Abstract
Purpose
The abundance of nitrogen (N)-cycling genes is frequently used to indicate N cycling and predict N2O emissions. However, it remains difficult to clearly define how soil N-cycling genes in different ecosystems respond to anthropogenic N additions.
Methods
We applied a meta-analysis approach to examine data about N-cycling genes (nifH, ammonia-oxidizing archaea (AOA), ammonia-oxidizing bacteria (AOB), nirK, nirS, and nosZ) in different ecosystems from 119 peer-reviewed articles.
Results
In the ecosystems examined, the patterns of change in the abundances of the target genes, apart from AOA, varied considerably. This variation reflects the distinctive soil characteristics of ecosystems that develop when different forms of N are applied at different rates and over different durations. The nifH abundance decreased significantly, by 32.79%, in forests but did not change in grasslands and croplands. The AOB abundance increased in all three ecosystems, by 193.06% in grasslands, 73.26% in forests, and 151.86% in croplands, respectively. The denitrification gene abundances, namely the nirK, nirS, and nosZ, in croplands also increased significantly, by 60.74%, 47.42%, and 69.54%, respectively.
Conclusion
In general, climate factors and long-term applications of organic N at high rates had significant effects on the gene abundances in different ecosystems, through their influence on soil properties. An enhanced understanding of how N additions influence the abundance of other N-cycling functional genes can help us improve our ability to model the populations and activities of microbial functional communities and predict N fluxes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrous oxide (N2O) is a by-product of the complex microbial nitrogen (N) cycle in the environment. The element N undergoes the processes of N-fixation, nitrification, and denitrification, thus producing N2O (Canfield et al. 2010; Kuypers et al. 2018). These reactions are mainly driven by the physiological metabolic activities of microorganisms in the soil. Soil microorganisms are susceptible to external interference, especially applications of chemical and organic N fertilizers, which change the N availability in soil and substantially influence the biochemical processes of the N cycle (Makowski 2019). Therefore, the responses of soil microorganisms to external N additions can strongly influence N-cycling processes (Ouyang et al. 2017).
During N-cycling processes, microorganisms synthesize corresponding enzymes through the expression of specific genes. The nitrogen fixation gene (nifH) encodes nitrogenase reductase and reduces nitrogen gas (N2) to ammonium (NH4+). The nitrification genes (AOA and AOB) synthesize ammonium monooxygenase and promote the transformation of ammonia (NH3) or NH4+ to hydroxylamine, which then undergoes further cycling. Denitrification involves the reduction of nitrate (NO3−) to nitrite (NO2−). After NO3− is reduced to NO2−, it may be partitioned along two pathways, either to generate NH4+ by dissimilatory reduction, or to generate N2O (Canfield et al. 2010). Thus, various denitrification genes have important roles in regulating N2O emissions and, depending on the N management approach, the resulting N2O emissions will vary. NirS and nirK indicate denitrifiers that convert NO2− to nitric oxide (NO), while nosZ encodes nitrous oxide reductase and then the enzyme catalyzes the transformation of N2O to N2, the final step of denitrification (Canfield et al. 2010). These functional genes are frequently used to describe the richness and diversity of microbial communities in specific N transformation processes (Gaby and Buckley 2012; Henry et al. 2006; Morales et al. 2010; Ouyang et al. 2018).
Many scholars have studied how and why the abundances of N-cycling genes change under N additions. Different ecosystems have unique soil characteristics, which impact the gene abundance. Carey et al. (2016) determined that, in different ecosystems (croplands, pastures, and unmanaged wildlands), nitrification genes, namely ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB), all responded differently to N fertilizer, and that N applications had most effect on AOA in croplands and on AOB in wildlands. Also, different N forms have different effects. When organic N is applied, carbon (C) inputs are higher than under inorganic N applications, which then increases the metabolic activity of microorganisms and the gene abundance (Schmidt et al. 2011; Sulman et al. 2014). Sulman et al. (2014) found that microorganisms had differing abilities to obtain C and that the gene abundances differed among ecosystems. Carey et al. (2016) reported that the N addition rates and the duration of the N additions (the period from the start of the N application to the end) could affect ammonia oxidizers in different ecosystems. Furthermore, environmental factors may have a strong influence on N additions. For example, under high rates of atmospheric N deposition, the available N in soil may increase. Under the increased levels of available N, the soil microbial biomass and respiration may be suppressed (Phillips and Fahey 2007; Yang et al. 2021), which will in turn affect the microbial composition and the metabolic potential (Hallin et al. 2009; Yang et al. 2021; Zhao et al. 2020). High N inputs can cause increases in the soil exchangeable aluminium (Al3+) content and decreases in the soil pH (Bowman et al. 2008; Van Breemen et al. 1983). The aluminium will have toxic effects on microorganisms (Kuperman and Edwards 1997) and may also affect the expression of functional genes (Sun et al. 2021). Zhang et al. (2021) also reported that the abiotic factors played key roles in regulating the ammonia oxidizer and denitrifier abundances under N loading.
The findings discussed so far are from field experiments or indoor cultivation experiments that were performed in small areas for specific research purposes. The results of these studies are inconsistent, so researchers have carried out meta-analyses of the data from the individual studies to identify overall patterns from the different studies. In their meta-analysis, Carey et al. (2016) examined 98 sets of measurement data from 33 articles to gain insights into how N additions affected the abundances of AOA and AOB during nitrification. While they found that AOB responded more positively than AOA, they did not explore other functional genes or environmental factors that could have affected the gene abundances in their analysis. Ouyang et al. (2018) explored how the abundances of N cycling genes changed in agricultural ecosystems treated with N fertilizers. They found that, apart from nifH, the functional gene abundances increased during nitrification (amoA) and denitrification (nirK, nirS, and nosZ) when fertilized with N. Ouyang et al. (2018) concluded that the N form and the soil pH were the main drivers behind the responses of functional genes to N additions. Data from numerous studies and the World Bank, (https://data.worldbank.org.cn/indicator), show that grassland, forest, and cropland, which account for a large proportion of terrestrial ecosystems worldwide, suffer from over-applications of N (Bonan 2008; Hovenden et al. 2019; IPCC, 2013). An analysis that compares N-cycling genes among different N-treated ecosystems, and highlights how they are related to soil properties, will provide support for evaluations of gene-based models of N cycling in soil.
