Abstract
Aims
Abscisic acid (ABA) has been shown to prime rice seedlings for enhanced tolerance to alkaline stress under both greenhouse and field conditions. This study aimed to understand the mechanism of the ABA priming effect.
Methods
Rice seedlings were grown hydroponically and pretreated with ABA (10 μM) for 24 h, and then subjected to alkaline stress simulated by 15 mM Na2CO3 solution (pH 10.87). Paraquat (2.5–25 μM) was used as an intracellular generator of reactive oxygen species (ROS).
Results
Compared to the control treatment, pretreatment with ABA significantly mitigated root damage and improved the survival rate of rice seedlings under alkaline conditions. Furthermore, ABA pretreatment increased the antioxidant enzyme activities of superoxide dismutase, catalase, peroxidase and ascorbate peroxidase, and reduced the alkalinity- and paraquat-induced ROS accumulation (O2·- and H2O2) and seedling injury and mortality. In addition, the expression of the ABA-responsive genes SalT and OsWsi18, as well as the stress tolerance-related genes OsJRL, OsPEX11, OsNAC9, OsAKT1 and OsHKT1, was superinduced by ABA pretreatment under alkaline conditions.
Conclusions
ABA priming enhances tolerance to alkaline stress by upregulating the antioxidant defense system and stress tolerance-related genes in the roots of rice seedlings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saline-alkaline (SA) stress is a major limiting factor in world crop production. The FAO (FAO 2016) has estimated that globally, more than 830 million ha of land suffers from soil salinization and/or alkalization. Due to its high salinity and inordinately high alkalinity (high pH), the combined SA stress is considerably more damaging to plants than either stress alone (Peng et al. 2008; Lv et al. 2013; An et al. 2016). The alkalinity of SA soil can range from pH 8.5 to pH 11 (Ma and Liang 2007; Hossner 2008; Wang et al. 2009; Amini et al. 2016). Consequently, plants growing in SA soil suffer not only from osmotic stress (Munns and Tester 2008; Li et al. 2017) and high ion toxicity (James et al. 2006; Lin et al. 2012; Huang et al. 2017), but also high pH stress resulting from the alkaline conditions that can severely inhibit plant growth by directly damaging root cells and disturbing physiological metabolism (Patil et al. 2012; Lv et al. 2013, 2014; An et al. 2016; Zhang et al. 2017). In addition, high soil pH also result in deficiencies of nutritional minerals like iron and phosphorus (Rehman et al. 2012; Nandal and Hooda 2014; Tian et al. 2016; Guo et al. 2017). However, the physiological and molecular mechanisms underlying the plant response and adaptation to SA stress remain largely elusive.
Plants generate reactive oxygen species (ROS) like superoxide anions (O2·-) and hydrogen peroxide (H2O2) in response to various environmental stresses (Choudhury et al. 2017). These ROS can have dual roles during plant stress responses, acting as important cellular signaling molecules in the regulation of stress tolerance at low levels (Müller et al. 2009; Baxter et al. 2014; Dietz et al. 2016; Willems et al. 2016; Mittler 2017; Yu et al. 2017), but inducing serious oxidative injury at high levels, including damage to cellular membranes (lipid peroxidation), proteins, RNA, and DNA, resulting in irreversible cellular damage, and even cell death (Choudhury et al. 2017; Mittler 2017). The oxidative stress generated by ROS in plants is considered a major limitation to crop productivity (Sharma et al. 2017). Normally, a fine balance is maintained between ROS production and reduction in plants (Raja et al. 2017). To maintain ROS homeostasis upon ROS accumulation, plant cells activate an antioxidant defense system that consists of ROS-scavenging enzymes, including superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), and ascorbate peroxidase (APX), as well as antioxidants like ascorbic acid (AsA) and reduced glutathione (GSH) (Sewelam et al. 2016; Choudhury et al. 2017; Kao 2017; Zhang et al. 2017).
In rice, high pH conditions cause severe cellular damage to roots, resulting in root cell death and subsequent wilting of the whole plant, and even death (Lv et al. 2013; Wei et al. 2015). These damaging effects of alkaline stress on root cells have recently been associated with massive accumulation of ROS under alkaline stress (Zhang et al. 2017). Alkaline treatment resulted in significant accumulation of O2·- and H2O2 in roots of rice seedlings, whereas application of procyanidins, which are potent antioxidants, 24 h prior to alkaline treatment significantly reduced ROS accumulation, alleviating alkalinity-induced root damage and seedling growth inhibition (Zhang et al. 2017). Consistent with these results, Guo et al. (2014) showed that the alkaline tolerance of a rice mutant, alkaline tolerance 1 (alt1), is associated with reduced ROS accumulation under alkaline conditions. Furthermore, Guan et al. (2017) recently reported that overexpression of a chloroplast SOD gene (OsCu/Zn-SOD) in rice plants increased survival rate, growth performance and grain yield under alkaline conditions. These findings suggest that alkaline tolerance in rice plants is correlated with ROS-scavenging capability.
The plant hormone abscisic acid (ABA) plays vital roles in enabling plants to cope with various environmental stresses (Tuteja 2007; Sah et al. 2016; Vishwakarma et al. 2017; Dar et al. 2017). One important mechanism of ABA action in stress tolerance has been correlated with its priming effect on plants (Srivastava et al. 2010; Sah et al. 2016; Wei et al. 2015, 2017). Priming is a physiological process by which plants acquire an enhanced capacity to mount defense responses to subsequent impending stresses rapidly and effectively (Conrath 2009; Pastor et al. 2013; Aranega-Bou et al. 2014; Wei et al. 2017). The priming of rice seeds or seedlings by ABA improved seedling survival and growth, as well as the grain yield of plants in saline fields, by reducing the Na+/K+ ratio and accumulation of Na+ and Cl− (Gurmani et al. 2006; Li et al. 2010; Gurmani et al. 2011, 2013). However, only a limited number of studies have examined the involvement of ABA in plant responses to alkaline stress. We previously reported that ABA priming of rice seedlings significantly enhanced tolerance to alkaline stress, as shown by improved seedling survival rate, biomass accumulation, and root growth under alkaline conditions (Wei et al. 2015). These priming effects of ABA were further validated in a three-year field experiment, during which ABA priming of rice seedlings 24 h before transplantation to SA paddy fields significantly reduced leaf withering and seedling death and improved subsequent plant growth and the final grain yield (Wei et al. 2017). These results demonstrate that ABA priming of rice seedlings represents a significant and promising new approach for enhancing rice production in SA paddy fields. However, the mechanism by which ABA priming protects roots of rice seedlings from alkaline damage remains unclear.
This study aimed to explore the physiological and molecular mechanisms of ABA priming for alkaline tolerance in rice seedlings. We show that ABA primes rice seedlings for alkaline tolerance by reducing ROS accumulation and upregulating stress tolerance-related genes in roots.
