 6(27):eaba3756. https://doi.org/10.1126/sciadv.aba3756
Bernard-Verdier M, Navas M-L, Vellend M, Violle C, Fayolle A, Garnier E (2012) Community assembly along a soil depth gradient: Contrasting patterns of plant trait convergence and divergence in a Mediterranean rangeland. J Ecol 100:1422–1433. https://doi.org/10.1111/1365-2745.12003
Brundrett MC (2009) Mycorrhizal associations and other means of nutrition of vascular plants: Understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 320:37–77. https://doi.org/10.1007/s11104-008-9877-9
Canarini A, Kaiser C, Merchant A, Richter A, Wanek W (2019) Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front Plant Sci 10:422679. https://doi.org/10.3389/fpls.2019.00157
Chapin FS (1980) The mineral nutrition of wild plants. Annu Rev Ecol Syst 11:233–260. https://doi.org/10.1146/annurev.es.11.110180.001313
Claassen VP, Zasoski RJ (1992) A containerized staining system for mycorrhizal roots. New Phytol 121:49–51. https://doi.org/10.1111/j.1469-8137.1992.tb01091.x
Comas LH, Callahan HS, Midford PE (2014) Patterns in root traits of woody species hosting arbuscular and ectomycorrhizas: Implications for the evolution of belowground strategies. Ecol Evol 4:2979–2990. https://doi.org/10.1002/ece3.1147
Craine JM (2009) Resource strategies of wild plants. Princeton University Press, Princeton
Dallstream C, Weemstra M, Soper FM (2022) A framework for fine-root trait syndromes: Syndrome coexistence may support phosphorus partitioning in tropical forests. Oikos 1:e08908. https://doi.org/10.1111/oik.08908
Dray S, Dufour A-B (2007) The ade4 package: Implementing the duality diagram for ecologists. J Stat Softw 22:1–20. https://doi.org/10.18637/jss.v022.i04
Dray S, Chessel D, Thioulouse J (2003) Co-inertia analysis and the linking of ecological data tables. Ecology 84:3078–3089. https://doi.org/10.1890/03-0178
Eissenstat DM, Graham JH, Syvertsen JP, Drouillard DL (1993) Carbon economy of sour orange in relation to mycorrhizal colonization and phosphorus status. Ann Bot 71:1–10. https://doi.org/10.1006/anbo.1993.1001
Eissenstat DM, Kucharski JM, Zadworny M, Adams TS, Koide RT (2015) Linking root traits to nutrient foraging in arbuscular mycorrhizal trees in a temperate forest. New Phytol 208:114–124. https://doi.org/10.1111/nph.13451
Ericsson T, Rytter L, Vapaavuori E (1996) Physiology of carbon allocation in trees. Biomass Bioenergy 11:115–127. https://doi.org/10.1016/0961-9534(96)00032-3
Fitter AH (1991) Costs and benefits of mycorrhizas: implications for functioning under natural conditions. Experientia 47:350–355. https://doi.org/10.1007/BF01972076
Freschet GT, Valverde-Barrantes OJ, Tucker CM, Craine JM, McCormack ML, Violle C, Fort F, Blackwood CB, Urban-Mead KR, Iversen CM et al (2017) Climate, soil and plant functional types as drivers of global fine-root trait variation. J Ecol 105:1182–1196. https://doi.org/10.1111/1365-2745.12769
Freschet GT, Roumet C, Comas LH, Weemstra M, Bengough AG, Rewald B, Bardgett RD, De Deyn GB, Johnson D, Klimešová J et al (2021) Root traits as drivers of plant and ecosystem functioning: Current understanding, pitfalls and future research needs. New Phytol 232:1123–1158. https://doi.org/10.1111/nph.17072
Funayama-Noguchi S, Shibata M, Noguchi K, Terashima I (2021) Effects of root morphology, respiration and carboxylate exudation on carbon economy in two non-mycorrhizal lupines under phosphorus deficiency. Plant Cell Environ 44:598–612. https://doi.org/10.1111/pce.13925
Guilbeault-Mayers X, Turner BL, Laliberté E (2020) Greater root phosphatase activity of tropical trees at low phosphorus despite strong variation among species. Ecology 101:e03090. https://doi.org/10.1002/ecy.3090
Guilbeault-Mayers X, Laliberté E (2024) Root phosphatase activity is coordinated with the root conservation gradient across a phosphorus gradient in a lowland tropical forest. New Phytol NA:NA-NA. https://doi.org/10.1111/nph.19567
Guyonnet JP, Cantarel AAM, Simon L, Haichar FEZ (2018) Root exudation rate as functional trait involved in plant nutrient-use strategy classification. Ecol Evol 8:8573–8581. https://doi.org/10.1002/ece3.4383
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50:346–363. https://doi.org/10.1002/bimj.200810425
