Abstract
Background and aims
Plant-soil feedback may vary across host species and environmental gradients. The relative importance of these biotic versus abiotic drivers of feedback will determine the stability of plant and microbial communities across environments. If plant hosts are the main driver of soil microbial communities, plant-soil feedback may be stable across changing environments. However, if microbial communities vary with environmental gradients, feedback may also vary, limiting its capacity to predict plant distributions.
Methods
We characterized arbuscular mycorrhizal (AM) fungi across tree plantations and a primary Neotropical rainforest. We then performed a plant-soil feedback pot experiment of AM fungi from these plantations on three plant species and related feedback and AM fungal communities in the field.
Results
In the field, temporal and spatial variation in AM fungal composition was similar in magnitude to variation across plant host species. Composition of AM fungi in the pot experiment significantly differed from the field plots. Furthermore, differential feedback was explained by shifts in AM fungal composition only for one plant host species (Hyeronima alchorneoides) in the pot experiment.
Conclusions
Natural AM fungal communities were temporally and spatially heterogeneous and AM fungal communities in the greenhouse did not reflect natural soils. These factors led to heterogeneous and unpredictable feedback responses, which suggests that applying greenhouse derived plant-soil feedback trends to predict plant coexistence in natural systems may be misleading.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plants can cultivate soil abiotic and biotic conditions that in turn affect subsequent plant performance, termed plant-soil feedback (Bever et al. 1997). Despite the prominent role of microorganisms in plant-soil feedback, the drivers of microbial distributions are rarely connected to their feedback effects. However, understanding plant-soil feedback in the broader context of other factors is key to understanding plant and microbial community trajectories in natural ecosystems. Plant-soil feedback depends on both the host and soil microbial community (Bennett et al. 2017; Teste et al. 2017); therefore when microbial distributions are controlled by abiotic factors (e.g., Chagnon et al. 2013; Coince et al. 2014) more so than by host species (e.g., Klironomos 2002), we might expect feedback to become environmentally context-dependent. This is especially relevant when considering environmental change where plant community trajectories may depend on the resulting shifts in plant-microbe interactions (van der Putten et al. 2013; Classen et al. 2015; van der Putten et al. 2016).
Plant-soil feedback can be the result of relationships with both positive (e.g., mutualists) and negative (e.g., pathogens) microorganisms. Indeed, in a meta-analysis of plant-soil feedback, mostly conducted in temperate ecosystems, the majority of plant-soil feedback was negative (Kulmatiski et al. 2008). Despite the dearth of plant-soil feedback studies from tropical ecosystems, feedback dynamics are likely also important at low latitudes where benign environments create situations where negative biotic interactions are expected to regulate communities more than abiotic filtering (Janzen 1970). Recent work in Panamanian tropical forests supports a role for feedback in these hyper-diverse plant communities: plant-soil feedback was highly positively correlated with the abundance of adult trees (Mangan et al. 2010b). This trend was driven by varying degrees of negative interactions with soil microbiota and in particular arbuscular mycorrhizal (AM) fungi, which can differentially affect the growth of conspecific and heterospecific host plants (Mangan et al. 2010a). Because AM fungal symbiosis with plants can range from parasitism to mutualism (Johnson et al. 1997) and the majority of tropical forest plant species associate with AM-fungi (Averill et al. 2014), AM-fungi can be a causal driver of both positive and negative plant-soil feedback. Therefore, these results are consistent with AM fungi structuring tropical rainforest plant communities. However, the generality of these findings remains unknown. Understanding how feedback relates to the biotic and abiotic drivers of AM fungal distributions can potentially help us extend these findings across the landscape and through time. For instance, in temperate communities, soil resources, particularly phosphorus, are an important driver of AM fungal composition (Johnson 2010) and thus knowing the relationship between phosphorus, AM fungi, and host-AM feedback could help predict low and high resource locations where positive or negative feedback may be more likely (Revillini et al. 2016). In the tropics, however, there is substantial variation in how AM fungi respond to resources (e.g., Treseder and Vitousek 2001; Waring et al. 2016; Schappe et al. 2017) and other abiotic factors (e.g., Guadarrama and Alvarez-Sanchez 1999) and thus drivers of tropical AM fungal communities remain an area of active study.
Predicting plant-AM fungal interactions in the context of environmental and host variation remains challenging. Plant-soil feedback outcomes varied between greenhouse and field experiments based on a meta-analysis of 38 studies (Lekberg et al. 2018), but only four of the studies were done in the field, making it difficult to parse causal mechanisms. In large part this may be due to the frequent disconnect between the greenhouse conditions under which plant-soil feedback experiments are performed and the field conditions where feedback occurs. Greenhouse experiments, by necessity, rarely reflect environmental variation in climate or resources. Furthermore, AM fungal communities in the greenhouse may be a nested subset of true field diversity composed of disturbance-tolerant taxa (Mariadassou et al. 2015) or generalist taxa that thrive in a broad variety of habitats (Barberan et al. 2014). However, the likelihood of greenhouse environmental filtering of AM fungal taxa is unknown because AM fungal composition in greenhouse-based plant-soil feedback studies has never been compared to that of natural environments. When greenhouse experiments either accurately reflect field conditions or when plant-soil feedback is based solely on host species associations that are temporally consistent, plant-soil feedback may predict plant relative abundance in the field (Klironomos 2002; Mangan et al. 2010b). However, feedback to plant seedlings in the greenhouse may not predict plant abundance in field settings when for example, feedback changes over time (e.g., Kardol et al. 2006; Hawkes et al. 2013) or depends on fluctuating resources (Revillini et al. 2016; Van Nuland et al. 2017) or climate (Ren et al. 2015). Another possibility is that host-specific effects on AM fungi are diluted non-additively by diverse plant communities (Kivlin and Hawkes 2011), making it difficult to compare greenhouse experiments to real-world ecosystems with high plant diversity. A greater understanding of what factors control AM fungal distributions in the field and how these fungi relate to plant-soil feedback in the greenhouse will provide a framework for when feedback can influence natural ecosystems.
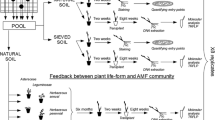
Here we addressed the biotic and abiotic drivers of AM fungal distributions, their feedback to plants in a Neotropical rainforest, and how the spatial and temporal heterogeneity of AM fungal communities relate to the observed feedback. Given the plant host-specific associations and high spatial and temporal variation of soil-borne saprotrophic fungi (Kivlin and Hawkes 2016a) and bacteria (Kivlin and Hawkes 2016b) at this experimental site, we expected that vegetation type and spatial and temporal heterogeneity would influence AM fungal composition in the field. We hypothesized strong and consistent plant-soil feedback if AM fungal composition in the field plots was largely determined by vegetation type, but weaker or inconsistent feedback if AM fungal composition was heterogeneous across space or time. Finally, we predicted different AM fungal composition and richness in the field versus pot samples given their different environmental conditions and the likelihood that not all AM taxa found in the field would be collected as spores or survive in pots.