Materials and methods
Plant material and growth conditions
Rice cultivar Dongdao-4 (D4), an elite cultivar in SA land area in Jilin province, China, was used in this study. D4 shows a moderate tolerance to SA conditions (Wei et al. 2015). It was bred by crossing ‘Akitakomachi’ with ‘Nongda-10’ in Da’an Sodic Land Experiment Station, Jilin, China (Yang et al. 2011). Seeds were surface-sterilized with 75% (v/v) alcohol for 5 min, and rinsed with deionized water five times. After immersing in water for 2 days, the seeds were sprinkled onto wet filter paper in a petri dish, and germinated for 24 h in an incubator under dark condition at 28 °C. Eighteen uniformly germinated seeds were transplanted onto a multi-well plate floating on a 320 ml cup containing deionized water for 7 days, and then grown for 7 days in half-strength Kimura B nutrient solutions (Miyake and Takahashi 1983) in a controlled growth chamber under the following conditions: 25 °C day/20 °C night, and 12-h photoperiods, 350 μmol photons m−2 s−1 light intensity.
Alkaline-stress treatment and ABA application
We used Na2CO3 at 15 mM (pH =10.87, EC = 2.72 mS/cm) to simulate alkaline stress (Lv et al. 2013; Wei et al. 2015; Zhang et al. 2017). The EC and pH of the solutions were measured using conductivity meter DDS-12 (Lida In., Shanghai, China) and pH meter PHS-25 (Baiyuan In., Beijing, China), respectively.
Abscisic acid (ABA) (Sigma, Inc., St, Louis, MO, USA) was dissolved in a small amount of absolute ethanol and then diluted with deionized water to the desired concentrations. The solvent (ethanol) did not exceed a final concentration of 0.1% in the solutions used for plant treatments and had no effect on the rice growth and gene expressions examined in this work. Seedlings at approximately three-leaf stage were pretreated with ABA at a concentration of 10 μM by root-drench for 24 h, and then transferred to deionized water (unstressed) or Na2CO3 solution that did not contain ABA (Lv et al. 2013; Wei et al. 2015). Rice roots of each treatment were sampled for the measurement of root vigor, malondialdehyde (MDA) content, reactive oxygen species (ROS) levels, enzyme activities and quantitative real-time PCR (qRT-PCR) at 0 h, 6 h, 12 h, 24 h, 48 h, and 72 h after alkaline stress, respectively.
Treatment of rice seedlings with paraquat
In this study, paraquat were used to examine the effect of ROS generation on ABA priming. Paraquat is a highly toxic compound for plants owing to the formation of ROS, such as the superoxide anion, hydrogen peroxide, and hydroxyl radical (Suntres 2002; Hung et al. 2002).
Two-week-old rice seedlings were pretreatment with or without 10 μM ABA for 24 h. Then transferred to solutions of deionized water (control), 2.5, 5, 7.5, 10, 12.5, 15, 20, 25 μM of paraquat, respectively. Seedlings survival rate, malondialdehyde (MDA) content, membrane injury (MI), and reactive oxygen species (ROS) levels were determined after 3 days of each treatment.
Measurement of survival rate and root browning ratio under alkaline stress
The survival rate of rice seedlings was determined after 3 d, 4 d, and 5 d of Na2CO3 treatment, respectively. Individual seedlings were classified as dead if all the leaves were dry and brown (Wei et al. 2015). The root browning ratio per seedling was determined after 24 h of Na2CO3 treatment with or without ABA pretreatment.
Measurement of membrane injury, root vigor, and malondialdehyde content
Membrane injury (MI) was measured by electrolyte leakage (Tantau and Dörffling 1991). Rice seedlings were randomly selected from each treatment group, washed with deionized water to remove surface-adhered electrolytes, and cut and divided into shoots and roots. Samples (2 g fresh weight) of the roots were submerged in 15 ml of deionized water in 50 ml conical tubes and kept at 20 °C for 1 h. The electrical conduction of the effusion was then measured (R1) with a conductivity meter DDS-12 (Lida In., Shanghai, China). The tissue samples were killed by heating tubes in a boiling bath for 40 min, cooled to 20 °C, and the electrical conduction of the effusion was measured again (R2). The MI was evaluated using the formula MI (%) = R1/R2 × 100%.
Root α-naphthylamine-oxidizing activity was used as an indicator of root vigor according to the method described by Ramasamy et al. (1997). Rice roots (0.1 g) were harvested in a 10 ml tube containing 5 ml of 0.1 mol/L phosphate buffer (pH 7.0) and 25 mg/ml of α-naphthylamine (V:V = 1:1) and incubated at room temperature for 10 min. A 200 μl aliquot of the solution was then taken as the reaction control, mixed with 1 ml of distilled water, 100 μl of 1% sulfanilic acid anhydrous, and 100 μl of 1 mg/ml sodium nitrite, and incubated for 5 min. After diluting the mixture with 1.1 ml of distilled water, the resulting color was measured using a spectrophotometer (UV-2700, Shimadzu, Kyoto, Japan) at 510 nm, and denoted as start value. The tube was further incubated at 25 °C for 1 h, and then a 200 μl aliquot of the solution was taken, and repeated the above-described steps to record the end value. The total oxidized and the auto-oxidation value was expressed by the start value minus the end and control values, respectively. Value of oxidized α-NA was expressed by total oxidized value minus the auto-oxidation value. Root vigor is expressed as micrograms oxidized α-NA per gram fresh weight (FW) per hour (μg α-NA g−1 FW h−1).
Malondialdehyde (MDA) is a decomposition product of polyunsaturated fatty acid hydroperoxides, and is often used as an indicator of lipid peroxidation due to oxidative stress. The MDA content was determined by the thiobarbituric acid reaction as described by Heath and Packer (1968). A fresh root sample (0.1 g) was homogenized in 1 ml of 50 mM phosphate buffer (pH 7.8) using a bench-top ball-mill (Scientz-48, Ningbo Scientz Biotechnology Co. Ltd., Ningbo, China) at 50 Hz for 30 s, and centrifuged at 12,000×g for 15 min. Subsequently, 400 μl of supernatant was mixed with 1 ml of 0.5% thiobarbituric acid, and the mixture was placed in a boiling water bath for 20 min. The mixture was then cooled and centrifuged, and the absorbance of the resulting supernatant was measured at 532, 600, and 450 nm using a spectrophotometer (UV-2700, Shimadzu, Kyoto, Japan). The MDA content was calculated using the following formula: 6.45× (A532 - A600) - 0.56 × A450.
Measurement of ROS levels and enzyme activities
The O2·- contents were measured as described by Elstner and Heupel (1976) by monitoring the nitrite formation from hydroxylamine in the presence of O2·-, with some modifications as described by Jiang and Zhang (2001). Absorbance values at 530 nm were calibrated to calculate the contents of O2·- from the chemical reaction of O2·- and hydroxylamine.
The H2O2 contents were measured as described by monitoring the A415 of the titanium-peroxide complex (Brennan and Frenkel 1977). Absorbance values were calibrated to a standard curve generated with known concentrations of H2O2. The analytical reagent used to measure the H2O2 and O2·- content were acquired from the determination kit, according to the manufacturer’s instructions (Comin biotechnology Co., ltd. Suzhou, China) (Zhang et al. 2017).
