Abstract
Aims
Sorghum (Sorghum bicolor) roots release biological nitrification inhibitors (BNIs) to suppress soil nitrification. Presence of NH4 + in the rhizosphere stimulates BNIs release and it is hypothesized to be functionally associated with plasma membrane (PM) H+-ATPase activity. However, whether the H+-ATPase is regulated at the transcriptional level, and if so, which isoforms of the H+-ATPases are involved in BNIs release are not known. Also, it is not clear whether the stimulation on BNIs release from roots is due to NH4 + uptake or its assimilation, which are addressed in this study.
Methods
Root exudates from intact sorghum plants were collected using aerated solutions of NH4 + or methyl-ammonium (MeA); and the BNI-activity release was determined. PM vesicles were isolated from fresh roots using a two-phase partitioning system; and the hydrolytic H+-ATPase activity was determined. All genes encoding PM H+-ATPases were searched in sorghum genome, and their expression in response to NH4 + or MeA were analyzed by quantitative RT-PCR in sorghum roots.
Results
BNIs release and PM H+-ATPase activity increased with NH4 + concentration (≤1.0 mM) in the root-exudate collection solutions, but at higher concentrations, it did not respond further or declined in case of the PM H+-ATPase activity. Twelve PM H+-ATPase genes were identified in sorghum genome; and these isoforms were designated SbA1 to SbA12. Five H+-ATPase genes were stimulated by NH4 + in the rhizosphere, and have similar expression pattern, which is consistent with the variation in H+-ATPase activity. MeA, a non-metabolizable analogue of NH4 +, had no significant effects on BNIs release, H+-ATPase activity, or expression of the H+-ATPase genes.
Conclusions
Our results suggest that the functional link between PM H+-ATPase activity and BNIs release is evident only at NH4 + levels of ≤1.0 mM in the rhizosphere. The variation in PM H+-ATPase activity by NH4 + is due to transcriptional regulation of five isoforms of the H+-ATPases. The stimulatory effect of NH4 + on BNIs release is functionally associated with NH4 + assimilation and not just with NH4 + uptake alone.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plasma membrane (PM) H+-ATPase is the universal electrogenic H+ pump, a functional monomer and has N- and C-terminal segments protruding into cytoplasm (Palmgren and Harper 1999; Palmgren 2001). The C-terminal region of the H+-ATPase contains a penultimate Thr known as autoinhibitory domain to keep the H+-ATPase in a low-activity state (Palmgren 2001). The phosphorylation of the penultimate Thr and subsequent binding of the 14-3-3 protein to the phosphorylated penultimate Thr results in the activation of the H+-ATPase (Olsson et al. 1998). Plant PM H+-ATPases are encoded by a multi-gene family. There are 12 members in Arabidopsis (Arabidopsis thaliana) (AHA1-AHA12) and 10 members in rice (Oryza sativa) (OSA1-OSA10) classified into five sub-families (Arango et al. 2003). PM H+-ATPases are involved in generating H+ electrochemical gradient to provide driving force for the active influx, efflux of ions and metabolites across the PM (Portillo 2000; Palmgren 2001; Arango et al. 2003; Duby and Boutry 2009).
Ammonium (NH4 +) is oxidized to nitrite (NO2 −) and subsequently to nitrate (NO3 −), by nitrifying bacteria (Nitrosomonas and Nitrobacter, respectively) in the soil. This biological process termed nitrification, however, is one of the major causes of nitrogen loss and associated environmental pollution due to NO3 − leaching and denitrification. If the nitrification process is repressed, NO3 − leaching and N2O emission are reduced, then, nitrogen recovery in agricultural system can be improved (Subbarao et al. 2015). The existence of plant-derived nitrification inhibitors are known (Moore and Waid 1971; Lata et al. 1999, 2004). The phenomenon of nitrification inhibitors (BNIs) produced and released from plant roots is termed “biological nitrification inhibition” (BNI) (Subbarao et al. 2006, 2015). Several BNIs have been isolated and characterized; e.g., brachialactone is isolated from root exudates of pasture grass Brachiaria humidicola; methyl 3-(4-hydroxyphenyl) propionate (MHPP), sakuranetin and sorgoleone are isolated from root exudates of sorghum (Sorghum bicolor) (Subbarao et al. 2006, 2009, 2012a, b, 2013; Zakir et al. 2008; Tesfamariam et al. 2014). Presence of NH4 + in the growth media or root exudate collection solution is reported to stimulate BNIs release from sorghum roots (Subbarao et al. 2007b, 2009; Zakir et al. 2008).