Materials and methods
Field experiment
All studies were conducted at La Selva Biological Station, Costa Rica (10°25′53.14”N, 84°0′10.51”W). Mean annual temperature at this site is 25.8 °C while mean annual precipitation is 4142 mm (Sanford Jr. et al. 1994; Clark et al. 2010). Soils and AM fungal communities were collected from monodominant stands of three tree species Hyeronima alchorneoides Allemao (Phyllantaceae), Pentaclethra macroloba (Willd.) Ktze (Fabaceae), and Virola koschnyi Warb. (Myristicaceae), and a mature, primary forest within 150 m. We will refer to the experimental tree plots and primary forests collectively as “vegetation types.” The location of the monodominant stands was slash-burned and planted with Panicum maxicum L. and Melinis minutifolia Pal. in 1955 and grazed until 1988 when the experiment was established (Fisher 1995). The three tree species were placed in a randomized block design with four blocks and primary forest samples were similarly acquired from four random blocks across the same distance. All blocks and primary forest plots are within 800 m of each other (see Kivlin and Hawkes 2016b for details). Plots within blocks are 0.25 ha in size and were planted with individual trees spaced 3 m apart (Fisher 1995). Soils are derived from andesitic lava flows (Alvarado 1990) and are classified as Mixed Haplic Haploperox (Kleber et al. 2007). Soils are generally acidic (~4 pH) and contain low levels of inorganic phosphorus, base cations, and nitrogen, but relatively high levels of soil organic matter (Russell et al. 2007); however, some of these properties also vary among the experimental tree species (Russell et al. 2010).
To examine AM fungi in the field, we collected soils and roots from five (2.5 cm wide × 10 cm deep) cores in September 2012, February 2013, and September 2013. We used a stratified random sampling design to avoid coring the same exact location over time. The five cores were combined in each plot and within 24 h of collection, roots were separated from soils and frozen at −20 °C for later analysis of field AM fungal communities. Soils were processed for biogeochemical pools as described below.
Feedback pot experiment
For the feedback experiment, we leveraged the vegetation types from the field as a 24-yr conditioning phase to serve as sources of both the AM fungi and soils. Varying different AM inocula addressed how host-specific AM fungal communities affect plant-soil feedback, whereas varying soil sources allowed us to examine the abiotic drivers of plant-soil feedback. The full design included three tree species grown with five AM fungal communities (three tree species, primary forests, and sterile) on three soil origins (from the three tree species) in a fully factorial design. With four replicates per treatment, there were a total of 180 pots. Seeds were collected from the canopy when ripe, stored dry for up to 2 months, and surface sterilized with 1% sodium hypochlorite prior to use. At the same time as the seeds matured, we collected soils from 0 to 15 cm depth separately from each vegetation type in the field. For each soil source, soils were combined across all four blocks of each monodominant tree species and across the four plots in the nearby primary forest plots.
To create the AM fungal treatment inoculum, AM fungal spores were isolated from 1 kg of fresh soil from each plot via sucrose centrifugation (Allen et al. 1979). The supernatant was sieved through a 45-μm sieve to isolate AM fungal spores and discard other saprotrophic fungi and bacteria. Spores isolated from the same vegetation type were combined across blocks for the final AM fungal inoculum treatments. The remaining soils were used to create the soil treatment: soils were combined by vegetation type, sieved to 4 mm, and sterilized by autoclaving three times at 121 °C for 1 h over three consecutive days. To improve pot drainage, sand (accumulated in a nearby alluvial flat) was similarly sterilized and soil and sand were mixed 1:1. Pots were constructed as follows: (1) ~2.5 L of the sterile soil-sand mix was added to a 2.65 L deep pot (10.2 cm wide × 34.3 cm tall; Stuewe and Sons, Inc., Tangent, OR, USA), (2) spores of AM fungi were added at a uniform density of 1000 spores/pot in 2 ml of sterile water and sterile controls were given the same volume of sterile water, placed approximately 2 cm below the soil surface, (3) one surface-sterilized seed was added to the top of the pot and lightly covered. The pots were also modified with a 20 cm tall clear vinyl ring around the top to prevent cross-pot transfer of AM fungal spores via rain splash.
We focused on matching the AM fungi and soil treatments to seed maturation dates in order to create treatments that seeds would experience. Thus, the timing of each experiment depended on seed maturation: Pentaclethra trials began in August 2012 and Hyeronima and Virola trials began in June 2013. Soil and AM fungal inoculum for each plant species were collected approximately 1 month before the feedback experiments started. All plants were allowed to grow for 6 months in a shadehouse at La Selva Biological station and watered as needed.
At the end of 6 months, we measured plant size as above- and belowground plant biomass. Fungal colonization of roots was quantified by microscopy for aseptate and septate hyphae, arbuscules, and vesicles; roots were cleared in 10% KOH, stained with acid fuchsin, and 100 fields of view were counted at 160× magnification (McGonigle et al. 1990). Extraradical hyphal lengths of both aseptate and septate fungi were determined by extracting hyphae from soil in a 5% sodium hexametaphosphate solution and visualizing under 160× magnification with a gridded reticle (Brundrett et al. 1994). Soils were also analyzed for biogeochemical pools as described below. Roots (~0.75 g) were frozen at −20 °C within 24 h for later examination of AM fungal community composition.
Biogeochemical methods
For both field and pot soils, we extracted ammonium, nitrate, and labile dissolved organic carbon (DOC) in 0.5 M K2SO4 using a 1:5 ratio of soil to extractant (Jones and Willett 2006; Makarov et al. 2013). Soluble inorganic phosphorus was extracted by shaking ~10 g of soil with anion resin strips (Membranes International, Ringwood NJ) in 100 ml of water and subsequently extracting the strips in 0.5 M HCl (Lajtha et al. 1999). Ammonium, nitrate, and phosphate concentrations were quantified colorimetrically (D’Angelo et al. 2001; Doane and Horwath 2003). In addition, we measured microbial biomass carbon (MBC) using chloroform fumigation and direct extraction in 0.5 M K2SO4 (Scott-Denton et al. 2006). Microbial biomass carbon was quantified by combustion (Apollo 9000 TOC Analyzer, Teledyne Tekmar) and calculated as the difference between the fumigated and unfumigated extractions adjusted for an extraction efficiency of 0.45 (Brookes et al. 1985). Soil moisture was measured for each soil sample by drying soils at 105 °C to constant weight. A 2-g subsample of soils was frozen at −20 °C for measurement of extracellular phosphatase activity as a metric of AM fungal function; enzyme activity was quantified fluorometrically following Allison et al. (2007). Statistical variation in soil biogeochemical parameters in field soils are presented in Table S1.
AM fungal community characterization
From the field vegetation types, DNA was extracted from adult plant roots picked from all soil cores and identified morphologically as belonging to the dominant tree species at three dates (September 2012, February 2013, and September 2013) (4 vegetation types × 4 blocks × 3 dates = 48 samples). From the feedback pot experiment, DNA was extracted from roots of each host species from a subset of replicates: we focused on the experimental treatments where each host plant was paired with AM fungi from conspecific tree stands and the mature forest (3 plant hosts × 2 AM fungal communities/host × 3 soil types × 4 replicates = 72 samples). We chose these treatments to compare the drivers of AM fungal composition under adult trees in the experimental field plots and seedlings in the shadehouse pots.