For the determination of antioxidant enzyme activities, the fresh roots (0.1 g) were loaded in a 2 ml tube and frozen in liquid nitrogen, then homogenized in 1 ml of 50 mM phosphate buffer (pH 7.8) using the bench-top ball-mill at 50 Hz for 30s. The homogenate was centrifuged at 12,000×g for 15 min at 4 °C, and the resulting supernatant was used for SOD, CAT, and POD assays.
SOD (EC 1.15.1.1) activity was determined using the nitro blue tetrazolium (NBT) method described by Giannopolitis and Ries (1977). One unit of SOD was defined as the amount of enzyme required to cause 50% inhibition of NBT reduction, as monitored at 560 nm. CAT (EC 1.11.1.6) activity was measured according to the method described by Aebi (1984). CAT activity was assayed by the decline in absorbance per minute at 240 nm as a consequence of H2O2 consumption. POD (EC 1.11.1.7) activity was determined by assessing the rate of guaiacol oxidation in the presence of H2O2. One unit of POD was defined as the increase in absorbance per minute at 470 nm (Klapheck et al. 1990).
To assay APX activity, samples were extracted with 1 ml of 50 mM phosphate buffer (pH 7.0) containing 1 mM ascorbic acid and 1 mM EDTA, and homogenized using the bench-top ball-mill at 50 Hz for 30s. The homogenate was centrifuged at 12,000×g for 15 min at 4 °C. Subsequently, the supernatant was mixed with phosphate buffer (pH 7.0), 15 mM ascorbic acid (ASA), and 0.3 mM H2O2. The reaction mixture was analyzed with a spectrophotometer at 290 nm. One unit of APX was defined as the variable quantity of absorbance per minute at 290 nm (Nakano and Asada 1981).
Relative indices of MDA (Fig. 2c), root vigor (Fig. 2d), O2·- (Fig. 4c), H2O2 (Fig. 4d), SOD (Fig. 5e), POD (Fig. 5e), CAT (Fig. 5f) and APX (Fig. 5f) were expressed by taking unpretreated control as 1, under alkaline stress conditions.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Rice roots (0.2 g) were sampled in liquid nitrogen and ground using the bench-top ball-mill at 50 Hz for 30s. Total RNA was extracted with TRIzol reagent (TaKaRa Bio Tokyo, Japan) and first-strand cDNA was synthesized using M-MLV reverse transcriptase (Thermo, Carlsbad, CA, USA), according to the manufacturer’s protocols. Quantitative real-time PCR (qRT-PCR) was performed to determine the transcriptional expression of genes. Gene-specific primers were designed using Primer 5.0 software (Table S1; Table S2). The housekeeping gene β-actin (GenBank ID: X15865.1) was used as an internal standard. PCR was conducted in a 20 μl reaction mixture containing 1.6 μl cDNA template (50 ng), 0.4 μl 10 mM specific forward primer, 0.4 μl 10 mM specific reverse primer, 10 μl 2× SYBR Premix Ex Taq (TaKaRa, Bio Inc.), and 7.6 μl double-distilled H2O in a PCRmax machine (Illimina, ECORT48, UK). The procedure was performed as follows: 1 cycle for 30 s at 95 °C, 40 cycles for 5 s at 95 °C, and 20 s at 60 °C, and 1 cycle for 60 s at 95 °C, 30 s at 55 °C, and 30 s at 95 °C for melting curve analysis. The level of relative expression was computed using the 2-△△CT method (Livak and Schmittgen 2001).
Experimental design and statistical analyses
All of the experiments were conducted in a controlled growth chamber with three biological replicates, each consisting of 3 cups of rice seedlings, with eighteen seedlings each cup. Statistical analyses were performed using the statistical software SPSS 21.0 (IBM Corp., Armonk, NY). Based on one-way analysis of variance (ANOVA), Duncan’s multiple range test (DMRT) was used to compare differences in the means among treatments.
Results
ABA priming mitigated root damage under alkaline stress conditions
Pretreatment with ABA rescued rice seedling from wilting and death under alkaline stress as shown by the higher survival rates of seedlings pretreated with ABA (Fig. 1a, c). Alkaline treatment normally causes rapid root browning, which is thought to be a symptom of cell necrosis associated with a reduced capacity for water and nutrient uptake (Palanjian et al. 2005; Vanterpool and Truscott 2011). Pretreatment with ABA significantly reduced the percentage of root browning resulting from alkaline stress (Fig. 1b, d). Pretreatment with ABA also significantly reduced MDA accumulation (Fig. 2a), and increased root vigor (Fig. 2b), under alkaline stress conditions. The relative MDA and relative root vigor levels of ABA-pretreated plants were lowest after 24 h (Fig. 2c) and 48 h (Fig. 2d) of alkaline treatment, respectively.
ABA priming rescued rice seedlings from wilting and death under alkaline stress. Two-week-old rice seedlings were root-drenched with (ABA) or without (CK) 10 μM ABA for 24 h, and then subjected either to a no-stress or an alkaline-stressed (Na2CO3) condition. Images of seedling growth (a) were taken at day 3 (3d), day 4 (4d), and day 5 (5d); and roots (b) at 24 h. Survival rate (c) of rice seedlings was recorded after 3, 4, and 5 d; root browning rate (d) per seedling was calculated after 24 h. Values are means ± SD, n = 3. Asterisks denote a significant difference compared to control plants (*P < 0.05, **P < 0.01)
ABA priming mitigated root damage under alkaline stress. Two-week-old rice seedlings were root-drenched with (ABA) or without (CK) 10 μM ABA for 24 h, and then subjected either to a no-stress or an alkaline-stressed (Na2CO3) condition. Malondialdehyde (MDA) content of roots (a) and root vigor (b) of rice seedlings were measured at the indicated treatment hours. Relative MDA (c) and root vigor (d) are expressed by taking unpretreated control as 1 under alkaline stress conditions (ABA-Na2CO3/Na2CO3). Values are means ± SD, n = 3. Different letters on the columns represent a significant difference (P < 0.05) at each time point based on Duncan’s test
ABA priming downregulated OsKOD1 and upregulated OsBI1 expression
We further analyzed the levels of two cell death-related genes: OsKOD1, an inducer of programmed cell death (PCD) (Blanvillain et al. 2011); and OsBI1, a cell death suppressor important for the modulation of PCD in response to abiotic and biotic stresses (Weis et al. 2013). As shown in Fig. 3a, OsKOD1 expression was significantly induced in response to alkaline treatment, and the expression levels increased with increased treatment duration. This alkalinity-induced OsKOD1 expression was significantly suppressed by ABA pretreatment throughout the alkaline treatment (Fig. 3a). In contrast, the downregulation of OsBI1 under alkaline stress was significantly mitigated by ABA pretreatment (Fig. 3b).