The stimulation of BNIs release by NH4 + is reported to be related to the PM H+-ATPase activity in sorghum (Zhu et al. 2012). The PM H+-ATPase activity was stimulated by NH4 + nutrition, and the BNIs release was repressed by pharmacological inhibitor, vanadate or stimulated by fusicossin, a known stimulant of PM H+-ATPase (Zhu et al. 2012). The release of BNIs is a tightly regulated physiological process where an interplay among NH4 + uptake, rhizosphere pH, and PM H+-ATPase regulate BNIs release in sorghum roots (Zhu et al. 2012). Whether the PM H+-ATPase genes respond to NH4 + presence in the rhizosphere and which isoforms are involved in BNIs release is not known; but such information is needed to understand the role of NH4 + nutrition on BNIs release. In this study, we investigated the effect of NH4 + nutrition on expression of PM H+-ATPase genes from sorghum genome. Also, it is not known whether NH4 + uptake alone or its assimilation is responsible for stimulating BNIs release; this is addressed by comparing the influence of methyl-ammonium (MeA) (a non-metabolizable analogue of NH4 +) (Ermilova et al. 2007; Kosola and Bloom 1994) and NH4 + on BNIs release, PM H+-ATPase activities and expression of PM H+-ATPase genes in sorghum roots.
Materials and methods
Plant cultivation
Sorghum seeds (Sorghum bicolor L. Moench var. hybrid sorgo) were germinated in trays containing vermiculite. After 1 week, the seedlings were transferred to aerated nutrient solution of 70 L tanks covered by a styrofoam board with 4 plants grown in each of the 45 holes. The seedlings were grown in a growth chamber under controlled conditions (photoperiod 14-h-light/10-h-dark at 30/28 °C, light intensity 300 μmol m−2 s−1). The composition of the nutrient solution is as follows: (NH4)2SO4 0.5 mM; KH2PO4 0.3 mM; K2SO4 0.2 mM; CaCl2 0.1 mM; MgSO4 0.15 mM; Fe-EDTA 0.2 mM; H3BO3 20 μM; CuSO4 0.3 μM; MnSO4 9.0 μM; Na2MoO4 0.5 μM; ZnSO4 0.8 μM. The nutrient solutions were replaced every 3 days. At the end of every day, the pH of the nutrient solution was adjusted to 5.0 using 1 N NaOH or H2SO4.
Root exudates collection
Two weeks after transplanting, the intact roots of sorghum seedlings (a sample size of 12 plants with three replications) were removed from the nutrient solution and rinsed with distilled water, then immersed for 4 h (from 10:00 AM to 14:00 PM) in 1 L aerated root exudates collection solution (200 μM CaCl2) containing NH4Cl or methyl-ammonium (tetra methyl-ammonium chloride); using a pH-stat system (NPH-660 NDE, Nissin, Japan), the pH of the root exudate solution was maintained at 5.0 during the 4 h collection period, as described earlier (Zhu et al. 2012). For vanadate experiment, 0.5 mM vanadate was used in the root exudates collection solution. After collection of root exudates, roots were separated from shoots, dried at 70 °C for 2 days in a forced air-circulating oven before determining the dry weight. For PM isolation, fresh roots were rinsed with distilled water and then ground immediately. For RNA isolation and gene expression analysis, fresh roots were frozen in liquid nitrogen for 4 h and then stored at −80 °C.