In this study, we carried out a meta-analysis based on 397 field observations from 119 articles related to N-cycling genes in grassland, forest, and cropland ecosystems. The aim of the analysis was to (1) compare the responses of functional genes in different ecosystems to N additions, and (2) identify the key drivers of change in the gene abundances and how they affected the responses of N-cycling functional genes to N additions in different ecosystems.
Materials and methods
Data extraction and collection
To obtain the data for this analysis, we searched for studies about N-cycling functional genes published before February 2022 under different keyword combinations (nitrogen fixation, ammonia oxidizing, nitrous oxide, functional gene, grassland, forest, wetland, and agriculture) in Google Scholar, the Web of Science, and the China Knowledge Resource Integrated Database (CNKI). We carried out a detailed examination of studies that reported the effects of N additions on N-cycling functional genes in soil and included those that met the following criteria: (1) the authors used real-time quantitative PCR (qPCR) to measure the abundance of functional genes; (2) the experiment had an established control group without N applications; (3) they were field studies; (4) at least one functional gene was determined; (5) the sample size, mean values, and standard errors or standard deviations of the experimental and control groups were provided in the article or could be calculated; (6) data with less than three replicates were excluded, and (7) the data could be obtained either directly from the text or through GET DATA GRAPH DIGITIZER software (version 2.26, http://www.getdatagraph-digitizer.com/download.php).
We conducted a meta-analysis of the following functional genes where there were sufficient observations: nifH, amoA (AOA and AOB), nirK, nirS, and nosZ. For studies without standard deviations or standard errors, we referred to other data sets to establish log equations for estimating the standard deviation and the mean of the experimental results (Marinho et al. 2003). For a meta-analysis, data must be independent of each other; we therefore selected only the data measured during the latest year from multi-year experiments, and for the top layer of the soil where an experiment involved adding the same treatment to different soil layers. To increase the research data, we also included multi-factor research results. For example, two treatments of phosphorus-potassium (PK) additions and nitrogen-phosphorus-potassium (NPK) additions were set up in an individual study. We used the PK treatment as the control group, and the NPK treatment as the experimental group.
As well as collating data for the gene abundance, we also recorded data for the mean annual temperature (MAT), the mean annual precipitation (MAP), the soil pH, the soil water content (WC), soil available phosphorus (AP), NH4+, NO3−, soil organic carbon (SOC), total nitrogen (TN), soil microbial carbon (MBC), soil microbial nitrogen (MBN), and the ratio of soil organic carbon to soil total nitrogen (C/N),. We also collected 73 observations of nitrification potential (NP) and 41 observations of N2O emissions, but there were insufficient data to permit robust analysis of each ecosystem. Overall, we collected 397 sets of observation data from 119 articles about N-cycling genes. Of these, 86 observations were from grassland, 57 were from forest, and 253 were from cropland (Fig. S1). To study the influence of other experimental or environmental factors on the functional gene abundance, we grouped the data into (1) ecosystem type (grassland ecosystems, forest ecosystems, and cropland ecosystems); (2) N form (inorganic, organic, and inorganic–organic mixed N; inorganic N forms included NH4+, NO3−, NO2−, and urea, and organic N forms, including compost, crop residues, and animal manure); (3) N addition rate (with 100 and 200 kg N ha−1 as the thresholds, giving different gradient divisions according to the amount of data and response ratios of each gene); and (4) N fertilization duration (with 5 and 10 years as the thresholds, resulting in different gradient divisions according to the amount of data and response ratios of each gene). For the two continuous variables, the N rate, duration, and gradient were partitioned based on the observations at each level and the gene abundance responses to each gradient in the preliminary analysis, in line with previous studies (Carey et al. 2016; Liu and Greaver 2010; Ouyang et al. 2018).
Data analysis
We used the natural logarithm of the response ratio (RR) as the effect size of the amount of N added on the abundances of the different N cycling genes (Hedges et al. 1999):
where \(\overline{{X }_{E}}\) and \(\overline{{X}_{C}}\) represent the mean results of the experimental and control groups, respectively. The following formula was used to calculate the corresponding variance (v):
where SE and SC represent the standard deviation of the experimental and control groups, and NE and NC represent the number of samples in the experimental and control groups, respectively.
We used the rma function in the metafor package (Viechtbauer 2010) to calculate the weighted effect size (RR++) and the 95% confidence interval (CI) with the mixed-effects model. When the 95% CI overlapped with 0, the difference between the results of the experimental and control groups was not significant; when the 95% CI did not overlap with the invalid line, the average effect sizes were considered significant. To show the impact more clearly, we converted the effect size (RR+) to a percentage expression:
The heterogeneity was calculated for each categorical group. We partitioned the total heterogeneity (QT) into the between-group heterogeneity (QM) and the residual error (QE). We used the QM to determine whether the values of the effects of the different groups differed noticeably. When P < 0.05, there was a significant difference in the response of the functional genes to N additions among gradients or categories (Rosenberg et al. 2000).
The relative effects of the climate and soil factors on the responses of the functional gene abundances to N additions were examined by calculating a relative importance value for each variable. The relative importance value was calculated as the sum of the Akaike weights for all the models that included this factor using the corrected Akaike’s Information Criteria with the glmulti package (Calcagno and de Mazancourt 2010). A cut-off of 0.8 was set for the most important predictors (sum of Akaike weights > 0.8) (Chen et al. 2020; Feng and Zhu 2019; Terrer et al. 2016). We used linear regression analysis to fit the relationships between the variables and calculated the corresponding R2 and P values. We used the R platform [version 4.0.3] (https://www.R-project.org) to perform all the statistical analysis and GraphPad Prism 8 (https://www.graphpad.com/) to draw the plots.