ABA priming reduced cell death in the roots under alkaline stress. Two-week-old rice seedlings were root-drenched with (ABA) or without (CK) 10 μM ABA for 24 h, and then subjected either to a no-stress or an alkaline-stressed (Na2CO3) condition. Expression levels of the cell death-related genes, OsKOD1 (a) and OsBI1 (b) in the roots were measured at the indicated treatment hours. A quantitative real-time PCR was performed using OsACT1 as an internal standard. The expression levels of unpretreated control (CK) at 0 h were set as the unit to calculate the expression levels, shown as fold changes relative to the CK at 0 h. Values are means ± SD, n = 3. Different letters on the columns represent a significant difference (P < 0.05) at each time point based on Duncan’s test
ABA priming reduced alkalinity-induced accumulation of O2 ·- and H2O2
A notable accumulation of O2·- and H2O2 was observed under alkaline stress conditions (Fig. 4a, b), but this accumulation was partially inhibited by ABA pretreatment (Fig. 4a, b). The relative O2·- and H2O2 levels were decreased to approximately 0.5 and 0.7, respectively, of the unpretreated control levels after 24 h and 48 h of alkaline stress (Fig. 4c, d).
ABA priming decreased ROS accumulation under alkaline stress. Two-week-old rice seedlings were root-drenched with (ABA) or without (CK) 10 μM ABA for 24 h, and then subjected either to a no-stress or an alkaline-stressed (Na2CO3) condition. Accumulation of O2·- (a) and H2O2 (b) in the roots was measured at the indicated treatment hours. Relative O2·- (c) and H2O2 (d) are expressed by taking unpretreated control as 1 under alkaline stress conditions (ABA+Na2CO3/Na2CO3). Values are means ± SD, n = 3. Different letters on the columns represent a significant difference (P < 0.05) at each time point based on Duncan’s test
ABA priming increased antioxidant enzyme activity under alkaline stress
Antioxidant enzymes play a key role in ROS homeostasis in plant cells. The enzyme activities of SOD, POD, CAT, and APX were significantly induced in response to alkaline treatment (Fig. 5a–d). Enzyme activity increased with increased treatment duration until the end of the time course. Interestingly, ABA pretreatment further increased the activities of these antioxidant enzymes under alkaline stress (Fig. 5a, d). The relative antioxidant enzymes activities reached their highest level after 24 h of alkaline treatment, and were subsequently maintained at these levels (Fig. 5e, f). The activity of POD in the ABA-pretreated plants was lower than in unpretreated plants after 72 h of alkaline stress (Fig. 5b), possibly due to the onset of senescence in the seedlings (Veljovic-jovanovic et al. 2006). Pretreatment with ABA also resulted in the increased expression of 17 genes related to ROS-scavenging under alkaline stress (Fang et al. 2015; Fig. S1).
ABA priming enhanced antioxidant enzymes activity under alkaline stress. Two-week-old rice seedlings were root-drenched with (ABA) or without (CK) 10 μM ABA for 24 h, and then were subjected either to a no-stress or an alkaline-stressed (Na2CO3) condition. Enzyme activities of (a) superoxide (SOD), (b) peroxidase (POD), (c) catalase (CAT), and (d) ascorbate peroxidase (APX) were measured at the indicated treatment hours. The relative antioxidant enzyme activities of SOD and POD (e), and CAT and APX (f) are expressed by taking unpretreated control as 1 under alkaline stress conditions (ABA+Na2CO3/Na2CO3). Values are means ± SD, n = 3. Different letters on the columns represent a significant difference (P < 0.05) at each time point based on Duncan’s test
ABA priming enhanced tolerance to paraquat-induced oxidative stress
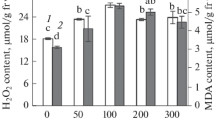
The results showing that ABA pretreatment significantly upregulated antioxidant enzyme activity (Fig. 5) and reduced ROS accumulation (Fig. 4) suggest that the ABA priming effect for alkaline tolerance may be associated with ROS-scavenging capacity. To test this, we determined the effect of paraquat (2.5–25 μM), a ROS-generating chemical, on the priming effect of ABA. As shown in Fig. 6, paraquat treatment caused seedling wilt and death (Fig. 6a, b), and cellular damage in both roots (Fig. 6c, e) and shoots (Fig. 6d, f). Paraquat-induced cellular damage and seedling mortality increased in a concentration-dependent manner (5 to 25 μM), and paraquat at levels ≥10 μM resulted in more than 60% of seedling mortality (Fig. 6b). In contrast, ABA pretreatment significantly mitigated the seedling injury and mortality resulting from moderate levels (≤ 15 μM) of paraquat treatment (Fig. 6a–f), while higher concentrations of paraquat (≥ 20 μM) almost completely inhibited the priming effect of the ABA pretreatment (Fig. 6a–f). Consistent with these results, ABA pretreatment reduced the paraquat-induced ROS accumulation in both roots (Fig. 6g, i) and shoots (Fig. 6h, j). Only a limited capability of ABA-priming for ROS reduction was observed in seedlings treated with high levels of paraquat (≥ 20 μM), which was not sufficient to reduce the seedling injury and mortality.
ABA priming enhanced tolerance to paraquat-induced oxidative stress. Two-week-old rice seedlings were root-drenched with or without 10 μM ABA for 24 h, and then subjected to a no-stress (CK), an alkaline-stressed (Na2CO3), a paraquat-induced oxidative stressed (2.5–25 μM) condition. Seedling survival rate (b), MI (c, d), MDA content (e, f), O2‧- content (g, h), and H2O2 content (i, j) of ABA-unprimed (ABA, −; open columns) and ABA-primed (filled columns) seedlings were measured after 3 d of each treatment. Seedling images (a) were obtained after 3 d of each treatment. Values are means ± SD, n = 3. Asterisks denote a significant difference compared to control plants (*P < 0.05, **P < 0.01)
ABA priming superinduced stress tolerance-related genes under alkaline stress
Two ABA-responsive genes, SalT (Rabbani et al. 2003) and OsWsil18 (Joshee et al. 1998), were induced by ABA as well as by alkaline stress (Fig. 7a, b), indicating that the ABA signaling pathway is indeed activated in response to alkaline stress. Under alkaline conditions, ABA pretreatment further greatly increased the expression of these genes. The induced expression levels peaked after 24 h of alkaline stress, with observed 29-fold and 16-fold increases in SalT (Fig. 7a) and OsWsil18 (Fig. 7b) expression, respectively, compared to the unpretreated control.
ABA priming upregulated the expression of the ABA-responsive genes SalT (a) and OsWsi18 (b), and the stress tolerance genes, OsJRL (c), OsPEX11 (d), OsNAC9 (e), OsAKT1 (f), and OsHKT1 (g) in rice roots under alkaline stress. Two-week-old rice seedlings were root-drenched with (ABA) or without (CK) 10 μM ABA for 24 h, and then subjected either to a no-stress or an alkaline-stressed (Na2CO3) condition for the indicated treatment hours. A quantitative real-time PCR was performed using OsACT1 as an internal standard. The expression levels of the unpretreated control (CK) at 0 h were set as the unit to calculate the expression levels, shown as fold changes relative to the CK at 0 h. Values are means ± SD, n = 3. Different letters on the columns represent a significant difference (P < 0.05) at each time point based on Duncan’s test
To gain further insights into the mechanism of the ABA priming for alkaline stress, the expression of five additional stress tolerance-related genes were analyzed: OsJRL, a gene that plays an important role in cell protection and stress signal transduction (overexpression of OsJRL in rice enhances salt resistance) (He et al. 2017); OsPEX11, a gene that modulates the Na+/K+ transporter and reduces ROS accumulation in rice under salt stress (Cui et al. 2016); OsNAC9, a gene coding for the NAC domain-containing protein 9 that enhances the root diameter and uptake in roots under drought conditions (Redillas et al. 2012); OsAKT1, a gene encoding the main inward rectifying K+ channel in rice roots (Li et al. 2014; Ahmad et al. 2016); and OsHKT1, a gene that codes for a Na+ transporter (Garciadeblãs et al. 2003; Kader et al. 2006).