Root DNA was extracted from two 0.25 g subsamples per replicate with MoBio PowerPlant Pro DNA extraction kits (MoBio, Carlsbad, CA, USA) and quantified fluorometrically (Qubit Fluorometer, Life Technologies, Carlsbad, CA, USA). Duplicate extractions were combined and standardized to ~10 ng μl−1. A ~350 b region of 28S AM fungal DNA was amplified with primers that consisted of Illumina TruSeq V3 indices (Illumina, San Diego, CA, USA) ligated to 454 barcodes (Roche, Basel, Switzerland) linked to AM-fungal specific FLR3-FLR4 primers (Gollotte et al. 2004). Each reaction contained: 21.5 μl of Platinum PCR Supermix (Invitrogen, Carlsbad, CA, USA), 1.25 μl of each primer (10 μM), 0.5 μl of BSA (20 mg ml−1), and 2 μl (~20 ng) of DNA. PCR reactions were run in triplicate with an initial denaturing step of 93 °C for 5 min, 35 cycles of 93 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, followed by a final extension step of 72 °C for 10 min. Triplicate reactions were combined, cleaned with Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA), and quantitated fluorometrically (Qubit Fluorometer, Life Technologies, Carlsbad, CA, USA). Samples were then pooled in equal amounts into eight libraries (four for the field experiment and four from the pot experiment) and sequenced as 2 × 300 b reads to an average depth of 25,000 reads/sample on 1/8th of a flowcell of an Illumina MiSeq v3 sequencer at the University of Texas Genome Sequencing and Analysis Facility.
Contigs of forward and reverse sequences (478,633 in total) were created using the mothur v. 1.33.3 pipeline (Schloss et al. 2009). All sequences were quality filtered using the default settings in the QIIME pipeline v. 1.8.0–20,140,103 (Caporaso et al. 2010) and discarded if they were less than 300 bases in length, had more than six ambiguous bases, contained any ambiguous bases in the barcode, or were chimeric based on UCHIME with default parameters (Edgar et al. 2011). This excluded 48% of the run leaving 231,600 sequences. Sequences were clustered into operational taxonomic units (OTUs) at a 97% similarity cutoff using UCLUST with default parameters (Edgar 2010). OTUs that only occurred once in the entire dataset and OTUs that only occurred in one sample were discarded as probable sequencing artifacts (Dickie 2010). This removed ~30% of the OTUs, leaving 680. The remaining OTUs were taxonomically classified using the Ribosomal Database Project (RDP) classifier against the RDP 28S fungal database. Non-AM fungal OTUs were discarded when they did not match known Glomeromycotina with 97% or greater confidence, leaving 649 full-length AM fungal OTUs. This method may have excluded some novel tropical AM fungi.
We then used the DeSeq2 algorithm (Love et al. 2014) to transform the relative abundance of each OTU. This algorithm provides the most sensitivity to detect treatment differences across a range of sample sizes (McMurdie and Holmes 2014). An alignment and maximum likelihood phylogeny were created with one representative sequence from each of the 649 AM fungal OTUs using the Practical Alignment using SATé and TrAnsitivity (PASTA) algorithm (Mirarab et al. 2015) with default settings. To ensure that OTU assignments were accurate, we re-defined OTUs using this alignment, which collapsed 7 OTUs. Sequences were deposited in the NCBI Sequence Read Archive (SRA) under accession number (SUB1665085).
Statistics
Composition of AM fungi in experimental field plots
AM fungal diversity and composition were assessed both phylogenetically and taxonomically. We first calculated alpha diversity for each sample as richness based on the number of unique OTUs that occurred in each replicate. We calculated Faith’s PD of AM fungal communities in each replicate as the sum of branch lengths connecting all AM fungal species in an ultrametric phylogeny using the Picante package (v. 1.6–2, Kembel et al. 2010) in R (v. 3.1.1, R Development Core Team 2009). Community-wide phylogenetic signals were calculated as both mean pairwise distance (MPD) and mean nearest taxon distance (MNTD) using the Picante package. MPD assesses phylogenetic structure at the base of the phylogeny, whereas MNTD focuses on phylogenetic relatedness near the tips of the phylogeny.
To understand how vegetation type, date, block, and soil abiotic factors affected AM fungal community composition in adult trees in the field, we performed a PERMANOVA with vegetation type, block, and date as well as the two-way interactions of vegetation type with block and date. We also included continuous abiotic soil variables (ammonium, nitrate, phosphorus, DOC, and moisture) in the model. PERMANOVA was run using the Adonis function in the vegan package in R (R Core Team 2009). To obtain Type III sums of squares, rather than the default Type I, we repeated ran the full model with each variable added last. This did not change the model outcome compared to the typical Adonis output with Type I sums of squares (data not presented). We visualized the shifts in AM fungal community composition among vegetation types using non-metric multidimensional scaling (NMS) ordination in the metaMDS function in the vegan package in R (R Core Team 2009). Axis 1 of this ordination explained 51% of the variance in AM fungal composition and axis 2 explained 29% of the variance in AM fungal composition. The mean scores for each vegetation type from each of the NMS axes were used to represent AM fungal composition in the initial inoculum for the pot experiment. All community composition statistics were run in the vegan package in R (Oksanen et al. 2007).
Feedback pot experiment
The field-based vegetation types were used to represent the conditioning phase of the plant-soil feedback experiment. Therefore, for each AM fungal inoculum source and soil source treatment we calculated feedback to the plant as ln (conspecific treatment/heterospecific treatment), where conspecific AM fungi and soil source refer to growth of a focal plant species on AM fungal inoculum or soil from field plots where the tree species matched the pot focal plant and heterospecific refers to growth of a focal plant species on AM fungal inocula and soil from all other tree species that did not match, as well as the sterile control (Bever 2003; Brinkman et al. 2010). For each plant species, we calculated both an overall feedback effect of the combined feedback of all treatments. In addition, feedback was calculated separately for each inoculum (including sterile controls) and soil source to understand variation among conditioning plant species. We used total biomass of roots and shoots for each feedback calculation. For each plant species, student’s T test was used to examine if the overall plant-soil feedback effect was different from zero. For each plant species, we used two-way ANOVA to examine how feedback was affected by AM fungal community source, soil source, and their interactions. All factors were treated as fixed effects.
Drivers of feedback
To examine the contributions of abiotic and biotic mechanisms to overall observed feedback, we created three separate multiple regression models for each plant species including (1) all variables, (2) abiotic variables only, and (3) biotic variables only. The abiotic variables were soil moisture, ammonium, nitrate, phosphate, and DOC in all of the greenhouse pots. The biotic variables were microbial biomass carbon, phosphatase activity, intra-radical colonization by aseptate hyphae, septate hyphae, arbuscules, and vesicles, extra-radical aseptate and septate hyphal lengths in all of the greenhouse pots, and AM fungal community parameters from the field vegetation types: AM fungal species richness, AM fungal community composition, and AM fungal PD, MPD, and MNTD. The field AM fungal community composition was represented by the centroid for each vegetation type from NMS axes 1 and 2. NMS ordination centroids are non-metric, so correlations of plant-soil feedback with AM fungal composition should be interpreted as magnitude, agnostic of sign. The composition of AM fungi from the pot experiment was not included in these analyses as we only sequenced the AM fungal community from a subset of treatments. To meet assumptions of normality and homoscedasticity, phosphatase activities were log-transformed and fungal colonization rates were arcsine square root transformed for all multiple regression analyses.