Huang G, Hayes PE, Ryan MH, Pang J, Lambers H (2017) Peppermint trees shift their phosphorus-acquisition strategy along a strong gradient of plant-available phosphorus by increasing their transpiration at very low phosphorus availability. Oecologia 185:387–400. https://doi.org/10.1007/s00442-017-3961-x
Jager MM, Richardson SJ, Bellingham PJ, Clearwater MJ, Laughlin DC (2015) Soil fertility induces coordinated responses of multiple independent functional traits. J Ecol 103:374–385. https://doi.org/10.1111/1365-2745.12366
Jakobsen I, Rosendahl L (1990) Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plants. New Phytol 115:77–83. https://doi.org/10.1111/j.1469-8137.1990.tb00924.x
Jakobsen I, Abbott LK, Robson AD (1992) External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. New Phytol 120:371–380. https://doi.org/10.1111/j.1469-8137.1992.tb01077.x
Kattge J, Bönisch G, Diaz S, Lavorel S, Prentice IC, Leadley P, Tautenhahn S, Werner G, Günther A (2020) TRY plant trait database-enhanced coverage and open access. Glob Chang Biol 26:119–188. https://doi.org/10.1111/gcb.14904
Kendrick GW, Wyrwoll KH, Szabo BJ (1991) Pliocene-Pleistocene coastal events and history along the western margin of Australia. Quat Sci Rev 10:419–439. https://doi.org/10.1016/0277-3791(91)90005-F
Kramer-Walter KR, Bellingham PJ, Millar TR, Smissen RD, Richardson SJ, Laughlin DC (2016) Root traits are multidimensional: specific root length is independent from root tissue density and the plant economic spectrum. J Ecol 104:1299–1310. https://doi.org/10.1111/1365-2745.12562
Laliberté E (2017) Below-ground frontiers in trait-based plant ecology. New Phytol 213:1597–1603. https://doi.org/10.1111/nph.14247
Laliberté E, Lambers H, Burgess TI, Wright SJ (2015) Phosphorus limitation, soil-borne pathogens and the coexistence of plant species in hyperdiverse forests and shrublands. New Phytol 206:507–521. https://doi.org/10.1111/nph.13203
Lambers H (2022) Phosphorus acquisition and utilization in plants. Annu Rev Plant Biol 73:17–42. https://doi.org/10.1146/annurev-arplant-102720-125738
Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ (2006) Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Ann Bot 98:693–713. https://doi.org/10.1093/aob/mcl114
Lambers H, Hayes PE, Laliberté E, Oliveira RS, Turner BL (2015a) Leaf manganese accumulation and phosphorus-acquisition efficiency. Trends Plant Sci 20:83–90. https://doi.org/10.1016/j.tplants.2014.10.007
Lambers H, Martinoia E, Renton M (2015b) Plant adaptations to severely phosphorus-impoverished soils. Curr Opin Plant Biol 25:23–31. https://doi.org/10.1016/j.pbi.2015.04.002
Lambers H, Albornoz F, Kotula L, Laliberté E, Ranathunge K, Teste FP, Zemunik G (2018) How belowground interactions contribute to the coexistence of mycorrhizal and non-mycorrhizal species in severely phosphorus-impoverished hyperdiverse ecosystems. Plant Soil 424:11–33. https://doi.org/10.1007/s11104-017-3427-2
Lambers H, de Britto CP, Cawthray GR, Denton MD, Finnegan PM, Hayes PE, Oliveira RS, Power SC, Ranathunge K, Shen Q, Wang X, Zhong H (2022) Strategies to acquire and use phosphorus in phosphorus-impoverished and fire-prone environments. Plant Soil 476:133–161. https://doi.org/10.1007/s11104-022-05464-8
Lambers H, Poorter H (1992) Inherent variation in growth rate between higher plants: A search for physiological causes and ecological consequences. In Begon M, Fitter AH (Eds.) Advances in Ecological Research. Elsevier Academic Press, Amsterdam, 23:187–261. https://doi.org/10.1016/S0065-2504(08)60148-8
Lenth R (2020) Emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.5.1. https://CRAN.R-project.org/package=emmeans
Liu G, Freschet GT, Pan X, Cornelissen JHC, Li Y, Dong M (2010) Coordinated variation in leaf and root traits across multiple spatial scales in Chinese semi-arid and arid ecosystems. New Phytol 188:543–553. https://doi.org/10.1111/j.1469-8137.2010.03388.x
Lv C, Wang C, Li Y, Zhou Z (2023) Coordination among root exudation C, mycorrhizal colonization, and functional traits and their responses to drought in five temperate tree species. For Ecol Manag 546:121316. https://doi.org/10.1016/j.foreco.2023.121316
Lynch JP, Ho MD (2005) Rhizoeconomics: Carbon costs of phosphorus acquisition. Plant Soil 269:45–56. https://doi.org/10.1007/s11104-004-1096-4