Results
Effects of N additions on physical and chemical soil factors
In the different ecosystems, the responses of the physical and chemical soil properties to N additions differed significantly (Fig. 1). The N additions had significant effects on the soil pH in forests, croplands, and grasslands (P < 0.05), and the effect size was greatest for grasslands. The NH4+ and NO3− increased significantly across all the ecosystems; the effect size of NH4+ was greatest for forests and least for croplands, while the effect size of NO3− was greatest for grasslands and least for forests (Fig. 1). The N additions had significant effects on SOC and TN in croplands (P < 0.05), TN in forests (P < 0.05), but had no significant effects on TN or SOC in the grasslands (P > 0.05). The MBC and MBN had contrasting responses to the N additions across the ecosystems. The responses of MBC and MBN in croplands were positive and significant (MBC = 40%, MBN = 45%, respectively, (P < 0.05)) but were insignificant in forests (P > 0.05). MBN had a positive response, while MBC had insignificant response, in grasslands.
Effect size of N additions on soil physical and chemical variables. Error bars represent the 95% confidence intervals. The results with significant responses are indicated by hollow dots. The numbers in parentheses represent the number of studies included for each variable. SOC, soil organic carbon; TN, soil total nitrogen; MBC, soil microbial carbon; MBN, soil microbial nitrogen
Effects of N additions on N-cycling functional genes
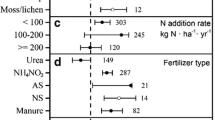
The N application rate had a significant effect on the response of the nifH abundance in grasslands, but the N application duration and N form were the main influences of change in forests and croplands, respectively (P < 0.05) (Fig. 2, Table 1). In grasslands, the nifH abundance increased significantly (by 58%) when the N application rate was low (< 100 kg N ha−1) but decreased by 45% when the N application rate was high (> 200 kg N ha−1) (P < 0.05). The nifH abundance in forests decreased significantly when the N application duration was less than 5 years (− 27%). In croplands, the nifH abundance decreased under inorganic N applications but increased under organic N and mixed N applications (P < 0.05).
The effect size of the N fixation gene (nifH) to N additions in different ecosystems. Error bars indicate the 95% confidence intervals. The results with significant responses are indicated by hollow dots. The numbers in brackets indicate the number of studies included for each variable. ‘Overall’ indicates the abundance of each functional gene
The response of AOA to N additions was mainly controlled by the N application duration in grasslands, and by the form of N and the pH in croplands (P < 0.01) (Fig. 3, Table 1). The AOA abundance in grasslands did not change significantly when the N application duration was less than 10 years (P > 0.05) but increased significantly when the N application duration exceeded 10 years (P < 0.05). In croplands, the AOA abundance decreased by 23% under inorganic N applications but increased by 99% under organic N applications (P < 0.05). When the pH was between 5.5 and 7.5, the AOA abundance decreased by 27% (P < 0.05). The AOB abundance was not significantly affected by the different variables in forests and grasslands, but was significantly affected by the N form (P < 0.01) and the N application duration (P = 0.013) in croplands (Fig. 3, Table 1). The AOB abundance showed the greatest increase in croplands for mixed N applications and when the N application duration was greater than 10 years (P < 0.05).
The effect size of the amoA nitrification genes (AOA and AOB) to N additions in different ecosystems. Error bars indicate the 95% confidence interval. The results with significant responses are indicated by hollow dots. The numbers in brackets indicate the number of studies included for each variable. ‘Overall’ indicates the abundance of each functional gene
The patterns of change in the denitrification genes varied significantly among the ecosystem types (Fig. 4, Table 1). NirK was mainly controlled by the N form in forests but was mainly controlled by the N form and N application duration in croplands. The nirK abundance increased significantly under inorganic N applications but decreased significantly under organic N applications in forests (P < 0.05). The nirS abundance in croplands was significantly affected by the N form and increased significantly under organic N applications and mixed N applications. The influences on the nosZ abundance varied among the ecosystems. The nosZ abundance was significantly affected by the N application rate in grasslands, by the N form in forests, and by the N form, N rate, and pH in croplands.
The effect size of N additions on the denitrification genes (nirK, nirS, and nosZ) in different ecosystems. The error bars indicate the 95% confidence intervals. The results with significant responses are indicated by hollow dots. The numbers in brackets indicate the number of studies included in each variable. ‘Overall’ indicates the abundance of each functional gene
Correlations between climate and soil properties and N-cycling functional genes
We examined the effects of climate factors (MAT and MAP) and soil properties on the genes (Table 2). The nifH and nosZ were closely correlated to the climate variables in grasslands and croplands. AOA was closely related to N fractions in grasslands, while AOA and AOB were both strongly related to the pH in croplands. In addition, NO3− was important for the denitrifiers in croplands. Overall, there were strong relationships between TN and SOC and most genes in croplands.
The results from the model-averaged relative importance showed that different variables were important for the gene abundances in the different ecosystems (Fig. 5). There was no clear pattern for grasslands, with only a few important variables for a limited number of functional genes. In forests, the SOC was important for all the functional genes except nosZ. The cropland showed most effects from the N applications, with numerous important variables. SOC was important for all the functional genes, and pH was important for AOA, AOB, nirS, and nosZ. MAT, NO3−, and TN were important for nifH, AOA, and AOB.
The model-averaged relative importance of the predictors of N additions on different N-cycling functional genes in different ecosystems. The cut-off between important (red squares) and nonessential predictors (white squares) was 0.8. The importance was based on the sum of the Akaike weights derived from model selection using the corrected Akaike’s Information Criteria
We also examined the correlations between the effect sizes of the functional genes in the different ecosystems (Table S3). We found that, in grasslands, there were no significant correlations between the nitrification genes and denitrification genes, while nifH was significantly correlated with the nitrification genes. The nitrification genes were significantly correlated with denitrification genes in forests and croplands. The denitrification genes were positively correlated with each other in the three ecosystems.