For each species and all species together, separate models were created for plant total biomass, aboveground biomass, AM fungal feedback, and soil feedback. Regressions were run using the MASS package in R (R Core Team 2009) with model fit optimized using AIC selection. None of the biotic or abiotic factors co-varied by more than 75% when compared with Pearson correlations and thus all independent variables were retained in the analyses.
Composition of AM fungi in feedback pot experiment
We used PERMANOVA to analyze AM fungal community composition in the pot experiment as a function of plant species, AM fungal inoculum source, soil source, their interactions, and the continuous abiotic soil variables (ammonium, nitrate, phosphorus, labile carbon, and moisture). PERMANOVAs were performed in R as described for field AM fungi.
Comparing AM fungal composition in the field and feedback pot experiments
We compared the composition of AM fungal OTUs in the field and in pots with a PERMANOVA that included plant species, soil nutrients, and soil moisture. PERMANOVA was chosen because it is robust to the unbalanced design between the field and feedback pots. AM fungi were only sequenced from the three focal plant species in the greenhouse, whereas AM fungi in the field were sequenced from these three plant species as well as the primary forest. This design precluded Procrustes analysis which is more common in community composition concordance studies (Jackson 1995). In addition, we characterized the overall and genus-specific percent overlap of AM fungal taxa in field and pots for each plant species. To understand the contributions of generalist and specialist AM fungi from the field to the pot AM fungal communities, we correlated the proportion of vegetation types from which each of the AM fungal OTUs were isolated with their abundance in the feedback pot experiment using Pearson correlations with Bonferroni correction for multiple comparisons (Bonferroni 1936). We also determined if the AM fungal community in the pots was a nested subset of the field AM fungal community using two approaches to consider both abundance and presence of taxa: (1) we examined relative abundance data following Rodriguez-Girones and Santamaria (2006) using the nestedtemp function in Vegan in R (Oksanen et al. 2007), and (2) we compared the nestedness ranks of pot versus field AM fungal communities with a Mann-Whitney U test (Mann and Whitney 1947).
Results
AM fungi in experimental field plots
Soils conditioned by the vegetation types in the field contained different AM fungal communities, except Hyeronima and Virola (R2 = 0.145, P < 0.001; Fig. 1a, Tables S2, S3). There was also significant spatial variation in AM fungal composition both among blocks (R2 = 0.093, P = 0.023; Fig. 1b) and based on the block interaction with vegetation type (R2 = 0.182, P = 0.011). There was no significant effect of purely temporal variation in AM fungal community composition (R2 = 0.052, P = 0.152; Fig. 1c), but the effect of vegetation type varied over time (R2 = 0.160, P = 0.006). Abiotic soil parameters did not explain any additional variation in AM fungal composition (Table S2). AM fungal species richness, PD, MPD, and MNTD did not vary with vegetation type, block, date, or their interactions (P > 0.05).
Feedback in the pot experiment
Because we were able to sample mono-dominant vegetation types and a nearby primary forest, inoculum and soil source in our experiment represent the conditioning phase of a plant-soil feedback experiment. Each of the three plant species experienced different biomass feedback to the AM fungal inoculum sources compared to its own AM fungal community. Across all AM fungal inoculum sources, Hyeronima plants experienced positive feedback (Fig. 2a), Pentaclethra neutral feedback (Fig. 2b) and Virola negative feedback (Fig. 2c). When broken down by AM fungal inoculum source, Hyeronima had positive feedback when growth on conspecific AM fungi was compared to growth with AM fungi from Pentaclethra (T = 5.968, P < 0.001), primary forests (T = 15.568, P < 0.001), and in sterile controls (T = 17.271, P < 0.001), but neutral feedback when compared to growth on AM fungi from Virola (P > 0.05, Fig. 2). Pentaclethra experienced negative feedback when growth on conspecific AM fungal inoculum was compared to growth on primary forest AM fungi (T = 2.814, P = 0.009) and neutral feedback when growth was compared on all other AM fungal treatments (P > 0.05; Fig. 2). Virola had negative feedback when grown on its own versus Pentaclethra AM fungi (T = 3.894, P < 0.001), but neutral feedback when growth on its own AM fungi was compared to growth on Hyeronima AM fungi, primary forest AM fungi, and sterile controls (P > 0.05; Fig. 2).
Feedback of experimental pots to AM fungal inoculum for (a) Hyeronima, (b) Pentaclethra, and (c) Virola plants. Gray bars represent the overall feedback of each plant species. Black bars represent average feedback for each treatment (± 1 SE; n = 16). Letters indicate treatments that were significantly different in Sidak posthoc comparisons. Asterisks designate treatments in which feedback was significantly different from zero. Note the 10× difference in scale for Hyeronima feedback
The three plant species also experienced differential biomass feedback to soils originating from the different vegetation types in the field. When all soil sources were considered together, Hyeronima (Fig. 3a) and Virola (Fig. 3c) plants experienced positive feedback when growth on conspecific soils was compared to that on heterospecific soils, whereas Pentaclethra plants experienced negative feedback in the same comparison (Fig. 3b). When broken down by soil sources, Hyeronima plants had positive feedback when growth on its soils was compared to growth on soils conditioned by all other plant species (Pentaclethra: T = 4.192, P < 0.001; Virola: T = 3.915, P < 0.001; Fig. 3). Pentaclethra plants had negative feedback when growth on its soil was compared to growth on soil conditioned by Hyeronima (T = 3.021, P = 0.004) and neutral feedback compared to growth on soil conditioned by Virola (P < 0.05; Fig. 3). Finally, Virola experienced neutral feedback when growth on its own soil was compared to growth on all soil treatments (P > 0.05; Fig. 3). There were no interactions of AM fungal inoculum source and soil source for biomass feedback in Hyeronima (F = 1.087, P = 0.390; Table S4), Pentaclethra (F = 0.422, P = 0.889), or Virola (F = 0.680, P = 0.706) plants. Consistent with the feedback response, the total biomass of Hyeronima plants was only affected by AM fungal inoculation source (F = 9.665, P < 0.001). Biomass of Pentaclethra and Virola were not affected by AM fungal inoculation source or soil source (P > 0.05; Tables S4, S5).
Feedback of experimental pots to soil source for (a) Hyeronima, (b) Pentaclethra, and (c) Virola plants. Gray bars represent the overall feedback of each plant species. Black bars represent average feedback for each treatment (± 1 SE; n = 24). Asterisks designate treatments that were significantly different from zero. Note the 10× difference in scale for Hyeronima feedback
Drivers of plant-soil feedback
Based on multiple regression models, the relationship between feedback and abiotic vs. biotic factors differed for each of the three plant species. The best-fit model for Hyeronima feedback to AM fungal inoculum source included only biotic factors: feedback was positively associated with AM fungal species richness in the field vegetation types and negatively associated with both AM fungal phylogenetic diversity in the field vegetation types and non-AM fungal hyphal colonization in the pots. Both Pentaclethra and Virola feedback to AM fungal inoculum source were best explained by abiotic factors alone, based only on positive associations with soil moisture in the pots. All plant species feedback to soil source was driven by the same variables as feedback to AM fungal inoculum source. However, regression models only explained between 15 and 39% of the observed variation in feedback to AM fungal and soil sources (Table 1). The drivers of plant biomass were similar to those for feedback for all three plant species, except the biomass of Hyeronima plants was also negatively associated with soil phosphatase activities in the pots (Table S6).