McKane RB, Johnson LC, Shaver GR, Nadelhoffer KJ, Rastetter EB, Fry B, Giblin AE, Kielland K, Kwiatkowski BL, Laundre JA et al (2002) Resource-based niches provide a basis for plant species diversity and dominance in arctic tundra. Nature 415:68–71. https://doi.org/10.1038/415068a
Meier IC, Tuckmantel T, Heitkotter J, Muller K, Preusser S, Wrobel TJ, Kandeler E, Marschner B, Leuschner C (2020) Root exudation of mature beech forests across a nutrient availability gradient: the role of root morphology and fungal activity. New Phytol 226:583–594. https://doi.org/10.1111/nph.16389
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P et al (2020) vegan: Community Ecology Package (Version 2.5–7). https://CRAN.R-project.org/package=vegan
Ordoñez JC, Van Bodegom PM, Witte J-PM, Wright IJ, Reich PB, Aerts R (2009) A global study of relationships between leaf traits, climate and soil measures of nutrient fertility. Glob Ecol Biogeogr 18:137–149. https://doi.org/10.1111/j.1466-8238.2008.00441.x
Peres-Neto PR, Jackson DA, Somers KM (2003) Giving meaningful interpretation to ordination axes: Assessing loading significance in principal component analysis. Ecology 84:2347–2363. https://doi.org/10.1890/00-0634
Pinheiro J, Bates D, Sarkar D, R Core Team (2020) Nlme: Linear and nonlinear mixed effects models. R package version 3.1–149. https://CRAN.R-project.org/package=nlme
Playford PE, Cockbain AE, Lowe GH (1976) Geology of the Perth Basin, Western Australia; Bulletin 124 of the Geological Survey of Western Australia. Geological Survey of Western Australia, Perth, Australia
R Core Team (2020) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Raven JA, Lambers H, Smith SE, Westoby M (2018) Costs of acquiring phosphorus by vascular land plants: Patterns and implications for plant coexistence. New Phytol 217(4):1420–1427. https://doi.org/10.1111/nph.14967
Read DJ, Leake JR, Perez-Moreno J (2004) Mycorrhizal fungi as drivers of ecosystem processes in heathland and boreal forest biomes. Canad J Bot 82:1243–1263. https://doi.org/10.1139/b04-123
Reich PB (2014) The world-wide ‘fast–slow’ plant economics spectrum: A traits manifesto. J Ecol 102:275–301. https://doi.org/10.1111/1365-2745.12211
Reich PB, Walters MB, Ellsworth DS (1992) Leaf life-span in relation to leaf, plant, and stand characteristics among diverse ecosystems. Ecol Monogr 62:365–392. https://doi.org/10.2307/2937116
Reich PB, Walters MB, Ellsworth DS (1997) From tropics to tundra: Global convergence in plant functioning. Proc Natl Acad Sci 94:13730–13734. https://doi.org/10.1073/pnas.94.25.13730
Sell M, Ostonen I, Rohula-Okunev G, Rusalepp L, Rezapour A, Kupper P (2022) Responses of fine root exudation, respiration and morphology in three early successional tree species to increased air humidity and different soil nitrogen sources. Tree Physiol 42:557–569. https://doi.org/10.1093/treephys/tpab118
Shane MW, Cramer MD, Funayama-Noguchi S, Cawthray GR, Millar AH, Day DA, Lambers H (2004) Developmental physiology of cluster-root carboxylate synthesis and exudation in Harsh Hakea. Expression of phosphoenolpyruvate carboxylase and the alternative oxidase. Plant Physiol 135:549–560. https://doi.org/10.1104/pp.103.035659
Smith JE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Elsevier Academic Press, Amsterdam
Sun L, Ataka M, Han M, Han Y, Gan D, Xu T, Guo Y, Zhu B (2021) Root exudation as a major competitive fine-root functional trait of 18 coexisting species in a subtropical forest. New Phytol 229:259–271. https://doi.org/10.1111/nph.16865
Tjoelker MG, Craine JM, Wedin D, Reich PB, Tilman D (2005) Linking leaf and root trait syndromes among 39 grassland and savannah species. New Phytol 167:493–508. https://doi.org/10.1111/j.1469-8137.2005.01428.x
Treseder KK, Allen MF (2002) Direct nitrogen and phosphorus limitation of arbuscular mycorrhizal fungi: a model and field test. New Phytol 155:507–515. https://doi.org/10.1046/j.1469-8137.2002.00470.x
Turner BL (2008) Resource partitioning for soil phosphorus: A hypothesis. J Ecol 96:698–702. https://doi.org/10.1111/j.1365-2745.2008.01384.x
Turner BL, Romero TE (2009) Short-term changes in extractable inorganic nutrients during storage of tropical rain forest soils. Soil Sci Soc Am J 73:1972–1979. https://doi.org/10.2136/sssaj2008.0407