Nitrification inhibition determination
The inhibition of nitrification was determined following the methods described previously (Subbarao et al. 2006; Zhu et al. 2012). For the extraction of biological nitrification inhibition (BNI) compounds, root exudates were evaporated to dryness using a rotary evaporator (Buchi, V-850, Flawil, Switzerland) under vacuum at 45 °C, followed by extraction with 20 mL of methanol. The methanol extract was then further evaporated to dryness using a rotary evaporator at 40 °C; and the residue was extracted with 50 μL of dimethyl sulphoxide (DMSO). The DMSO extract was then used to determine the BNI activity using a modified bioassay that employs recombinant luminescent N. europaea (Iizumi et al. 1998; Subbarao et al. 2006). The BNI activity of samples is expressed in units defined in terms of the action of a standard inhibitor, allylthiourea (AT); the inhibitory effect of 0.22 μM AT in an assay containing 18.9 mM of NH4 + is defined as one ATU (AT unit) of activity (Subbarao et al. 2006).
PM isolation and H+ ATPase activity analysis
PM isolation and H+-ATPase activity analysis were conducted using the methods as described previously (Yan et al. 2002; Zhu et al. 2012; Zeng et al. 2013).
Identification of PM H+-ATPase genes in sorghum
To identify novel PM H+-ATPase genes of sorghum in a genome-wide level, the amino acid sequence of Arabidopsis AHA2 (a well-characterized PM H+-ATPase member) was used to blast against the sorghum genome database (version 2.1) (http://www.phytozome.net/search.php?org=Org_Sbicolor_v2.1). In addition, sorghum genes containing a functional annotation of KOG0205 (PM H+-transporting ATPase) were also collected. The nucleotide and amino acid sequences, as well as other information of these putative PM H+-ATPase genes were obtained.
Phylogenetic tree construction and sequence analysis
Protein sequences of PM H+-ATPases of Arabidopsis, rice and sorghum were obtained from TAIR (http://www.arabidopsis.org/), NCBI (http://www.ncbi.nlm.nih.gov/) and Phytozome (http://www.phytozome.net/sorghum.php), respectively. The amino acid sequences of PM H+-ATPase proteins were aligned using ClustalW (BLOSUM series was used for Protein Weight Matrix), and a phylogenetic tree was constructed by the neighbor-joining method using the software MEGA4.0 (Tamura et al. 2007). Gene structure feature was analyzed by Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/index.php).
RNA extraction and gene expression analysis
Total RNAs were extracted with the RNeasy Plant Mini Kit (QIAGEN) and digested with DNase I (QIAGEN) to eliminate DNA contamination, and then 1 μg RNA was used for reverse transcription in a 20 μL reaction system with PrimeScript RT reagent kit (TAKARA). Quantitative real-time RT-PCR (qRT-PCR) was performed with SYBR Premix EX Taq (TAKARA), and amplification was real-time monitored on a miniOpticon real-time PCR system (Bio-Rad). Briefly, 2 μL of a 1/10 dilution of cDNA, 10 μL of the 2 × SYBR Premix Ex Taq (TaKaRa), 1 μL each primer of 5, and 6 μL water was mixed in a final volume of 20 μL. The reaction was amplified for 30s at 95 °C, followed by for 5 s at 95 °C and 30 s at 60 °C for 40 cycles. All reactions were run in triplicate and included no template or reverse transcription controls for each gene. PCR efficiency was determined by a series of 2-fold dilutions of cDNAs, and the calculated efficiency of all primers varied from 0.8 to 1.0. The primers used for amplification of sorghum H+-ATPase genes were as follows: 5′ CGCCTTATTGCGACGGA 3′ and 5′ CGCTTTACAACTAGGGCTGCT 3′ for SbA1; 5′ CCACATTCACCACCGAGC 3′ and 5′ CCCTTGCAGTTCCAGATTTATA 3′ for SbA2; 5′ TCAGGTTTCTTTTGGATTAGACA 3′ and 5′ AACTTACAAGGAGGGAGGAGG 3′ for SbA3;; 5′ TCACCACCAAGAAGGACTACG 3′ and 5′ GGTGCGGTCGTTGAGGAT 3′ for SbA5; 5′ GCCTCCGACCCTTCTTCT 3′ and 5′ GACGGTTTCGTTGGTGATG 3′ for SbA10; 5′ AGCAGCACAACACCATCTTC 3′ and 5′ CGCTTCAGCCTCATCACA 3′ for SbA11. Gene relative expression levels were normalized to one ACTIN gene (Sobic005g047100) and presented as 2-△△CT to simply the presentation of data. The specific primers for ACTIN were 5′CATCCCCACTTCGGCTCC 3′ and 5′ CCTCAATAGGCTGGCAATCTC 3′.