AM fungi in the pot experiment
The three plant species in the pot experiment were associated with different AM fungal communities (R2 = 0.060 P < 0.001, Fig. 4a; Tables S7, S8). AM fungal community composition in the pots also varied by AM inoculation source (R2 = 0.069, P < 0.001; Fig. 4b). There were no significant main or interaction effects of soil origin treatment on AM fungal communities in the pots (Fig. 4c; Table S6). However, AM fungal composition varied by AM inoculation sources differently across plant hosts (Table S7). Root colonization of AM fungi varied from ~4–31% depending on plant host, but did not vary among inoculated treatments. AM fungi were observed at low rates in sterile controls; abundance was consistently ~10 times lower compared to treatments inoculated with AM fungi (Table S8).
Comparing AM fungi in experimental field plots and pot experiment
The composition of AM fungi in the pots was weakly associated with AM fungal composition in conspecific field plots in the PERMANOVA analysis (R2 = 0.100, P < 0.001; Fig. 5a). At the individual pot level, the pot AM fungal communities did not capture the AM fungal species richness of roots in the field (Hyeronima pots 43 ± 9%; Pentaclethra pots 25 ± 5%; Virola pots 48 ± 6% of field richness). This was also seen across treatments as only 24% of Pentaclethra AM fungi and only 44% of both Hyeronima and Virola AM fungi overlapped between the field vegetation types and the pot experiment (Fig. 5b). When the fungi were partitioned by observed genera, the experimental field vegetation plots and pots shared 3–8% of Scutellospora, 14–15% of Rhizophagus, and 6–15% Glomus AM fungal taxa, with the proportions of shared taxa in each genus varying slightly by vegetation type. There was also one Acaulospora AM fungal taxon found in the field vegetation types, but never in the pots. The overall abundance of AM fungi in the feedback pot experiment was positively, but weakly correlated with their prevalence among vegetation types in the field (R2 = 0.121, P < 0.001, Fig. 5c) and this was true regardless of plant host in the pot (Table S9). While the entire AM fungal community in the study was nested (T = 6.818, P < 0.001), the AM fungal community in the pots was not a nested subset of the AM fungal community in the field for any plant species (P > 0.05; Fig. 5d).
a AM fungal community composition in field tree roots and in roots of feedback pots (lighter shaded symbols) plotted as average NMS scores (± 1 SE). b Overlap of AM fungal OTUs in the pot versus field samples for each host species visualized in Venn diagrams. c The abundance of AM fungal taxa in the feedback pot experiment as a function of the proportion of vegetation types colonized in the field. d Nestedness plot of AM fungal OTUs that occurred in each sample (black rectangles). The curve (black line) indicates the isocline of perfect nestedness. Blue columns are AM fungal communities from the field vegetation types and yellow columns are AM fungal communities from the feedback pot experiment. Orange rows noted by black hashes on the left side of the panel indicate generalist AM fungal OTUs that occurred in all five field vegetation types
When all plant hosts were considered simultaneously, the main effects of plant host and soil nutrients explained some of the variation in AM fungal composition in the field (R2host = 0.145; R2soils = 0.040) and in the pots (R2host = 0.060; R2soils = 0.035). However, AM fungal communities on individual plant species in the pot experiment varied with respect to host-specific AM fungal inoculum isolated from the field. The composition of AM fungi colonizing Pentaclethra (R2 = 0.055, P = 0.029) and Virola (R2 = 0.137, P < 0.001) plants varied among AM fungal inoculation sources, but AM fungi colonizing Hyeronima plants did not (R2 = 0.033, P = 0.512).
Discussion
In the field, spatial and temporal heterogeneity of AM fungal communities among plots was on par with differences among plant hosts, and interactions between them were dominant. Evidence from this study site (Kivlin and Hawkes 2016a, b) and other tropical rainforests (Waring et al. 2016) suggests that microbial communities are highly spatially variable in tropical forests. If so, then relating plant-soil feedback to plant abundance in the tropics (Mangan et al. 2010b) may overestimate the role of biotic interactions in predicting plant coexistence in these ecosystems. Moreover, in a recent survey of AM fungi from the same forest plots as the Mangan et al. (2010b) study, soil phosphorus, and not plant host, was the main driver of AM fungal composition (Schappe et al. 2017), suggesting that plant-soil feedback may be spatially variable or not rely on AM fungi in tropical ecosystems. In the current study, heterogeneity of AM fungal composition may explain why feedback based on field conditioning of AM fungal inocula and soils was affected by AM fungal inoculum source in only one plant species (Hyeronima) in the greenhouse plant-soil feedback experiment.
The degree of mycorrhizal dependence likely varies among tropical forest plant hosts (Janos 1980). We found that Hyeronima plant biomass was highly correlated with shifts in AM fungal diversity and non-AM fungal abundance and for the most part experienced positive feedback; Hyeronima plants performed better on their own versus foreign inoculum. Hyeronima plants also experienced the largest shifts in plant-soil feedback among inoculation sources. Conversely, the other two plants in our experiment, Pentaclethra and Virola, responded more to abiotic shifts in soil moisture and had feedbacks that were 1/10th in magnitude compared to Hyeronima. Recent evidence from surveys of AM fungi in Puerto Rico supports a changing functional role of AM fungi as forests age, with AM fungi negatively affecting early successional trees, but positively affecting late successional (Bachelot et al. 2018) and rare tree species (Bachelot et al. 2017). Here, the rarest species in our study, Hyeronima, a tree-fall gap colonizing species, may rely on AM fungal networks to successfully establish and acquire limiting soil nutrients, but then quickly be out-competed or accumulate pathogens as trees age. Conversely, Pentaclethra and Virola are later successional species and therefore may benefit from a wide range of AM fungi or be more able to withstand soil pathogens (Bagchi et al. 2014). In addition, Pentaclethra is a nitrogen-fixer and thus may rely on these symbionts more than AM fungi (Nasto et al. 2014), although we did not observe any nodules in our experiment.
The AM fungi in our field vegetation plots varied among vegetation types, with significant spatial and temporal heterogeneity. Across an approximately 400 × 800 m area of tropical rainforest, spatial and temporal variation in AM fungal composition was similar in magnitude to other soil microbial guilds (Kivlin and Hawkes 2016a, b), although with different patterns among vegetation types. This suggests that underlying variation in soil nutrients (Vandecar et al. 2011; Waring et al. 2016; Schappe et al. 2017), topography (Powers et al. 1997), and understory vegetation (Slik et al. 2013) in tropical forests influence a range of microbial taxa, in varying ways. Moreover, the temporal variation of AM fungal communities among vegetation types was substantial, despite the aseasonality of rainfall in this region. While plant community composition is correlated with belowground mycorrhizal communities in other tropical forests (Peay et al. 2013), fungi can also respond to temporal variation in precipitation, even across a plant diversity gradient (McGuire et al. 2012), highlighting the lack of generality in soil microbial drivers identified within and among tropical ecosystems.