Turner BL, Hayes PE, Laliberté E (2018) A climosequence of chronosequences in southwestern Australia. Eur J Soil Sci 69:69–85. https://doi.org/10.1101/113308
Valverde-Barrantes OJ, Freschet GT, Roumet C, Blackwood CB (2017) A worldview of root traits: The influence of ancestry, growth form, climate and mycorrhizal association on the functional trait variation of fine-root tissues in seed plants. New Phytol 215:1562–1573. https://doi.org/10.1111/nph.14571
Vierheilig H, Coughlan AP, Wyss U, Piché Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64:5004–5007. https://doi.org/10.1128/AEM.64.12.5004-5007.1998
Walker TW, Syers JK (1976) The fate of phosphorus during pedogenesis. Geoderma 15:1–19. https://doi.org/10.1016/0016-7061(76)90066-5
Walker LR, Wardle DA, Bardgett RD, Clarkson BD (2010) The use of chronosequences in studies of ecological succession and soil development. J Ecol 98:725–736. https://doi.org/10.1111/j.1365-2745.2010.01664.x
Wang Q, Xiao J, Ding J, Zou T, Zhang Z, Liu Q, Yin H (2021) Differences in root exudate inputs and rhizosphere effects on soil N transformation between deciduous and evergreen trees. Plant Soil 458:277–289. https://doi.org/10.1007/s11104-019-04156-0
Ward EB, Duguid MC, Kuebbing SE, Lendemer JC, Bradford MA (2022) The functional role of ericoid mycorrhizal plants and fungi on carbon and nitrogen dynamics in forests. New Phytol 235:1701–1718. https://doi.org/10.1111/nph.18307
Weemstra M, Mommer L, Visser EJW, van Ruijven J, Kuyper TW, Mohren GMJ, Sterck FJ (2016) Towards a multidimensional root trait framework: A tree root review. New Phytol 211:1159–1169. https://doi.org/10.1111/nph.14003
Weigelt A, Mommer L, Andraczek K, Iversen CM, Bergmann J, Bruelheide H, Fan Y, Freschet GT, Guerrero-Ramírez NR, Kattge J et al (2021) An integrated framework of plant form and function: The belowground perspective. New Phytol 232:42–59. https://doi.org/10.1111/nph.17590
Wen Z, White PJ, Shen J, Lambers H (2022) Linking root exudation to belowground economic traits for resource acquisition. New Phytol 233:1620–1635. https://doi.org/10.1111/nph.17854
Williams A, Langridge H, Straathof AL, Muhamadali H, Hollywood KA, Goodacre R, de Vries FT (2022) Root functional traits explain root exudation rate and composition across a range of grassland species. J Ecol 110:21–33. https://doi.org/10.1111/1365-2745.13630
Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, Cavender-Bares J, Chapin T, Cornelissen JH, Diemer M et al (2004) The worldwide leaf economics spectrum. Nature 428:821–827. https://doi.org/10.1038/nature02403
Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM (2009) Mixed effects models and extensions in ecology with R. Springer, New York
Acknowledgements
Special thanks to Sharyn and Shaun Cody; without their kindness and advice, the project would not have been successful. We extend our sincere appreciation to David Poissant and Caroline Fink-Mercier for their invaluable assistance during fieldwork. We also express our profound gratitude for the invaluable comments and suggestions provided by two anonymous reviewers and Editor Dr. M. Luke McCormack.
Funding
Funding for this research was provided by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC; grant RGPIN-2014–06106 and RGPIN-2019–04537). XGM received additional support from the Fonds de recherche du Québec-Nature et technologies.
Author information
Authors and Affiliations
Contributions
XGM, EL and HL conceived the study; XGM and EL conceived the methodology; XGM conducted the data collection; XGM analyzed the data; XGM, HL and EL interpreted the results; XGM led the writing of the manuscript. All authors contributed to the draft versions.
Corresponding author
Ethics declarations
Competing interests
None that need to be declared.
Additional information
Responsible Editor: Michael Luke McCormack.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guilbeault-Mayers, X., Lambers, H. & Laliberté, E. Coordination among leaf and fine-root traits along a strong natural soil fertility gradient. Plant Soil (2024). https://doi.org/10.1007/s11104-024-06740-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11104-024-06740-5