Results
Presence of NH4 + in the root exudate solution on BNIs release and PM H+-ATPase activity in sorghum roots
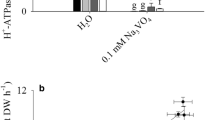
The release of BNI activity from sorghum roots increased by the presence of NH4 + in the concentration range of 0 to 1.0 mM, but no further increase was observed at concentration >1.0 mM (1.0 to 4.0 mM) (Fig. 1a). Root PM H+-ATPase activity also increased in the concentration range of 0 to 1.0 mM NH4 + in the root exudate solutions, but it declined subsequently at concentration >1.0 (1.0 to 4.0 mM) (Fig. 1b). Vanadate is a known inhibitor of PM H+-ATPases. After incubation with 0.5 mM vanadate, BNI release from intact sorghum plants was significantly suppressed at 0.5 mM and 4.0 mM NH4 + (Fig. 1c). These results suggest that PM H+-ATPase is functionally involved in the stimulation of BNIs release by NH4 + in the concentration range of ≤1.0 mM.
Effects of ammonium (AM) on the BNI release and the PM H+-ATPase activity. After growing in nutrient solution for 2 weeks, sorghum seedling roots were incubation with 1 L aerated root exudate collection solution containing NH4Cl of different concentrations (0, 0.1, 0.5, 1.0, 2.0, 4.0 mM) for 4 h, and then root exudates were collected for BNI determination (a), and PM vesicles were isolated from fresh roots for analysis of the activity of PM H+-ATPases (b). c To investigate the effect of vanadate (VA) on BNIs releases from sorghum roots, root exudates were collected after incubation for 4 h in 1 L aerated root exudate collection solution with 0.5 mM or 4.0 mM NH4Cl, and with or without 0.5 mM VA. The data of the BNI activity and the PM H+-ATPase activity at 0–1.0 mM NH4 + is adopted from Zhu et al. (2012). The bars represent means ± SE of three replications. Different letters indicate that the values are significantly different at P < 0.05 (based on one-way ANOVA)
Genome-wide identification of sorghum PM H+-ATPase genes
To investigate whether the transcriptional levels of the H+-ATPase genes are affected by NH4 +, we first identified novel PM H+-ATPase genes by blasting against sorghum genome database using the amino acid sequences of Arabidopsis AHA2 as query. A total of 12 homologous genes encoding putative PM H+-ATPases were found in sorghum genome, and they showed high sequence identity with AHA2, arranged between 67 and 84 %. These genes were sequentially named SbA1-SbA12, based on their amino acid identity to AHA2 (Table 1). Similar to their counterparts in other plants, sorghum H+-ATPase are relatively big proteins with lengths ranging from 874 to 1004 aa. The coding region of sorghum H+-ATPase genes is interrupted by 1–20 introns (Table 1, Fig. 2), which is similar to that in Arabidopsis and rice (Arango et al. 2003). With the exception of SbA12, all the other 11 isoforms possess a penultimate Thr or Ser, and conserved region I and region II in the C-terminal regions (Fig. 3). Because of the lack of C-terminal auto-inhibitory domain, SbA12 was considered to be a probable pseudogene. Region I and region II in the C-terminal region have been identified to be important for the auto-inhibitory effects on the H+-ATPase (Palmgren et al. 1991; Axelsen et al. 1999). The phosphorylation of penultimate Thr or Ser is required for 14-3-3 protein binding, which results in the activation of H+-ATPases (Olsson et al. 1998; Maudoux et al. 2000; Kinoshita and Shimazaki 2002). Phylogenetic analysis using full-length amino acid sequences indicated that sorghum PM H+-ATPase genes are localized in each of the five subfamilies (Fig. 4), similar to PM H+-ATPase genes in Arabidopsis and rice (Arango et al. 2003).