Given the high variability of AM fungi in the field plots, it is not surprising that plant-soil feedback observed in response to AM fungal inoculum in the feedback pot experiment was also variable. Recent studies have indicated that plant-soil feedback in the greenhouse is not consistent with plant-soil interactions in the field and therefore cannot explain patterns of coexistence (Lankau and Lankau 2014; Stanescu and Maherali 2017). Several factors could explain these findings. Plant coexistence may not rely on soil microorganisms if environmental filtering (Lavorel and Garnier 2002) or plant-plant competition (Tilman 1994) structures communities, and this may especially be true in forest ecosystems with long-lived tree species. Even if AM fungi do affect plant coexistence, when AM fungal communities differ with plant ontogeny (e.g., Husband et al. 2002a, b), feedback experiments of seedling and juvenile species in pots may never match the feedback that adult plants experience in natural ecosystems (Hawkes et al. 2013; Kardol et al. 2013). For example, in the only studies to track variation in AM fungi across plants over time in tropical forests, the abundance of individual AM fungal taxa varied 8-fold after 1 year (Husband et al. 2002a) and up to 55-fold over 5 years (Husband et al. 2002a). Therefore, it is likely that the current study and all other greenhouse-based plant-soil feedback studies also exhibit some degree of discordance owing to different plant life stages. In addition, differences in AM fungal composition in plant-soil feedback pots compared to field communities may be one mechanism contributing to the lack of transferability of feedback patterns, especially when plants are highly responsive to AM fungi. These differences may arise because AM fungal inoculum in greenhouse experiments is often added as spores, not a mycorrhizal network (e.g., the current study, Mangan et al. 2010a), AM fungi are added in isolation without other animal and microbial taxa that may modify their composition (Klironomos and Ursic 1998), or the fact that plant-soil feedback experiments usually only grow one plant host in isolation, which can affect AM fungal communities (Burrows and Pfleger 2002; Kivlin and Hawkes 2011) and create misleading coexistence predictions when plant-soil feedback is non-additive (Hawkes et al. 2013).
Perhaps because of these inherent differences in greenhouse versus field conditions, greenhouse roots contained vastly different, and in some cases more diverse AM fungal communities compared to field-collected analogs. However, despite the broad differences in AM fungal communities between field and pot experiments, AM fungi that were more widespread in the field vegetation types were also more abundant in the pots, suggesting that some generalists were at an advantage. At the global scale, some AM fungi tend to have widespread distributions across climates and plant communities (Kivlin et al. 2011; Davison et al. 2015), suggesting that this long-lived lineage can be environmentally plastic. However, local adaptation with respect to soil nutrients (Johnson et al. 2010) and climate (Antunes et al. 2011) supports the existence of many specialist, low abundance AM fungal taxa, which were the majority of AM taxa in the current experiment. Greenhouse plant-soil feedback experiments may have larger transferability to environmental plant-soil feedback when AM fungi drive feedback outcomes in a plant-specific manner, as in Mangan et al. (2010a, b). However, in more diverse and heterogeneous AM fungal communities, environmental interactions with specialist, low abundance AM fungal species may moderate plant-soil feedback. Such context-dependent feedback will be difficult to predict when AM fungal community composition differs along environmental gradients in underlying resources and climate. Current efforts to describe large-scale surveys of conditions favoring local adaptation of AM fungal taxa (e.g., Rua et al. 2016), and their relative effects on plant-soil feedback along environmental gradients (e.g., Smith-Ramesh and Reynolds 2017) are a strong way forward to creating an inclusive and generalizable framework of plant-soil feedback.
Our study has several caveats that could preclude broad applicability of these patterns to other tropical forests. First, our experimental design only allowed us to address plant-soil feedback in three species. While feedback varied among the three plant hosts, it is difficult to extrapolate these findings to entire primary forests. Furthermore, many of the biotic and abiotic drivers of plant-soil feedback were weak and this experiment only captured the AM fungal component of feedback. Future studies disentangling mycorrhizal versus decomposer and pathogen-driven feedback in isolation and combination are necessary to elucidate the belowground portion of feedback dynamics (Klironomos 2002). In addition, the control pots in our experiment only received water, and not sterilized AM fungal spores, which may have reduced their nutrient content given that spores are rich in lipids (Beilby 1980; Beilby and Kidby 1980). However, because the pots used in this experiment were so large, the inoculum only added ~8% more spores than would have been found in the sterilized background soil. There was also a low level of AM fungal and non-AM fungal colonization in our control pots, most likely via airborne dispersal or rain splash within the relatively open shadehouse environment. Control pots consistently contained <1/10th of the AM fungal hyphal lengths in soil compared to inoculated treatments and exhibited minimal intraradical root colonization, whereas colonization rates and extraradical hyphal lengths of AM fungi in inoculated pots in the greenhouse mimicked those of tree roots and soils in the field (Kivlin and Hawkes 2016b, Kivlin and Hawkes, unpublished data). The low level of non-AM fungal colonization observed in the greenhouse may have affected the outcome of plant-soil feedback, but to a minor extent. Non-AM fungal colonization explained less than 10% of the variation in plant-soil feedback in Hyeronima plants and none of the variation in feedback in the other two plant species. Nevertheless, these caveats do not undermine our finding that high spatial and temporally heterogeneity of AM fungal communities within vegetation types in tropical forests constrains the potential generality of plant-soil feedback derived from pot experiments.
Conclusions
Our study provides evidence that AM fungal communities in a Neotropical forest can be spatially and temporally heterogeneous and this heterogeneity may be a reason that observed greenhouse plant-soil feedback is not consistently influenced by AM fungi. Instead, plant growth in natural systems may be a function of AM fungal communities as well as a diverse array of biotic and environmental conditions. More expansive, field-based plant-soil feedback studies that occur across spatial and temporal scales are necessary to determine when feedback is consistent within a plant host and when it is environmentally context-dependent. Characterizing when and where soil microbial communities are spatially and temporally variable across ecosystems is the first step to understanding context-dependent plant-soil feedback. However, to capture truly long-term trends, natural experiments, such as plant invasions or climate change-induced range expansions may provide the most tractable platform, especially when time since establishment is known, allowing a temporally explicit look at the buildup of plant-soil feedback. Ultimately, approaches that compare the strength of plant-soil feedback to other biotic and abiotic processes (e.g., competition or herbivory; van der Putten et al. 2016) will provide a more expansive and predictive framework for understanding the role of plant-soil feedback in structuring natural plant communities.