As shown in Fig. 7c–g, all these stress tolerance-related genes displayed an expression pattern similar to SalT and OsWsil18 (Fig. 7a, b); overall, they were induced by both ABA and alkaline stress, and considerably higher induction levels were achieved in the stressed plants pretreated with ABA. The expression levels of these genes peaked at 24 h after alkaline stress, and then declined to pre-alkaline stress levels (Fig. 7c–g).
Discussion
Saline-alkaline (SA) stress represents a complex effect of ion toxicity from high salinity, alkalinity (high pH), and high osmotic pressure (low water availability); of these, alkaline stress was identified as the primary factor inhibiting rice seedling growth (Lv et al. 2013). Therefore, improving the alkaline stress tolerance of rice plants is considered critical for improving plant growth and productivity in SA soils. Alkaline stress results in severe cellular damage to the root system, as evidenced by the marked increase in cell injury and expression of cell death-related genes (Figs. 1, 2 and 3; Lv et al. 2013). These damaging effects on rice roots have been correlated with the overaccumulation of ROS (Guo et al. 2014; Zhang et al. 2017). Moreover, we previously reported that ABA pretreatment primed rice seedlings for enhanced tolerance to alkaline stress, both in the laboratory (Wei et al. 2015) and in SA field (Wei et al. 2017) experiments. However, the physiological and molecular mechanisms of ABA priming for alkaline tolerance in rice seedlings have remained unclear. In the present study, we showed that, under alkaline conditions, ABA pretreatment significantly increased the activity of antioxidant enzymes (Fig. 5), reduced ROS accumulation (Fig. 4), and upregulated stress tolerance-related genes (Fig. 7). Pretreatment with ABA also enhanced tolerance to paraquat-induced oxidative stress (Fig. 6). These data collectively suggest that preventing the overaccumulation of ROS and upregulating the expression levels of stress tolerance-related genes are important mechanisms of ABA priming for alkaline tolerance in rice seedlings.
Priming is a cost-effective and environment-friendly agrobiological technique widely used in agriculture to improve growth and stress tolerance in various crop plants because of its long-lasting effect on a broad range of stresses (Beckers and Conrath 2007; Conrath 2011; Savvides et al. 2016). The primed plants are physiologically conditioned or potentiated to mount a more rapid and/or stronger defense response upon exposure to subsequent stresses. It has been shown that ABA is a potent priming hormone for the enhancement of tolerance to various stresses in a range of crop plants (Gurmani et al. 2013; Sripinyowanich et al. 2013; Wang et al. 2013). However, to date, only a few studies have reported on the priming effect of ABA in the alkaline stress responses (Wei et al. 2015, 2017). In the present study, two ABA-response marker genes, SalT and OsWsil18, were significantly induced in response to alkaline treatment (Fig. 7a, b), indicating that the ABA signaling pathway was indeed activated in response to alkaline stress. This result implies that ABA plays a role in the defense against alkaline stress in rice seedlings. However, the levels of activation of the ABA signaling pathway may not be sufficient to activate downstream signals required to cope effectively with the stress. Neversness, ABA pretreatment greatly increased the induction levels of SalT and OsWsil18 (Fig. 7a, b), as well as the levels of antioxidant enzyme activity (Fig. 5), and ROS-scavenging genes (Fig. S1) and stress tolerance-related genes (Fig. 7c-g), under alkaline conditions. Additionally, we previously reported that ABA pretreatment decreased the Na+/K+ ratio under alkaline conditions (Wei et al. 2015). Taken together, these results suggest that ABA priming confers rice seedling with enhanced alkaline tolerance by potentiating the response of multiple downstream defense pathways upon alkaline exposure.
Reactive oxygen species exhibit dual roles in plant stress response; being both beneficial and deleterious, depending on the equilibrium between ROS production and scavenging (Gill and Tuteja 2010; Raja et al. 2017; Nikalje et al. 2018). At low levels, they serve as signaling messengers in a multitude of physiological processes required for stress tolerance. However, high levels of ROS accumulation can induce oxidative cellular damage and lead to cell death (Sewelam et al. 2016; Choudhury et al. 2017; Mittler 2017). The overproduction of ROS has been identified as a key causal factor for cellular damage in the roots of rice seedlings under alkaline stress (Guo et al. 2014; Guan et al. 2017; Zhang et al. 2017). In this study, rice seedlings presented a significant activation of the ROS scavenging enzymes SOD, POD, CAT and APX (Fig. 5), as well as a number of genes related to ROS scavenging (Fig. S1) under alkaline conditions, indicating that the antioxidant defense system was activated in response to alkaline stress in rice seedlings. However, this response mechanism may be overwhelmed by the oxidative burst (Fig. 4), resulting in oxidative damage to root cells (Figs. 1, 2 and 3). Nevertheless, ABA priming significantly strengthened the antioxidant defense system as shown by the greatly increased response of the ROS scavenging enzymes and related genes under alkaline conditions (Fig. 5, Fig. S1). Consistent with this concept, ABA pretreatment reduced ROS accumulation, and seedling injury and mortality resulting from application of moderate paraquat treatment levels (Fig. 6). These data demonstrate that increasing the ROS scavenging capacity is an important mechanism of ABA priming for alkaline tolerance. These findings provide useful insights for the molecular engineering for alkaline stress tolerance in rice plants by reducing oxidative stress (Kerchev et al. 2015). It has been shown that the transgenic overexpression of antioxidant enzyme-coding genes enhance ROS-scavenging activity and salt tolerance in rice plants (Tanaka et al. 1999; Zhao and Zhang 2006; Motohashi et al. 2010; Kumar et al. 2014). In future studies, it would be interesting to test whether these transgenic rice plants have a “cross-tolerance” to both salt and alkaline stresses, given that the ROS production is a common feature of plant responses to different stresses.
Plants respond to environmental stresses by reprograming gene expression to increase their ability to cope with the stresses. In this study, all the tested stress tolerance-related genes were significantly upregulated by ABA priming under alkaline conditions (Fig. 7), representing another molecular mechanism for the ABA-priming effect in addition to ROS modulation. Among the tested stress tolerance-related genes, OsPEX11 (Cui et al. 2016), OsAKT1 (Li et al. 2014; Ahmad et al. 2016), and OsHKT1 (Garciadeblãs et al. 2003; Kader et al. 2006) are involved in Na+ and/or K+ transport. These results may be in line with our previous finding that ABA-priming decreased the Na+/K+ ratio under alkaline conditions (Wei et al. 2015).