Discussion
Effect of climate factors on the responses of N-cycling functional genes to N additions
We found that the responses of the nifH and nosZ genes were significantly and positively correlated with the MAT and MAP in grasslands and croplands. These strong relationships suggest they were sensitive to changes in the external environment and may reflect the important role of MAT and MAP in controlling the exchange of N gases, namely NH3 and N2O. Chen et al. (2020) found that the response of the microbial activities to warming was minimal, and was closely related to the soil humidity. Homyak et al. (2017) found that the soil microbial biomass increased under small decreases in precipitation but decreased under large reductions in precipitation, with the changes related to the NH4+ concentrations. They also suggested that an ecosystem receiving N applications could lose N via a hydrological pathway when the precipitation subsequently increased, which would then reduce the production of N2O. Thus, microorganisms are sensitive to changes in temperature and humidity in the environment, which will affect their gene expression (Cantarel et al. 2012; Klarenberg et al. 2021). MAT and MAP must be considered as predictors of functional genes because of their impact on the microbial activity and composition (Hu et al. 2020; Tang et al. 2019; Verma and Sagar 2020).
While the nitrification and denitrification genes and the climate factors were not closely related, the relative importance analysis indicated that temperature was important for the nitrification genes in croplands. Zhang et al. (2022) found that increases in temperature had a minimal effect on functional genes associated with N cycling in soil, reflecting the insignificant change on the plant biomass. However, warming and N additions combined caused noticeable increases in the abundances of amoA. Even if a temperature change causes a significant response in the soil substrate, the biological characteristics of AOA and AOB in autotrophic organisms could mean that their gene expression abundance does not change or even decreases (Kowalchuk and Stephen 2001; Tang et al. 2019). It could affect subsequent denitrification processes and the N2O production (Cantarel et al. 2012; Ouyang et al. 2018).
Effect of N additions on the responses of the N-cycling functional genes
Nitrogenase, dominated by nifH, fixes N2 to NH4+, which is then followed by nitrification. However, this is not the only source of NH4+. The soil NH4+ may increase when inorganic NH4+-N is added from an external source or through mineralization of organic N by microorganisms (Nelson et al. 2016). This may help to explain why the nifH abundance does not change, and may even decrease, when inorganic NH4+-N is applied to forests.
During nitrification, we found that the AOA abundance did not respond significantly to N additions in these ecosystems. This differs from the results reported by other researchers (Carey et al. 2016; Ouyang et al. 2018) and could reflect the small number of observations included in the other studies (Carey et al. 2016; Ouyang et al. 2018). Furthermore, the ammonia monooxygenase that is encoded by AOA reaches saturation at a low NH3 concentration. Studies have shown that, under low N applications, AOA dominates the nitrification process but, when the N application rate is high, the soil microbial activity decreases because of acidification (Di et al. 2009; Ouyang et al. 2018), and so the response of AOA to N additions is not significant (Prosser and Nicol 2012). The model-averaged relative importance also showed that the soil pH was an important control on the amoA abundance in croplands. Thus, AOB are more adaptable than AOA and their response increases as the N contents increase (Prosser and Nicol 2012; Wang et al. 2016).
AOB were most abundant in grasslands, which had the highest inorganic N inputs and longest N application duration of the three ecosystems. This result is consistent with the physiological characteristics and NH4+ oxidation pathways of AOB (Hu and He 2018; Ouyang et al. 2017; Prosser and Nicol 2012). The response of AOB reached a minimum in forests, suggesting that the N fixation rate was suppressed in forests (Dynarski and Houlton 2017). The increase in the AOB abundance was less in croplands (+ 151.86%) than in grasslands. Croplands receive more organic N than grasslands, and the complexity of the organic matter has little effect on nutrient decomposition and use of nutrients by microorganisms (Zhao et al. 2016). As a result, inorganic N has more effect on AOB than AOA at the same application rate and duration because AOB has a stronger affinity for NH3 than AOA (Carey et al. 2016; Hu and He 2018; Ouyang et al. 2018).
The results show that the three denitrification genes were promoted most when organic N was added, with the promotion closely related to the nutrient content, and that SOC was an important driver of the responses of nirK, nirS, and nosZ to N additions (Fig. 5). Zhang et al. (2021) found that organic N had stronger effects on denitrifier abundances than inorganic N. Abdalla et al. (2019) reported that, as organic N applications to cropland under rotation increased, the NO3− leaching decreased and SOC retention improved. Thus, when supplied in large quantities, SOC and TN can promote the growth and physiological activities of soil microorganisms, especially heterotrophic denitrifying bacteria, causing the denitrification rate to increase (Morley and Baggs 2010) and enhancing the production of N2O. Other studies reported similar results (Kallenbach and Grandy 2011; Kramer et al. 2006; Ouyang et al. 2018). Studies have shown that, as the rate and duration of the N applications increase, the denitrification rate and the production of N2O will also change (Aronson and Allison 2012; Bai et al. 2014).
How soil properties affect the response of N-cycling functional genes to N additions
The soil pH has a strong influence on the microbial composition and gene abundance (Lauber et al. 2009). Functional communities each have unique optimum pH ranges and, for example, when the soil pH is greater than 5, the soil microbial biomass may increase significantly under N additions. External N applications have no effect on severely acidified soil (Geisseler and Scow 2014; Lauber et al. 2009; Ouyang et al. 2018). For example, unlike the grassland and cropland study sites, 84% of the soil in forest study sites examined in this analysis were already relatively acidic and had pH values between 3.7 and 5 before N applications commenced (Supplementary Dataset). Previous studies have reported that forest soils are frequently acidic (Friedland and Miller 1999). Researchers showed that, in forests, the proportion of AOA was higher than that of AOB, and that AOA preferred environments that had lower NH3 contents and soil pH values (Assémien et al. 2017; Prosser and Nicol 2012; Srikanthasamy et al. 2018). When N is applied, the available N increases, which promotes the release of H+ during litter decomposition and nitrification (Rao et al. 2009). Thus, the metabolism of microbes is suppressed in acidic soil. When analysing the roles of AOB and AOA in an acidified soil, Song et al. (2016) reported that AOA rather than AOB were responsible for nitrification in the acidic soil.