References
Allen MF, Moore TS, Christensen M, Stanton N (1979) Growth of vesicular-arbuscular mycorrhizal and nonmycorrhizal Bouteloua gracilis in a defined medium. Mycologia 71:666–669
Allison SD, Gartner TB, Holland K, Weintraub M, Sinsabaugh RL (2007) Soil enzymes, linking proteomics and ecological process. In: Manual of environmental microbiology, 3rd edn. ASM Press, Washington, D.C., pp 704–711
Alvarado G (1990) Caracteristicas geologicas de la Estacion Biologica La Selva, Costa Rica. Tecnologia en marcha 10:11–22
Antunes PM, Koch AM, Morton JB, Rillig MC, Klironomos JN (2011) Evidence for functional divergence in arbuscular mycorrhizal fungi from contrasting climatic origins. New Phytol 189:507–514
Averill C, Turner BL, Finzi AC (2014) Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 505:543–545
Bachelot B, Uriarte M, McGuire KL, Thompson J, Zimmerman J (2017) Arbuscular mycorrhizal fungal diversity and natural enemies promote coexistence of tropical tree species. Ecology 98:712–720
Bachelot B, Uriarte M, Muscarella R, Forero-Montana J, Thompson J, McGuire K, Zimmerman J, Swenson NG, Clark JS (2018) Associations among arbuscular mycorrhizal fungi and seedlings are predicted to change with tree successional status. Ecology 99:607–620
Bagchi R, Gallery RE, Gripenberg S, Gurr SJ, Narayan L, Addis CE, Freckleton RP, Lewis OT (2014) Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature 506:85–88
Barberan A, Ramirez KS, Leff JW, Bradford MA, Wall DH, Fierer N (2014) Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol Lett 17:794–802
Beilby JP (1980) Fatty acid and sterol composition of ungerminated spores of the vesicular-arbuscular mycorrhizal fungus Acaulospora laevis. Lipids 15:949–952
Beilby JP, Kidby DK (1980) Biochemistry of ungerminated and germinated spores of the vesicular-arbuscular mycorrhizal fungus, Glomus caledonius: changes in the neutral and polar lipids. J Lipid Res 21:739–750
Bennett JA, Maherali H, Reinhart KO, Lekberg Y, Hart MM, Klironomos J (2017) Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355:181–184
Bever JD (2003) Soil community feedback and the coexistence of competitors: conceptual frameworks and empirical tests. New Phytol 157:465–473
Bever JD, Westover KM, Antonovics J (1997) Incorporating the soil community into plant population dynamics: The utility of the feedback approach. J Ecol 85:561–573
Bonferroni CE (1936) Teoria statistica delle classi e calcolo delle probabilita. Publicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8:1–62
Brinkman EP, van der Putten WH, Bakker E-J, Verhoeven KJF (2010) Plant-soil feedback: experimental approaches, statistical analyses and ecological interpretations. J Ecol 98:1063–1073
Brookes PC, Landman A, Pruden G, Jenkinson DS (1985) Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soils. Soil Biol Biochem 17:837–842
Brundrett M et al (eds) (1994) Practical methods in mycorrhizal research. Mycologue, Waterloo, ON
Burrows RL, Pfleger FL (2002) Arbuscular mycorrhizal fungi respond to increasing plant diversity. Can J Bot 80:120–130
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chagnon PL, Bradley RL, Maherali H, Klironomos JN (2013) A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci 18:484–491
Clark DB, Clark DA, Oberbauer SF (2010) Annual wood production in a tropical rain forest in NE Costa Rica linked to climatic variation but not to increasing CO2. Glob Chang Biol 16:747–759
Classen AT, Sundqvist MK, Henning JA et al (2015) Direct and indirect effects of climate change on soil microbial-plant interactions: What lies ahead? Ecosphere 6:1–21
Coince A, Cordier T, Lengelle J et al (2014) Leaf and root-associated fungal assemblages do not follow similar elevational diversity patterns. PLoS One 9:e100668
D’Angelo E, Crutchfield J, Vandiviere M (2001) Rapid, sensitive, microscale determination of phosphate in water and soil. J Environ Qual 30:2206–2209
Davison J, Moora M, Opik M et al (2015) Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349:970–973
Dickie IA (2010) Insidious effects of sequencing errors on perceived diversity in molecular surveys. New Phytol 188:916–918
Doane TA, Horwath WR (2003) Spectrophotometric determination of nitrate with a single reagent. Anal Lett 36:2713–2722
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Fisher RF (1995) Amelioration of degraded rain forest soils by plantations of native trees. Soil Sci Soc Am J 59:544–549
Gollotte A, van Tuinen D, Atkinson D (2004) Diversity of arbuscular mycorrhizal fungi colonizing roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 14:111–117
Guadarrama P, Alvarez-Sanchez FJ (1999) Abundance of arbuscular mycorrhizal fungi spores in different environments in a tropical rain forest, Veracruz, Mexico. Mycorrhiza 8:267–270
Hawkes CV, Kivlin SN, Du J, Eviner VT (2013) The temporal development and additivity of plant-soil feedback in perennial grasses. Plant Soil 369:141–150
Husband R, Herre EA, Turner SL et al (2002a) Molecular diversity of arbuscular mycorrhizal fungi and patterns of host association over time and space in a tropical forest. Mol Ecol 11:2669–2678
Husband R, Herre EA, Young JP (2002b) Temporal variation in the arbuscular mycorrhizal communities colonizing seedlings in a tropical forest. FEMS Microbiol Ecol 42:131–136
Jackson DA (1995) PROTEST: A PROcrustean randomization TEST of community environment concordance. Ecoscience 2:297–303
Janos DP (1980) Vesicular-arbuscular mycorrhizae affect lowland tropical rain forest plant growth. Ecology 61:151–162
Janzen DH (1970) Herbivores and the number of tree species in tropical forests. Am Nat 104:501–528
Johnson NC (2010) Resource stoichiometry elucidates the structure and function of arbuscular mycorrhizas across scales. New Phytol 185:631–647
Johnson NC, Graham JH, Smith FA (1997) Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol 135:575–585
Johnson NC, Wilson GWT, Bowker MA et al (2010) Resource limitation is a driver of local adaptation in mycorrhizal symbioses. Proc Natl Acad Sci U S A 107:2093–2098
Jones DL, Willett VB (2006) Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biol Biochem 38:991–999
Kardol P, Bezemer TM, van der Putten WH (2006) Temporal variation in plant-soil feedback controls succession. Ecol Lett 9:1080–1088
Kardol P, De Deyn GB, Laliberte E et al (2013) Biotic plant–soil feedbacks across temporal scales. J Ecol 101:309–315
Kembel SW, Cowan PD, Helmus MR et al (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464
Kivlin SN, Hawkes CV (2011) Differentiating between effects of invasion and diversity: impacts of aboveground plant communities on belowground fungal communities. New Phytol 189:526–535
Kivlin SN, Hawkes CV (2016a) Temporal and spatial variation of soil bacteria richness, composition, and function in a Neotropical rainforest. PLoS One 11:e0159131
Kivlin SN, Hawkes CV (2016b) Tree species, spatial heterogeneity, and seasonality drive soil fungal abundance, richness, and composition in Neotropical rainforests. Environ Microbiol 18:4662–4673
Kivlin SN, Hawkes CV, Treseder KK (2011) Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biol Biochem 43:2294–2303
Kleber M, Schwendenmann L, Veldkamp E et al (2007) Halloysite versus gibbsite: silicon cycling as a pedogenetic process in two lowland neotropical rain froest soils of La Selva, Costa Rica. Geoderma 138:1–11
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70