Alignment of the C-terminal regions of AHA2 and 12 sorghum PM H+-ATPases with ClustalW (http://www.genome.jp/tools/clustalw/) and BoxShade (http://www.ch.embnet.org/software/BOX_form.html). Black blocks indicate highly conserved residues. Dashes indicate gaps introduced to allow for optimal alignment of the sequences. The 9th and 10th transmembrane segments (M9 and M10), the region I, the region II and the 14-3-3 protein binding site within the C-terminal regions are indicated by lines. SbA12 is a probable pseudogene which lacks C-terminal auto-inhibitory domain
Phylogenetic tree of PM H+-ATPase proteins from Arabidopsis, rice and sorghum. The full-length amino acid sequences of the H+-ATPase proteins were aligned using ClustalW, and a phylogenetic tree was constructed by the neighbor-joining method using the software MEGA4.0. Sorghum PM H+-ATPase proteins are marked with a blue circle before the protein names. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch. Roman numerals designate the subfamilies. The locus names of SbA1-SbA12 are listed in Table 1. The AGI (Arabidopsis Genome Initiative) number of AtAHA1-AtAHA12 is At2g18960, At4g30190, At5g57350, At3g47950, At2g24520, At2g07560, At3g60330, At3g42640, At1g80660, At1g17260, At5g62670, and At4g11730, respectively. The NCBI gene accession numbers of OsA1-OsA10 are AJ439999, AJ440000, AJ440001, AJ440002, AJ440216, AJ440217, AJ440218, AJ440219, AJ440220, and AJ440221, respectively. AHA12, OsA10 and SbA12 are probable pseudogenes which lack C-terminal auto-inhibitory domains (Axelsen and Palmgren 2001; Arango et al. 2003)
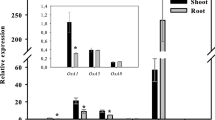
Expression of PM H+-ATPase genes in response to NH4 + in sorghum roots
Expression levels of sorghum H+-ATPase genes in response to NH4 + were analyzed by qRT-PCR using specific primers. At least six of these 12 H+-ATPase genes were successfully detected in sorghum roots, but no expression was detected for the other eight isoforms, which could be expressed in specific tissue or under specific conditions. Among the six genes, SbA3 was only marginally affected by NH4 +, whereas the other five genes (SbA1, SbA2, SbA5, SbA10 and SbA11) were found to respond to NH4 + nutrition and their expression patterns were similar (Fig. 5). The expression of the five H+-ATPase genes was increased by NH4 +, and the highest expression occurred at concentration of 0.5 mM or 1.0 mM. But their expression levels were decreased by high NH4 + concentration (4.0 mM). The expression patterns of H+-ATPase genes were largely consistent with the response of PM H+-ATPase activities (Fig. 1b), although post-translational regulation could not be excluded.
Expression of six sorghum PM H+-ATPase genes in response to NH4 + nutrition. After growing in nutrient solution for 2 weeks, sorghum seedling roots were incubated with 1 L aerated root exudate collection solution containing NH4Cl of different concentrations (0, 0.1, 0.5, 1.0, 2.0, 4.0 mM) for 4 h, and then total RNA was isolated from roots for expression analysis. Relative expression level of each individual PM H+-ATPase gene was normalized to that of SbACTIN (Sobic005g047100). The relative expression levels were normalized to 1 in control (without ammonium). The bars represent means ± SE of three replicates
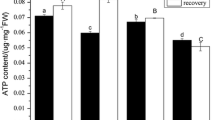
Influence of methyl ammonium (MeA) on BNIs release and PM H+-ATPase activity in sorghum roots
To investigate whether the effect of NH4 + nutrition on PM H+-ATPase activity of sorghum roots is related to NH4 + uptake or assimilation, we applied MeA, a non-metabolizable analogue of NH4 + to the root exudate collection solution (Kosola and Bloom 1994; Ermilova et al. 2007), and examined the influence of MeA on BNIs release and PM H+-ATPase activity in sorghum roots. No significant change was observed for either of these processes following MeA treatment (Fig. 6a, b). But the H+-ATPase activity decreased slightly at 4.0 mM of MeA, which may be caused by its toxicity at higher concentration. Consistent with the H+-ATPase activity, the expression of most PM H+-ATPase genes were not activated by MeA treatment (Fig. 6b). But the expression of some SbA5 and SbA10 were repressed by MeA at higher concentration and SbA11 was marginally increased by MeA. Thus, unlike NH4 +, MeA has little or no stimulatory effect on PM H+-ATPase activity and BNIs release.