SOC and TN also influenced the gene abundance. Soil fertility in croplands can be maintained through crop rotation or planting ground cover during the slack season (Smith et al. 2008; Tilman et al. 2002; Zak et al. 2003). N applications could promote inputs of C and other exudates by stimulating the plant productivity and litter mass (Abdalla et al. 2019; Acosta-Martínez et al. 2007; Deng et al. 2020; Dodor and Ali Tabatabai 2005; Schmidt et al. 2011). A high C environment promotes the dissimilation and reduction of NO3− to NH4+, which is then easily used by plants (Schmidt et al. 2011; Zhu et al. 2014). As the SOC and TN increase, the supply of NH4+ comes from directly improving the physiological metabolism of microbes and accelerating the decomposition of organic matter rather than through N2 fixation (Cannavo et al. 2004; Cao et al. 2021). Therefore, as the SOC and TN increased, the nifH abundance either did not change or decreased. The nitrification functional genes, AOA and AOB, are also regulated by SOC and TN. The mineralization rate and soil organic matter are higher in forests than grasslands, which may have benefits for nitrifier growth (Srikanthasamy et al. 2018). However, the microorganism activity and AOB abundance in forests may be low, because of the low inputs of organic fertiliser, and therefore SOC and TN (Cavicchioli et al. 2019). The microorganisms involved in denitrification are usually heterotrophic, and so compete for SOC (Tiedje 1988), which means that the denitrification genes are very sensitive to changes in SOC (Levy-Booth et al. 2014). Our analysis showed that SOC, which is important for plant productivity and microbial respiration (Fernández-Martínez et al. 2014), did not change significantly in grassland and forest ecosystems (Fig. 1), which helps to explain the lack of significant change in the denitrification gene abundance.
In general, the climate factors and N together influence the soil physical and chemical properties, and so affect the abundance of the various functional genes and their contributions to N cycling. Because of the unique properties of soil and microbial communities in different ecosystems, the degree of change in the soil properties and in the abundances and responses of the functional genes will differ. We summarized the results from our analysis and developed a conceptual model of how N additions and climate influence soil properties and N-cycling functional genes (Fig. 6). We believe our model will be mainly valid for temperate soils based on the sources of our data (Fig. S1). Some of the soil properties, such as NO3−, pH, and NH4+, and functional genes (AOB) changed considerably under N additions across all ecosystem types. The abundance of nifH decreased significantly (by 32.79%) in forests and the abundances of nirK, nirS, and nosZ increased significantly in croplands. The results demonstrate the role of climate and, in future, we suggest that MAT and MAP should be considered as predictors of functional genes because of their impact on the microbial activity, and the composition and concentrations of the reaction substrate. We advise careful nutrient budgeting and monitoring of the pH and NH4+, to avoid over applications of N, that drive increases in emissions, and consequent acidification of the soil and pollution of nearby waterways by NO3−.
Conclusion
This study applied meta-analysis techniques to data from 119 articles, to identify how N additions caused changes in the N-cycling functional genes in three different ecosystems. We found that the responses of the N-cycling functional genes to N additions varied among the ecosystems. Under N applications, the AOA abundance did not change significantly in the three ecosystems, the AOB abundance in grasslands increased by 193.06%, the nifH and AOB in forests changed significantly, and the AOB and denitrification gene (nirK, nirS, and nosZ) abundances in croplands increased significantly. The N-cycling genes were influenced by different variables among the ecosystems with most effects in the croplands. There were no strong overall patterns for the N-cycling genes in grasslands. The N-cycling genes in forests were mainly influenced by SOC, but in croplands, were influenced by the MAT, pH, NO3−, SOC, and TN. Through changing the reaction substrate, and affecting the microbial activity, the climate, N additions, and soil properties impacted the expression abundance of the functional genes. Quantifying the differences in the responses of the N-cycling gene abundances to N additions across three terrestrial ecosystems in this way can produce useful information about gene-based models of N cycling in soil, especially the populations and activities of microbial functional communities.
Data availability
All the data presented in this manuscript are available in the supporting information.
References
Abdalla M, Hastings A, Cheng K, Yue Q, Chadwick D, Espenberg M, Truu J, Rees RM, Smith P (2019) A critical review of the impacts of cover crops on nitrogen leaching, net greenhouse gas balance and crop productivity. Glob Change Biol 25:2530–2543. https://doi.org/10.1111/gcb.14644
Acosta-Martínez V, Mikha MM, Vigil MF (2007) Microbial communities and enzyme activities in soils under alternative crop rotations compared to wheat-fallow for the Central Great Plains. Appl Soil Ecol 37:41–52. https://doi.org/10.1016/j.apsoil.2007.03.009
Aronson E, Allison S (2012) Meta-analysis of environmental impacts on nitrous oxide release in response to N amendment. Frontiers in Microbiology 3. https://doi.org/10.3389/fmicb.2012.00272
Assémien FL, Pommier T, Gonnety JT, Gervaix J, Le Roux X (2017) Adaptation of soil nitrifiers to very low nitrogen level jeopardizes the efficiency of chemical fertilization in west african moist savannas. Sci Rep 7:10275. https://doi.org/10.1038/s41598-017-10185-5
Bai E, Li W, Li S, Sun J, Peng B, Dai W, Jiang P, Han S (2014) Pulse increase of soil N2O emission in response to N addition in a temperate forest on Mt Changbai. Northeast China Plos One 9:e102765. https://doi.org/10.1371/journal.pone.0102765
Bonan GB (2008) Forests and climate change: forcings, feedbacks, and the climate benefits of forests. Science 320:1444. https://doi.org/10.1126/science.1155121
Bowman WD, Cleveland CC, Halada Ĺ, Hreško J, Baron JS (2008) Negative impact of nitrogen deposition on soil buffering capacity. Nat Geosci 1:767–770. https://doi.org/10.1038/ngeo339
Calcagno V, de Mazancourt C (2010) glmulti: An R package for easy automated model selection with (generalized) linear models. Journal Of Statistical Software 34. https://doi.org/10.18637/jss.v034.i12.