In summary, in this study, rice seedlings showed significant activation of ABA signaling, and ROS-scavenging pathways, and stress tolerance-related genes in response to alkaline stress. However, these activation levels were likely not sufficient to effectively cope with the stress, and eventually resulted in seedling death. Nevertheless, ABA priming greatly upregulated not only ABA signaling, but also ROS-scavenging activity and stress tolerance-related gene expressions under alkaline conditions. Taken together, we conclude that ABA priming confers enhanced tolerance to alkaline stress in rice seedlings via a mechanism that potentiates the downstream antioxidant defense system and stress tolerance-related gene expression for an increased adaptive response to alkaline stress.
References
Aebi H (1984) Catalase in vitro. Methods Enzymol 105:121–126. https://doi.org/10.1016/S0076-6879(84)05016-3
Ahmad I, Mian A, Maathuis FJM (2016) Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J Exp Bot 67:2689–2698. https://doi.org/10.1093/jxb/erw103
Amini S, Ghadiri H, Chen CR, Marschner P (2016) Salt-affected soils, reclamation, carbon dynamics, and biochar: a review. J Soils Sediments 16:939–953. https://doi.org/10.1007/s11368-015-1293-1
An YM, Song LL, Liu YR, Shu YJ, Guo CH (2016) De novo transcriptional analysis of alfalfa in response to saline-alkaline stress. Front Plant Sci 7:931. https://doi.org/10.3389/fpls.2016.00931
Aranega-Bou P, Leyva MDLO, Finiti I, García-Agustín P, González-Bosch C (2014) Priming of plant resistance by natural compounds. Hexanoic acid as a model. Front Plant Sci 5:488. https://doi.org/10.3389/fpls.2014.00488
Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65:1229–1240. https://doi.org/10.1093/jxb/ert375
Beckers GJ, Conrath U (2007) Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol 10:425–431. https://doi.org/10.1016/j.pbi.2007.06.002
Blanvillain R, Young B, Cai YM, Hecht V, Varoquaux F, Delorme V, Lancelin JM, Delseny M, Gallois P (2011) The Arabidopsis peptide kiss of death is an inducer of programmed cell death. EMBO J 30:1173–1183. https://doi.org/10.1038/emboj.2011.14
Brennan T, Frenkel C (1977) Involvement of hydrogen peroxide in the regulation of senescence in pear. Plant Physiol 59:411–416. https://doi.org/10.1104/pp.59.3.411
Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90:856–867. https://doi.org/10.1111/tpj.13299
Conrath U (2009) Priming of induced plant defense responses. Adv Bot Res 5:362–384. https://doi.org/10.1016/S0065-2296(09)51009-9
Conrath U (2011) Molecular aspects of defence priming. Trends Plant Sci 16:524–531. https://doi.org/10.1016/j.tplants.2011.06.004
Cui P, Liu HB, Islam F, Li L, Farooq MA, Ruan SL, Zhou WJ (2016) OsPEX11, a Peroxisomal biogenesis factor 11, contributes to salt stress tolerance in Oryza sativa. Front Plant Sci 7:1357. https://doi.org/10.3389/fpls.2016.01357
Dar NA, Amin I, Wani W, Wani SA, Shikari AB, Wani SH, Masoodi KZ (2017) Abscisic acid: a key regulator of abiotic stress tolerance in plants. Plant Gene 11:106–111. https://doi.org/10.1016/j.plgene.2017.07.003
Dietz KJ, Turkan I, Krieger-Liszkay A (2016) Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol 171:1541–1550. https://doi.org/10.1104/pp.16.00375
Elstner EF, Heupel A (1976) Inhibition of nitrite formation from hydroxylammoniumchloride: a simple assay for superoxide dismutase. Anal Biochem 70:616–620. https://doi.org/10.1016/0003-2697(76)90488-7
Fang YJ, Liao KF, Du H, Xu Y, Song HZ, Li XH, Xiong LZ (2015) A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot 66:6803–6817. https://doi.org/10.1093/jxb/erv386
FAO (2016) Management of Salt Affected Soils. Food and Agriculture Organization of the United Nations http://www.fao.org/soils-portal/soil-management/management-of-some-problem-soils/salt-affected-soils/more-information-on-salt-affected-soils/en/
Garciadeblãs B, Senn ME, Bañuelos MA, Rodrãguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34:788–801. https://doi.org/10.1046/j.1365-313X.2003.01764.x
Giannopolitis CN, Ries SK (1977) Superoxide Dismutases: I. Occurrence in higher plants. Plant Physiol 59:309–314. https://doi.org/10.1104/pp.59.2.309
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Guan QJ, Liao X, He ML, Li XF, Wang ZY, Ma HY, Yu S, Liu SK (2017) Tolerance analysis of chloroplast OsCu/Zn-SOD overexpressing rice under NaCl and NaHCO3 stress. PLoS One 12:e0186052. https://doi.org/10.1371/journal.pone.0186052
Guo MX, Wang RC, Wang J, Hua K, Wang YM, Liu XQ, Yao SG (2014) ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS One 9:e112515. https://doi.org/10.1371/journal.pone.0112515
Guo R, Shi LX, Yan CR, Zhong XL, Gu FX, Liu Q, Xia X, Li HR (2017) Ionomic and metabolic responses to neutral salt or alkaline salt stresses in maize (Zea mays L.) seedlings. BMC Plant Biol 17:41. https://doi.org/10.1186/s12870-017-0994-6
Gurmani AR, Bano A, Salim M (2006) Effect of growth regulators on growth, yield and ions accumulation of rice (Oryza sativa L.) under salt stress. Pak J Bot 38:1415–1424. https://doi.org/10.1110/ps.062292106
Gurmani AR, Bano A, Khan SU, Din J, Zhang JL (2011) Alleviation of salt stress by seed treatment with abscisic acid (ABA), 6-benzylaminopurine (BA) and chlormequat chloride (CCC) optimizes ion and organic matter accumulation and increases yield of rice (Oryza sativa L.). Aust J Crop Sci 5:1278–1285
Gurmani AR, Bano A, Ullah N, Khan H, Jahangir M, Flowers TJ (2013) Exogenous abscisic acid (ABA) and silicon (Si) promote salinity tolerance by reducing sodium (Na+) transport and bypass flow in rice (Oryza sativa indica). Aust J Crop Sci 7:1219–1226
He X, Li L, Xu H, Xi J, Cao X, Xu H, Rong S, Dong Y, Wang C, Chen R, Xu J, Gao X, Xu Z (2017) A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice. Plant Biol 19:257–267. https://doi.org/10.1111/plb.12514
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198. https://doi.org/10.1016/0003-9861(68)90654-1
Hossner L (2008) “Field pH,” in encyclopedia of soil science, ed. W. Chesworth (Berlin: Springer), 271–272
Huang LH, Liu X, Wang ZC, Liang ZW, Wang MM, Liu M, Suarez DL (2017) Interactive effects of pH, EC and nitrogen on yields and nutrient absorption of rice (Oryza sativa L.). Agric Water Manag 194:48–57. https://doi.org/10.1016/j.agwat.2017.08.012