Effect of methyl-ammonium (MeA) on the BNIs release, the H+-ATPase activity and the expression of the H+-ATPase genes in sorghum roots. After growing in nutrient solution for 2 weeks, sorghum seedling roots were incubated with 1 L aerated root exudate collection solution containing MeA of different concentrations (0, 0.5, 1.0, 4.0 mM) for 4 h, root exudates were collected for BNI determination (a), and PM vesicles were isolated from fresh roots for analysis of the activity of H+-ATPases (b), and then total RNA was isolated from roots for expression analysis (c). Relative expression level of each individual PM H+-ATPase gene was normalized to that of SbACTIN (Sobic005g047100). The relative expression levels were normalized to 1 in control (without MeA). The bars represent means ± SE of three replicates
Discussion
Root exudates from sorghum plants were reported to inhibit nitrification (Alsaadawi et al. 1986), and their direct suppressive effect on Nitrosomonas bacteria has been validated recently (Subbarao et al. 2007a, 2013, 2015; Zakir et al. 2008; Tesfamariam et al. 2014). The presence of NH4 + in the rhizosphere has been shown to stimulate BNIs release in sorghum roots (Subbarao et al. 2007b, 2009, 2012a). It is hypothesized that PM H+-ATPase is functionally linked to BNIs release and is stimulated by the presence of NH4 + (≤1.0 mM in the rhizosphere; the concentration range where high affinity AMT1-type ammonium-transporters operate in PM) (Yuan et al. 2007). At concentrations of >1.0 mM, an apparent disconnect was observed between the BNIs release and the PM H+-ATPase activity, indicating alternative BNIs release mechanisms associated with mass-flow of NH4 +, which is open to future research.
Vanadate, a known inhibitor of PM H+-ATPase (O’Neill and Spanswick 1984), suppressed the release of BNIs in the presence of NH4 + in rhizosphere (Fig. 1c), providing further evidence to the functional link of PM H+-ATPase to BNIs release. Such functional link between PM H+-ATPase and BNI release was however disrupted at >2.0 mM NH4 + concentration, suggesting that other BNIs release mechanisms that do not require PM H+-ATPase activity may be operational. The BNI compounds are suggested to be anionic substances (Subbarao et al. 2007b). It is thus assumed that BNIs release mechanisms may be akin to mechanisms associated with the release of other organic anions, such as citrate and malate. For example, the aluminum-activated root citrate exudation is mediated by a PM-localized citrate transporter belonging to the multi-drug and toxic compound extrusion (MATE) family (Furukawa et al. 2007); and this physiological process is linked to the activity of PM H+-ATPase (Shen et al. 2005). During the release of organic compounds, the PM H+-ATPase activity seem crucial for providing the driving force and maintaining the charge balance in plant cells.
In the present study, we identified 12 PM H+-ATPase genes from sorghum genome (Table 1, Fig. 1); and most of the encoded H+-ATPases possess a conserved C-terminal region, with the exception of SbA12 (Fig. 3). Arabidopsis AHA12 and rice OsA10 also lack such C-terminal regions, and they are considered to be probable pseudogenes (Axelsen and Palmgren 2001; Arango et al. 2003). All the 12 sorghum H+-ATPases show high identity to AHA2, which is a well-characterized PM H+-ATPase member in Arabidopsis (Palmgren 2001); and they are closely related to the H+-ATPases from Arabidopsis and rice based on the phylogenetic analysis (Fig. 4), suggesting their functions are similar in plant species.