Klironomos JN, Ursic M (1998) Density-dependent grazing on the extraradical hyphal network of the arbuscular mycorrhizal fungi, Glomus intraradices, by the collembolan, Folsomia candida. Biol Fertil Soils 26:250–253
Kulmatiski A, Beard KH, Stevens JR, Cobbold SM (2008) Plant-soil feedbacks: a meta-analytical review. Ecol Lett 11:980–992
Lajtha K, Driscoll CT, Jarrell WM, Elliott ET (1999) Soil phosphorus: Characterization and total elemental analysis. In: Robertson G et al (eds) Standard soil methods for long-term ecological research. Oxford Univ. Press, New York, pp 124–127
Lankau EW, Lankau RA (2014) Plant species capacity to drive soil fungal communities contributes to differential impacts of plant-soil legacies. Ecology 95:3221–3228
Lavorel S, Garnier E (2002) Predicting changes in community composition and ecosystem functioning from plant traits: revisiting the Holy Grail. Funct Ecol 16:545–556
Lekberg Y, Bever JD, Bunn RA, Callaway RM, Hart MM, Kivlin SN, Klironomos J, Larken BG, Maron JL, Reinhart KO, Remke M, van der Putten WH (2018) Relative importance of competition and plant-soil feedback, their synergy, context dependency and implications for coexistence. Ecol Lett. https://doi.org/10.1111/ele.13093
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Makarov MI, Shuleva MS, Malysheva TI, Menyailo OV (2013) Solubility of the labile forms of soil carbon and nitrogen in K2SO4 of different concentrations. Eurasian Soil Sci 46:369–374
Mangan SA, Herre EA, Bever JD (2010a) Specificity between Neotropical tree seedlings and their fungal mutualists leads to plant-soil feedback. Ecology 91:2594–2603
Mangan SA, Schnitzer SA, Herre EA et al (2010b) Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466:752–755
Mann HB, Whitney DR (1947) On a test of whether one or two random variables is stochastically larger than the other. Ann Math Stat 18:50–60
Mariadassou M, Pichon S, Ebert D (2015) Microbial ecosystems are dominated by specialist taxa. Ecol Lett 18:974–982
McGonigle TP, Miller MH, Evans DG et al (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115:495–501
McGuire KL, Fierer N, Bateman C et al (2012) Fungal community composition in neotropical rain forests: the influence of tree diversity and precipitation. Microb Ecol 63:804–812
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Computat Biol 10:e1003531
Mirarab S, Nguyen N, Guo S et al (2015) PASTA: Ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol 22:377–386
Nasto MK, Alvarez-Clare S, Lekberg Y et al (2014) Interactions among nitrogen fixation and soil phosphorus acquisition strategies in lowland tropical rain forests. Ecol Lett 17:1282–1289
Oksanen J, Kindt R, Legendre P, et al. (2007) The vegan package. Community Ecology Package
Peay KG, Baralato C, Fine PV (2013) Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J 7:1852–1861
Powers JS, Haggar JP, Fisher RF (1997) The effect of overstory composition on understory woody regeneration and species richness in 7-year-old plantation in Costa Rica. For Ecol Manag 99:43–54
R Development Core Team (2009) R: A language and environment for statistical computing. Austria R Foundation for Statistical Computing, Vienna
Ren H, Xu Z, Huang J et al (2015) Increased precipitation induces a positive plant-soil feedback in a semi-arid grassland. Plant Soil 389:211–223
Revillini D, Gehring CA, Johnson NC (2016) The role of locally adapted mycorrhizas and rhizobacteria in plant-soil feedback systems. Funct Ecol 30:1086–1098
Rodriguez-Girones MA, Santamaria L (2006) A new algorithm to calculate the nestedness temperature of presence-absence matrices. J Biogeogr 33:924–935
Rua MA, Antoninka A, Antunes PM et al (2016) Home-field advantage? Evidence of local adaptation among plants, soils, and arbuscular mycorrhizal fungi through meta-analysis. BMC Evol Biol 16:122
Russell AE, Raich JW, Valverde-Barrante OJ, Fisher RF (2007) Tree species effects on soil properties in experimental plantations in tropical moist forest. Soil Sci Soc Am J 71:1389–1397
Russell AE, Raich JW, Bedoya Arrieta A et al (2010) Impacts of individual tree species on carbon dynamics in a moist tropical forest environment. Ecology 20:1087–1100
Sanford RL Jr, Paaby P, Luvall JC, Phillips E (1994) Climate geomorphology, and aquatic systems. Pgs 19–33. In: McDade LA et al (eds) La Selva Book: Ecology and natural history of a Neotropical rain forest. University of Chicago Press. Chicago, Illinois
Schappe T, Albornoz FE, Turner BL et al (2017) The role of soil chemistry and plant neighborhoods in structuring fungal communities in three Panamanian rainforests. J Ecol 105:569–579
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Scott-Denton LE, Rosenstiel TN, Monson RK (2006) Differential controls by climate and substrate over the heterotrophic and rhizospheric components of soil respiration. Glob Chang Biol 12:205–216
Slik JWF, Paoli G, McGuire K et al (2013) Large trees drive forest aboveground biomass variation in moist lowland forests across the tropics. Glob Ecol Biogeogr 22:1261–1271
Smith-Ramesh LM, Reynolds HL (2017) The next frontier of plant-soil feedback research: unraveling context dependence across biotic and abiotic gradients. J Veg Sci 28:484–494
Stanescu S, Maherali H (2017) Mycorrhizal feedback is not associated with the outcome of competition in old-field perennial plants. Oikos 126:248–258
Teste FP, Kardol P, Turner BL et al (2017) Plant-soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science 355:173–176
Tilman D (1994) Competition and biodiversity in spatially structured habitats. Ecology 75:2–16
Treseder KK, Vitousek PM (2001) Effects of soil nutrient availability on investment in acquisition of N and P in Hawaiian rain forests. Ecology 82:946–954
van der Putten WH, Bardgett RD, Bever JD et al (2013) Plant-soil feedbacks: the past, the present and future challenges. J Ecol 101:265–276
van der Putten WH, Bradford MA, Brinkmann EP et al (2016) Where, when and how plant-soil feedback matters in a changing world. Funct Ecol 30:1109–1121
Van Nuland ME, Bailey JK, Schweitzer JA (2017) Divergent plant-soil feedbacks could alter future elevation ranges and ecosystem dynamics. Nat Ecol Evol 1:1050
Vandecar K, Lawrence D, Clark D (2011) Phosphorus sorption dynamics of anion exchange resin membranes in tropical rain forest soils. Soil Sci Soc Am J 75:1520–1529
Waring BG, Adams R, Branco S, Powers JS (2016) Scale-dependent variation in nitrogen cycling and soil fungal communities along gradients of forest composition and age in regenerating tropical dry forests. New Phytol 209:845–854
Acknowledgments
The authors thank B.G. Waring, F. Cascante, M. Hernandez, E. Paniagua, and M. Sanchez for assistance with shadehouse and fieldwork. Previous versions of this manuscript were improved by comments from the Bailey and Schweitzer lab groups at the University of Tennessee, Y.A. Chung and two anonymous reviewers. The material is based upon work supported by the National Science Foundation under Grant No. DEB-1119169 to CVH. Soils were collected under Conagebio permit No. R-005-2012.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Thom W. Kuyper.
Electronic supplementary material
ESM 1
(DOCX 66 kb)
Rights and permissions
About this article
Cite this article
Kivlin, S.N., Bedoya, R. & Hawkes, C.V. Heterogeneity in arbuscular mycorrhizal fungal communities may contribute to inconsistent plant-soil feedback in a Neotropical forest. Plant Soil 432, 29–44 (2018). https://doi.org/10.1007/s11104-018-3777-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-018-3777-4