Canfield DE, Glazer AN, Falkowski PG (2010) The evolution and future of earth’s nitrogen cycle. Science 330:192–196. https://doi.org/10.1126/science.1186120
Cannavo P, Richaume A, Lafolie F (2004) Fate of nitrogen and carbon in the vadose zone: in situ and laboratory measurements of seasonal variations in aerobic respiratory and denitrifying activities. Soil Biol Biochem 36:463–478. https://doi.org/10.1016/j.soilbio.2003.10.023
Cantarel AAM, Bloor JMG, Pommier T, Guillaumaud N, Moirot C, Soussana J-F, Poly F (2012) Four years of experimental climate change modifies the microbial drivers of N2O fluxes in an upland grassland ecosystem. Glob Change Biol 18:2520–2531. https://doi.org/10.1111/j.1365-2486.2012.02692.x
Cao Y, He Z, Zhu T, Zhao F (2021) Organic-C quality as a key driver of microbial nitrogen immobilization in soil: A meta-analysis. Geoderma 383. https://doi.org/10.1016/j.geoderma.2020.114784
Carey CJ, Dove NC, Beman JM, Hart SC, Aronson EL (2016) Meta-analysis reveals ammonia-oxidizing bacteria respond more strongly to nitrogen addition than ammonia-oxidizing archaea. Soil Biol Biochem 99:158–166. https://doi.org/10.1016/j.soilbio.2016.05.014
Cavicchioli R, Ripple WJ, Timmis KN, Azam F, Bakken LR, Baylis M, Behrenfeld MJ, Boetius A, Boyd PW, Classen AT, Crowther TW, Danovaro R, Foreman CM, Huisman J, Hutchins DA, Jansson JK, Karl DM, Koskella B, Mark Welch DB, Martiny JBH, Moran MA, Orphan VJ, Reay DS, Remais JV, Rich VI, Singh BK, Stein LY, Stewart FJ, Sullivan MB, van Oppen MJH, Weaver SC, Webb EA, Webster NS (2019) Scientists’ warning to humanity: microorganisms and climate change. Nat Rev Microbiol 17:569–586. https://doi.org/10.1038/s41579-019-0222-5
Chen Y, Feng J, Yuan X, Zhu B (2020) Effects of warming on carbon and nitrogen cycling in alpine grassland ecosystems on the Tibetan Plateau: A meta-analysis. Geoderma 370:114363. https://doi.org/10.1016/j.geoderma.2020.114363
Deng L, Huang C, Kim D-G, Shangguan Z, Wang K, Song X, Peng C (2020) Soil GHG fluxes are altered by N deposition: New data indicate lower N stimulation of the N2O flux and greater stimulation of the calculated C pools. Glob Change Biol 26:2613–2629. https://doi.org/10.1111/gcb.14970
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624. https://doi.org/10.1038/ngeo613
Dodor DE, Ali Tabatabai M (2005) Glycosidases in soils as affected by cropping systems. J Plant Nutr Soil Sci 168:749–758. https://doi.org/10.1002/jpln.200521761
Dynarski K, Houlton B (2017) Nutrient limitation of terrestrial free-living nitrogen fixation. New Phytologist 217. https://doi.org/10.1111/nph.14905
Feng J, Zhu B (2019) A global meta-analysis of soil respiration and its components in response to phosphorus addition. Soil Biol Biochem 135:38–47. https://doi.org/10.1016/j.soilbio.2019.04.008
Fernández-Martínez M, Vicca S, Janssens IA, Sardans J, Luyssaert S, Campioli M, Chapin Iii FS, Ciais P, Malhi Y, Obersteiner M, Papale D, Piao SL, Reichstein M, Rodà F, Peñuelas J (2014) Nutrient availability as the key regulator of global forest carbon balance. Nat Clim Chang 4:471–476. https://doi.org/10.1038/nclimate2177
Friedland AJ, Miller EK (1999) Major-element cycling in a high-elevation adirondack forest: patterns and changes, 1986–1996. Ecol Appl 9:958–967. https://doi.org/10.1890/1051-0761(1999)009[0958:MECIAH]2.0.CO;2
Gaby JC, Buckley DH (2012) A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE 7:e42149. https://doi.org/10.1371/journal.pone.0042149
Geisseler D, Scow KM (2014) Long-term effects of mineral fertilizers on soil microorganisms - A review. Soil Biol Biochem 75:54–63. https://doi.org/10.1016/j.soilbio.2014.03.023
Zhu Y, Wang X, Yang X, Xu H, Jia Y (2014) Key microbial processes in nitrous oxide emissions of agricultural soil and mitigation strategies. Environmental Science 35: 9. https://doi.org/10.13227/j.hjkx.2014.02.008.
Hedges, Larry V, Gurevitch, Jessica (1999) The meta-analysis of response ratios in experimental ecology. Ecology 80: 1150-1156. https://doi.org/10.1890/0012-9658(1999)080[1150:TMAORR]2.0.CO;2.