NH4 + nutrition acidifies the rhizosphere in higher plants (Schubert and Yan 1997; Zhu et al. 2009, 2012); the induction of PM H+-ATPase activity by NH4 + nutrition could partly be due to acidification of rhizosphere (Yan et al. 1998; Zhu et al. 2009). In the present study, the pH of root exudate collection solutions (containing varying levels of NH4 +) were kept at 5.0 using pH-stat system, thereby removing the secondary effects of rhizosphere-acidification from ammonium uptake and assimilation on BNI release. The stimulatory effect from NH4 + on BNI release thus seems the direct effect from its assimilation and not from the secondary effects (i.e., rhizosphere-acidification) associated with its uptake.
Consistent with the activation of the PM H+-ATPase activity in sorghum roots, at least five PM H+-ATPase genes responded to moderate concentration of NH4 + similarly in expression pattern (Figs. 1b and 5). Thus, variation in the H+-ATPase activity with NH4 + nutrition is at least partly due to transcriptional regulation of H+-ATPase genes. The H+-ATPase activity was also activated by NH4 + nutrition in the roots of barley and rice (Yamashita et al. 1995; Zhu et al. 2009; Zeng et al. 2012). Recently, it was shown that the H+-ATPase activity is stimulated by NH4 + treatment in potassium (K)-deficient sorghum roots, where two H+-ATPase genes SbA1 and SbA2 were induced by NH4 + compared to NO3 − (Alvarez-Pizarro et al. 2014). In the present study, we have extended the members of NH4 +-responsive H+-ATPase genes of sorghum to five (SbA1, SbA2, SbA5, SbA10 and SbA11) (Fig. 5). In addition to transcriptional regulation, the rapid activation of the H+-ATPase activity in sorghum roots byNH4 + nutrition could be related to post-transcriptional regulation, such as phosphorylation of penultimate Thr and subsequent binding of 14-3-3 regulatory protein. Most sorghum isoforms of H+-ATPase possess penultimate Thr or Ser in the C-terminal region (Fig. 3). The phosphorylation of penultimate Thr or Ser enables the binding of 14-3-3 protein, which is needed for the activation of PM H+-ATPase in plants (Olsson et al. 1998; Maudoux et al. 2000; Kinoshita and Shimazaki 2002). The H+-ATPase activity can be modulated at transcriptional- and post-translational level under aluminum stress in soybean roots (Shen et al. 2005). Further research is thus needed to determine whether H+-ATPase activity is also modulated at the post-translational level when sorghum roots are fed with NH4 +.
Methyl ammonium (MeA) is a non-metabolizable analogue to NH4 + (Kosola and Bloom 1994; Ermilova et al. 2007) and is taken up by plant roots through NH4 + transporters (Kosola and Bloom 1994; Ninnemann et al. 1994). However, unlike in NH4 + treatment, there was no significant variation in PM H+-ATPase activity and in BNIs release following MeA treatment (Fig. 6a). In addition, a lack of transcriptional response for the NH4 +-responsive H+-ATPase genes under MeA (Fig. 6b) suggests that variation in PM H+-ATPase and BNIs release from NH4 + nutrition is not just due to NH4 + uptake, but from NH4 + assimilation (Fig. 7).
A hypothesis involving schematic description of ammonium (NH4 +)-induced BNIs release in relation to the PM H+-ATPase in sorghum roots. After NH4 + is taken up by high affinity ammonium transporters located on PM of the cell (in the concentration range of ≤1.0 mM), NH4 + is incorporated into glutamate to produce glutamine and other amino acids, which facilitates the generation of H+ in cytoplasm; to pump these additional H+ out of the cell, the PM H+-ATPase activity is activated and this facilitates the release of anionic BNIs possibly through anionic channels on the cell PM
Although the uptake of NH4 + or MeA can lead to depolarization of cell membrane potential because of positive inward currents across PM (Walker et al. 1979; Ullrich et al. 1984), the membrane potential change or the depolarization of cell membrane potential caused by NH4 + or MeA uptake is not sufficient to activate PM H+-ATPase activity. It has been suggested that the uptake and assimilation of NH4 + are closely synchronized in plant roots (Ishiyama et al. 2003; Loqué and von Wirén 2004); and the assimilation of NH4 + is a proton-generating process (Van Beusichem et al. 1988; Xu et al. 2012). In order to pump additional H+ out of cytoplasm and maintain the electrochemical potential gradient necessary for ion uptake, the H+-ATPase activity is thus stimulated to further BNIs release. With the results presented in this study, we propose a hypothesis that NH4 + assimilation and not just uptake alone sustains H+ supply for the continued functioning of PM-H+-pumps, which is critical for BNIs release; and that is probably mediated through anion-channels when rhizosphere NH4 + concentration is ≤1.0 mM (Fig. 7).