Hung KT, Chang CJ, Kao CH (2002) Paraquat toxicity is reduced by nitric oxide in rice leaves. J Plant Physiol 159:159–166. https://doi.org/10.1078/0176-1617-00692
James RA, Munns R, Caemmerer SV, Trejo C, Miller C, Condon TA (2006) Photosynthetic capacity is related to the cellular and subcellular partitioning of Na+, K+ and cl− in salt-affected barley and durum wheat. Plant Cell Environ 29:2185–2197. https://doi.org/10.1111/j.1365-3040.2006.01592.x
Jiang MY, Zhang JH (2001) Effect of abscisic acid on active oxygen species, Antioxidative Defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol 42:1265–1273. https://doi.org/10.1093/pcp/pce162
Joshee N, Kisaka H, Kitagawa Y (1998) Isolation and characterization of a water stress-specific genomic gene, pwsi 18, from Rice. Plant Cell Physiol 39:64–72. https://doi.org/10.1093/oxfordjournals.pcp.a029290
Kader MA, Seidel T, Golldack D, Lindberg S (2006) Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot 57:4257–4268. https://doi.org/10.1093/jxb/erl199
Kao CH (2017) Mechanisms of salt tolerance in rice plants:reactive oxygen species scavenging-systems. Taiwan Nong Ye Yan Jiu 66:1–8. https://doi.org/10.6156/JTAR/2017.06601.01
Kerchev P, De Smet B, Waszczak C, Messens J, Van Breusegem F (2015) Redox strategies for crop improvement. Antioxid Redox Signal 23:1186–1205. https://doi.org/10.1089/ars.2014.6033
Klapheck S, Zimmer I, Cosse H (1990) Scavenging of hydrogen peroxide in the endosperm of Ricinus communis by ascorbate peroxidase. Plant Cell Physiol 31:1005–1013. https://doi.org/10.1093/oxfordjournals.pcp.a077996
Kumar M, Lee SC, Kim JY, Kim SJ, Aye SS, Kim SR (2014) Over-expression of dehydrin gene, OsDhn1 , improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J Plant Biol 57:383–393. https://doi.org/10.1007/s12374-014-0487-1
Li XJ, Yang MF, Chen H, Qu LQ, Chen F, Shen SH (2010) Abscisic acid pretreatment enhances salt tolerance of rice seedlings: proteomic evidence. Biochim Biophys Acta 1804:929–940. https://doi.org/10.1016/j.bbapap.2010.01.004
Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26:3387–3402. https://doi.org/10.1105/tpc.114.123455
Li Q, Yang A, Zhang WH (2017) Comparative studies on tolerance of rice genotypes differing in their tolerance to moderate salt stress. BMC Plant Biol 17:141. https://doi.org/10.1186/s12870-017-1089-0
Lin JX, Li XY, Zhang ZJ, Yu XY, Gao ZW, Wang Y, Wang JF, Li ZL, Mu CS (2012) Salinity-alkalinity tolerance in wheat: seed germination, early seedling growth, ion relations and solute accumulation. Afr J Agric Res 7:467–474. https://doi.org/10.5897/AJAR11.1417
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC T method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lv BS, Li XW, Ma HY, Sun Y, Wei LX, Jiang CJ, Liang ZW (2013) Differences in growth and physiology of Rice in response to different saline-alkaline stress factors. Agron J 105:1119–1128. https://doi.org/10.2134/agronj2013.0017
Lv BS, Li XW, Ma HY, Yang HY, Wei LX, Lv HY, Jiang CJ, Liang ZW (2014) Different modes of proline accumulation in response to saline-alkaline stress factors in rice (Oryza sativa L.). Research on Crops 15:14–21. https://doi.org/10.5958/j.2348-7542.15.1.002
Ma HY, Liang ZW (2007) Effects of different soil pH and soil extracts on the germination and seedling growth of Leymus chinensis. Chin Bull Bot 24:181–188
Mittler R (2017) ROS are good. Trends Plant Sci 22:11–19. https://doi.org/10.1016/j.tplants.2016.08.002
Miyake Y, Takahashi E (1983) Effect of silicon on the growth of solution-cultured cucumber plant. Soil Sci Plant Nutr 29:71–83. https://doi.org/10.1080/00380768.1983.10432407
Motohashi T, Nagamiya K, Prodhan SH et al (2010) Production of salt stress tolerant rice by overexpression of the catalase gene, katE, derived from Escherichia coli. AsPac J Mol Biol Biotechnol 18:37–41
Müller K, Linkies A, Vreeburg RAM, Fry SC, Krieger-Liszkay A, Leubner-Metzger G (2009) In vivo Cell Wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol 150:1855–1865. https://doi.org/10.1104/pp.109.139204
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880. https://doi.org/10.1093/oxfordjournals.pcp.a076232
Nandal M, Hooda R (2014) Salt tolerance and physiological response of plants to salinity: a review. Int J Sci Eng Res 4:44–67
Nikalje GC, Mirajkar SJ, Nikam TD, Suprasanna P (2018) Multifarious role of ROS in halophytes: signaling and defense. In: Zargar S, Zargar M (eds) Abiotic stress-mediated sensing and signaling in plants: An omics perspective. Springer, Singapore, pp 207–223
Palanjian K, Valenzuela L, Neilsen D, Neilsen G, Eissenstant (2005) (321) rapid and differential rates of root browning in apple trees under different irrigation treatments. Hort Science 40:1038–1038
Pastor V, Luna E, Mauch-Mani B, Ton J, Flors V (2013) Primed plants do not forget. Environ Exp Bot 94:46–56. https://doi.org/10.1016/j.envexpbot.2012.02.013
Patil NS, Apradh VT, Karadge BA (2012) Effects of alkali stress on seed germination and seedlings growth of Vigna aconitifolia (Jacq.) Marechal. Pharm J 4:77–80. https://doi.org/10.5530/pj.2012.34.13
Peng YL, Gao ZW, Gao Y, Liu GF, Sheng LX, Wang DL (2008) Eco-physiological characteristics of alfalfa seedlings in response to various mixed salt-alkaline stresses. J Integr Plant Biol 50:29–39. https://doi.org/10.1111/j.1744-7909.2007.00607.x
Rabbani MA, Maruyama K, Abe H, Khadri MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring Expression Profiles of Rice Genes under Cold, Drought, and High-salinity Etresses and Abscisic Acid Application Using cDNA Microarray and RNA Gel-Blot Analyses. Plant Physiol 133:1755–1767. https://doi.org/10.1104/pp.103.025742
Raja V, Majeed U, Kang H, Andrabi KI, John R (2017) Abiotic stress: interplay between ROS, hormones and MAPKs. Environ Exp Bot 137:142–157. https://doi.org/10.1016/j.envexpbot.2017.02.010
Ramasamy S, TenBerge HFM, Purushothaman S (1997) Yield formation in rice in response to drainage and nitrogen application. Field Crop Res 51:65–82. https://doi.org/10.1016/S0378-4290(96)01039-8
Redillas MCFR, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805. https://doi.org/10.1111/j.1467-7652.2012.00697.x