Hallin S, Jones CM, Schloter M, Philippot L (2009) Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J 3:597–605. https://doi.org/10.1038/ismej.2008.128
Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol 72:5181–5189. https://doi.org/10.1128/AEM.00231-06
Homyak PM, Allison SD, Huxman TE, Goulden ML, Treseder KK (2017) Effects of drought manipulation on soil nitrogen cycling: A meta-analysis. J Geophys Res Biogeosci 122:3260–3272. https://doi.org/10.1002/2017JG004146
Hovenden MJ, Leuzinger S, Newton PCD, Fletcher A, Fatichi S, Lüscher A, Reich PB, Andresen LC, Beier C, Blumenthal DM, Chiariello NR, Dukes JS, Kellner J, Hofmockel K, Niklaus PA, Song J, Wan S, Classen AT, Langley JA (2019) Globally consistent influences of seasonal precipitation limit grassland biomass response to elevated CO2. Nature Plants 5:167–173. https://doi.org/10.1038/s41477-018-0356-x
Hu R, Wang X, Xu J, Zhang Y, Pan Y, Su X (2020) The mechanism of soil nitrogen transformation under different biocrusts to warming and reduced precipitation: From microbial functional genes to enzyme activity. Sci Total Environ 722:137849. https://doi.org/10.1016/j.scitotenv.2020.137849
Hu H, He J (2018) Manipulating the soil microbiome for improved nitrogen management. Microbiology Australia 39. https://doi.org/10.1071/MA18007
Kallenbach C, Grandy S (2011) Controls over soil microbial biomass responses to carbon amendments in agricultural systems: A meta-analysis. Agr Ecosyst Environ 144:241–252. https://doi.org/10.1016/j.agee.2011.08.020
Klarenberg IJ, Keuschnig C, Russi Colmenares AJ, Warshan D, Jungblut AD, Jónsdóttir IS, Vilhelmsson O (2021) Long-term warming effects on the microbiome and nifH gene abundance of a common moss species in sub-Arctic tundra. New Phytologist. https://doi.org/10.1111/nph.17837
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529. https://doi.org/10.1146/annurev.micro.55.1.485
Kramer SB, Reganold JP, Glover JD, Bohannan BJM, Mooney HA (2006) Reduced nitrate leaching and enhanced denitrifier activity and efficiency in organically fertilized soils. Proc Natl Acad Sci 103:4522. https://doi.org/10.1073/pnas.0600359103
Kuperman RG, Edwards CA (1997) Effects of acidic deposition on soil invertebrates and microorganisms. In: W G.W., N H.N., B A. (eds) Reviews of Environmental Contamination & Toxicology. Springer, New York.
Kuypers MMM, Marchant HK, Kartal B (2018) The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. https://doi.org/10.1038/nrmicro.2018.9
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Levy-Booth DJ, Prescott CE, Grayston SJ (2014) Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol Biochem 75:11–25. https://doi.org/10.1016/j.soilbio.2014.03.021
Li Z, Zeng Z, Tian D, Wang J, Fu Z, Zhang F, Zhang R, Chen W, Luo Y, Niu S (2020) Global patterns and controlling factors of soil nitrification rate. Glob Change Biol 26:4147–4157. https://doi.org/10.1111/gcb.15119
Liu L, Greaver TL (2010) A global perspective on belowground carbon dynamics under nitrogen enrichment. Ecol Lett 13:819–828. https://doi.org/10.1111/j.1461-0248.2010.01482.x
Makowski D (2019) N2O increasing faster than expected. Nat Clim Chang 9:909–910. https://doi.org/10.1038/s41558-019-0642-2
Marinho V, Higgins J, Logan S, Sheiham A (2003) Topical fluoride (toothpastes, mouthrinses, gels, varnishes) for preventing dental caries in children and adolescents. Cochrane database of systematic reviews (Online) 4: CD002782. https://doi.org/10.1002/14651858.CD002782.
Morales SE, Cosart T, Holben WE (2010) Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J 4:799–808. https://doi.org/10.1038/ismej.2010.8
Morley N, Baggs EM (2010) Carbon and oxygen controls on N2O and N2 production during nitrate reduction. Soil Biol Biochem 42:1864–1871. https://doi.org/10.1016/j.soilbio.2010.07.008
Nelson MB, Martiny AC, Martiny JBH (2016) Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci 113:8033. https://doi.org/10.1073/pnas.1601070113
Ouyang Y, Norton JM, Stark JM (2017) Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol Biochem 113:161–172. https://doi.org/10.1016/j.soilbio.2017.06.010
Ouyang Y, Evans SE, Friesen ML, Tiemann LK (2018) Effect of nitrogen fertilization on the abundance of nitrogen cycling genes in agricultural soils: A meta-analysis of field studies. Soil Biol Biochem 127:71–78. https://doi.org/10.1016/j.soilbio.2018.08.024
Phillips RP, Fahey TJ (2007) Fertilization effects on fineroot biomass, rhizosphere microbes and respiratory fluxes in hardwood forest soils. New Phytol 176:655–664. https://doi.org/10.1111/j.1469-8137.2007.02204.x
Prosser J, Nicol G (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531. https://doi.org/10.1016/j.tim.2012.08.001
Rao L, Parker D, Bytnerowicz A, Allen E (2009) Nitrogen mineralization across an atmospheric nitrogen deposition gradient in Southern California deserts. J Arid Environ 73:920–930. https://doi.org/10.1016/j.jaridenv.2009.04.007
Rosenberg M, Adams D, Gurevitch J (2000) MetaWin: Statistical software for meta-analysis. Version 2.0. Sinauer Associates.
Schmidt M, Torn M, Abiven S, Dittmar T, Guggenberger G, Janssens I, Kleber M, Kögel-Knabner I, Lehmann J, Manning D, Nannipieri P, Rasse D, Weiner S, Trumbore S (2011) Persistence of soil organic matter as an ecosystem property. Nature 478:49–56. https://doi.org/10.1038/nature10386
Smith RG, Gross KL, Robertson GP (2008) Effects of crop diversity on agroecosystem function: crop yield response. Ecosystems 11:355–366. https://doi.org/10.1007/s10021-008-9124-5
Song H, Che Z, Cao W, Huang T, Wang J, Dong Z (2016) Changing roles of ammonia-oxidizing bacteria and archaea in a continuously acidifying soil caused by over-fertilization with nitrogen. Environ Sci Pollut Res 23:11964–11974. https://doi.org/10.1007/s11356-016-6396-8
Srikanthasamy T, Leloup J, N’Dri AB, Barot S, Gervaix J, Koné AW, Koffi KF, Le Roux X, Raynaud X, Lata J-C (2018) Contrasting effects of grasses and trees on microbial N-cycling in an African humid savanna. Soil Biol Biochem 117:153–163. https://doi.org/10.1016/j.soilbio.2017.11.016
Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM Carbon and other biogeochemical cycles. In Climate change 2013: the physical science basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, United Kingdom and New York, NY, USA.
Sulman BN, Phillips RP, Oishi AC, Shevliakova E, Pacala SW (2014) Microbe-driven turnover offsets mineral-mediated storage of soil carbon under elevated CO2. Nat Clim Chang 4:1099–1102. https://doi.org/10.1038/nclimate2436
Sun R, Wang F, Hu C, Liu B (2021) Metagenomics reveals taxon-specific responses of the nitrogen-cycling microbial community to long-term nitrogen fertilization. Soil Biol Biochem 156:108214. https://doi.org/10.1016/j.soilbio.2021.108214
Tang L, Zhong L, Xue K, Wang S, Xu Z, Lin Q, Luo C, Rui Y, Li X, Li M, Liu W-t, Yang Y, Zhou J, Wang Y (2019) Warming counteracts grazing effects on the functional structure of the soil microbial community in a Tibetan grassland. Soil Biol Biochem 134:113–121. https://doi.org/10.1016/j.soilbio.2019.02.018
Terrer C, Vicca S, Hungate BA, Phillips RP, Prentice IC (2016) Mycorrhizal association as a primary control of the CO2 fertilization effect. Science 353:72. https://doi.org/10.1126/science.aaf4610
Tiedje J (1988) Ecology of denitrification and dissimilatory nitrate reduction to ammonium. In: Z A.J.B. (ed) Biology of Anaerobic Microorganisms. John Wiley & Sons, New York, USA.
Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S (2002) Agricultural sustainability and intensive production practices. Nature 418:671–677. https://doi.org/10.1038/nature01014
Van Breemen N, Mulder J, Driscoll CT (1983) Acidification and alkalization of soils. Plant Soil 75:283–308. https://doi.org/10.1007/BF02369968
Verma P, Sagar R (2020) Effect of nitrogen (N) deposition on soil-N processes: a holistic approach. Sci Rep 10:10470. https://doi.org/10.1038/s41598-020-67368-w
Viechtbauer W (2010) Conducting meta-analyses in R with the metafor package. J Stat Softw 36. https://doi.org/10.18637/jss.v036.i03.
Wang Q, Zhang L-M, Shen J-P, Du S, Han L-L, He J-Z (2016) Nitrogen fertiliser-induced changes in N2O emissions are attributed more to ammonia-oxidising bacteria rather than archaea as revealed using 1-octyne and acetylene inhibitors in two arable soils. Biol Fertil Soils 52:1163–1171. https://doi.org/10.1007/s00374-016-1151-3
Yang F, Zhang Z, Barberán A, Yang Y, Hu S, Guo H (2021) Nitrogen-induced acidification plays a vital role driving ecosystem functions: Insights from a 6-year nitrogen enrichment experiment in a Tibetan alpine meadow. Soil Biol Biochem 153:108107. https://doi.org/10.1016/j.soilbio.2020.108107
Zak DR, Holmes WE, White DC, Peacock AD, Tilman D (2003) Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 84:2042–2050. https://doi.org/10.1890/02-0433
Zhang Q, Zhou J, Li X, Zheng Y, Xie L, Yang Z, Liu X, Xu C, Lin H, Yuan X, Liu C, Zhu B, Chen Y, Yang Y (2022) Contrasting effects of warming and N deposition on soil microbial functional genes in a subtropical forest. Geoderma 408:115588. https://doi.org/10.1016/j.geoderma.2021.115588
Zhang Y, Zhang F, Abalos D, Luo Y, Hui D, Hungate BA, García-Palacios P, Kuzyakov Y, Olesen JE, Jørgensen U, Chen J (2021) Stimulation of ammonia oxidizer and denitrifier abundances by nitrogen loading: Poor predictability for increased soil N2O emission. Global Change Biology n/a. https://doi.org/10.1111/gcb.16042
Zhao J, Ni T, Li J, Lu Q, Fang Z, Huang Q, Zhang R, Li R, Shen B, Shen Q (2016) Effects of organic–inorganic compound fertilizer with reduced chemical fertilizer application on crop yields, soil biological activity and bacterial community structure in a rice–wheat cropping system. Appl Soil Ecol 99:1–12. https://doi.org/10.1016/j.apsoil.2015.11.006
Zhao Z, He J, Quan Z, Wu C, Sheng R, Zhang L, Geisen S (2020) Fertilization changes soil microbiome functioning, especially phagotrophic protists. Soil Biol Biochem 148:107863. https://doi.org/10.1016/j.soilbio.2020.107863
Acknowledgements
We would like to thank the reviewers and editor for proofreading and providing helpful suggestions on the manuscript. Sincere appreciation to the scientists whose research studies were included in this meta-analysis. This study was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) [Grant No. 2019QZKK0608] and the National Natural Science Foundation of China [Grant No. 31770519].
Author information
Authors and Affiliations
Contributions
All the authors contributed to the study conception and design. Yinghui Liu and Jingyi Dong conceived the ideas and designed the methodology; Jingyi Dong, Jiaqi Zhang, and Haichao Jing collected the data; Jingyi Dong and Jiaqi Zhang analysed the data, and Jingyi Dong wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Luca Bragazza.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, J., Zhang, J., Liu, Y. et al. How climate and soil properties affect the abundances of nitrogen-cycling genes in nitrogen-treated ecosystems: a meta-analysis. Plant Soil 477, 389–404 (2022). https://doi.org/10.1007/s11104-022-05420-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05420-6