Rehman H-u, Aziz T, Farooq M, Wakeel A, Rengel Z (2012) Zinc nutrition in rice production systems: a review. Plant Soil 361:203–226. https://doi.org/10.1007/s11104-012-1346-9
Sah SK, Reddy KR, LiX J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571. https://doi.org/10.3389/fpls.2016.00571
Savvides A, Ali S, Tester M, Fotopoulos V (2016) Chemical priming of plants against multiple abiotic stresses: Mission possible? Trends Plant Sci 21:329–340. https://doi.org/10.1016/j.tplants.2015.11.003
Sewelam N, Kazan K, Schenk PM (2016) Global plant stress signaling: reactive oxygen species at the cross-road. Front Plant Sci 7:187. https://doi.org/10.3389/fpls.2016.00187
Sharma M, Gupta SK, Deeba F, Pandey V (2017) Effects of reactive oxygen species on crop productivity: boon or bane-revisiting the role of ROS. Reactive Oxygen Species in Plants:117–136
Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu HY, Buaboocha T, Chadchawan S (2013) Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot 86:94–105. https://doi.org/10.1016/j.envexpbot.2010.01.009
Srivastava AK, Lokhande VH, Patade VY, Suprasanna P, Sjahril R, D'Souza SF (2010) Comparative evaluation of hydro-, chemo- , and hormonal-priming methods for imparting salt and PEG stress tolerance in Indian mustard (Brassica juncea L.). Acta Physiol Plant 32:1135–1144. https://doi.org/10.1007/s11738-010-0505-y
Suntres ZE (2002) Role of antioxidants in paraquat toxicity. Toxicology 180:65–77. https://doi.org/10.1016/S0300-483X(02)00382-7
Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Kishitani S, Takabe T, Yokota S, Takabe T (1999) Salt tolerance of transgenic rice overexpressing yeast mitochondrial Mn-SOD in chloroplasts. Plant Sci 148:131–138. https://doi.org/10.1016/S0168-9452(99)00133-8
Tantau H, Dörffling K (1991) In vitro-selection of hydroxyproline-resistant cell lines of wheat (Triticum aestivum): accumulation of proline, decrease in osmotic potential, and increase in frost tolerance. Physiol Plant 82:243–248. https://doi.org/10.1111/j.1399-3054.1991.tb00088.x
Tian ZJ, Li JP, Jia XY, Yang F, Wang ZC (2016) Assimilation and translocation of dry matter and phosphorus in rice genotypes affected by salt-alkaline stress. Sustainability 8:568. https://doi.org/10.3390/su8060568
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2:135–138. https://doi.org/10.4161/psb.2.3.4156
Vanterpool TC, Truscott JHL (2011) Studies on browning root rot of cereals: II.Some parasitic species of Pythium and their relation to the disease. Can J Res 6:68–93. https://doi.org/10.1139/cjr32-005
Veljovic-Jovanovic S, Kukavica B, Stevanovic B, Navari-Izzo F (2006) Senescence- and drought-related changes in peroxidase and superoxide dismutase isoforms in leaves of Ramonda serbica. J Exp Bot 57:1759–1768. https://doi.org/10.1093/jxb/erl007
Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M, Sharma S (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci 8:161. https://doi.org/10.3389/fpls.2017.00161
Wang L, Seki K, Miyazaki T, Ishihama Y (2009) The causes of soil alkalinization in the Songnen plain of Northeast China. Paddy Water Environ 7:259–270. https://doi.org/10.1007/s10333-009-0166-x
Wang GJ, Miao W, Wang JY, Ma DR, Li JQ, Chen WF (2013) Effects of exogenous abscisic acid on antioxidant aystem in weedy and cultivated rice with different chilling sensitivity under chilling stress. J Agron Crop Sci 199:200–208. https://doi.org/10.1111/jac.12004
Wei LX, Lv BS, Wang MM, Ma HY, Yang HY, Liu XL, Jiang CJ, Liang ZW (2015) Priming effect of abscisic acid on alkaline stress tolerance in rice (Oryza sativa L.) seedlings. Plant Physiol Biochem 90:50–57. https://doi.org/10.1016/j.plaphy.2015.03.002
Wei LX, Lv BS, Li XW, Wang MM, Ma HY, Yang HY, Yang RF, Piao ZZ, Wang ZH, Lou JH, Jiang CJ, Liang ZW (2017) Priming of rice (Oryza sativa L.) seedlings with abscisic acid enhances seedling survival, plant growth, and grain yield in saline-alkaline paddy fields. Field Crop Res 203:86–93. https://doi.org/10.1016/j.fcr.2016.12.024
Weis C, Pfeilmeier S, Glawischnig E, Isono E, Pachl F, Hahne H, Kuster B, Eichmann R, Hückelhoven R (2013) Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions. Mol Plant Pathol 14:791–802. https://doi.org/10.1111/mpp.12050
Willems P, Mhamdi A, Stael S, Storme V, Kerchev P, Noctor G, Gevaert K, Breusegem FV (2016) The ROS wheel: refining ROS transcriptional footprints. Plant Physiol 171:1720–1733. https://doi.org/10.1104/pp.16.00420
Yang F, Liang ZW, Wang ZC (2011) Breeding and cultivation techniques of new rice variety of Dongdao 4. Crops 2:111. https://doi.org/10.16035/j.Issn.1001-7283.2011.02.027
Yu SX, Feng QN, Xie HT, Li S, Zhang Y (2017) Reactive oxygen species mediate tapetal programmed cell death in tobacco and tomato. BMC Plant Biol 17:76. https://doi.org/10.1186/s12870-017-1025-3
Zhang H, Liu XL, Zhang RX, Yuan HY, Wang MM, Yang HY, Ma HY, Liu D, Jiang CJ, Liang ZW (2017) Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front plant Sci 8:1580. https://doi.org/10.3389/fpls.2017.01580
Zhao FY, Zhang H (2006) Salt and paraquat stress tolerance results from co-expression of the Suaeda salsa glutathione S-transferase and catalase in transgenic rice. Plant Cell Tissue Organ Cult 86:349–358. https://doi.org/10.1007/s11240-006-9133-z
Acknowledgements
This work is supported by National Key Research and Development Program of China (SQ2018YFD020224); Chinese Academy of Sciences STS network Foundation (No. KFJ-SW-STS-141-01); the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA080X0X0X); the foundation of Innovation team International Partner Program of Chinese Academy of Sciences (No. KZZD-EW-TZ-07-08).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Wieland Fricke.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 146 kb)
Rights and permissions
About this article
Cite this article
Liu, XL., Zhang, H., Jin, YY. et al. Abscisic acid primes rice seedlings for enhanced tolerance to alkaline stress by upregulating antioxidant defense and stress tolerance-related genes. Plant Soil 438, 39–55 (2019). https://doi.org/10.1007/s11104-019-03992-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-03992-4