Significance
Physiological understanding of the mechanisms governing BNIs release has implications to practical agriculture, as this helps in determining suitable niches in agro-ecological regions where BNI expression is likely favored. For example, light-soils that have low-buffering capacity such as Alfisols of the SAT (Semi-arid Tropics, where sorghum is predominantly grown as a rainy-season crop) India or sandy-loams of West Africa, where sorghum is grown as a major staple crop, seem better suited to develop acidic-rhizosphere (pH <6.0) compared to Vertisols, which are of heavy-clay type and have an alkaline pH of >7.5 (Subbarao et al. 2013). Alfisols with a natural soil pH of <6.5, are better suited for the expression of BNI function in sorghum as NH4 + uptake and assimilation (results from the present study) further reduces the rhizosphere pH in the acidic range (i.e., pH <6.0), critical to sustain operations of PM H+-pumps for BNIs release (Zhu et al. 2012; Subbarao et al. 2013). In contrast, Vertisols, where sorghum is grown as a post-rainy season crop in parts of Asia may not allow BNI expression due to resistance to changes in rhizosphere pH; thus NH4 + uptake and assimilation will not result in rhizosphere pH reaching <6.0, a level that is conducive for BNIs release in sorghum (Zhu et al. 2012; Subbarao et al. 2013, 2015). Knowledge of the underlying physiological mechanisms governing BNIs release thus helps in choosing suitable agro-ecological niche production systems where BNI function is expressed to its genetic potential for controlling nitrification.
The results from the present study improve our understanding of the physiological processes operating in sorghum roots for the BNIs release, thus a step forward to deploy sorghum BNI function in practical agriculture for the benefit of small-holder farmers in dry areas of tropical regions. In addition, the use of slow-release fertilizers can facilitate keeping soil ammonium levels <1.0 mM, this coupled with the development of genetically modified crops with high PM H+-ATPase activity can accelerate BNI release to make production systems low-nitrifying that is beneficial to the environment.
References
Alsaadawi I, Al-Uqaili J, Alrubeaa A, Al-Hadithy S (1986) Allelopathic suppression of weed and nitrification by selected cultivars of Sorghum bicolor (L.) moench. J Chem Ecol 12:209–219
Alvarez-Pizarro JC, Gomes-Filho E, Prisco JT, Grossi-De-Sá MF, De Oliveira-Neto OB, Da Rocha Fragoso R (2014) Plasma membrane H+-ATPase in sorghum roots as affected by potassium deficiency and nitrogen sources. Biol Plant 58:507–514
Arango M, Gevaudant F, Oufattole M, Boutry M (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216:355–365
Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126:696–706
Axelsen K, Venema K, Jahn T, Baunsgaard L, Palmgren M (1999) Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38:7227–7234
Duby G, Boutry M (2009) The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Arch Eur J Physiol 457:645–655
Ermilova EV, Nikitin MM, Fernández E (2007) Chemotaxis to ammonium/methylammonium in Chlamydomonas reinhardtii: the role of transport systems for ammonium/methylammonium. Planta 226:1323–1332
Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091
Iizumi T, Mizumoto M, Nakamura K (1998) A bioluminescence assay using Nitrosomonas europaea for rapid and sensitive detection of nitrification inhibitors. Appl Environ Microbiol 64:3656–3662
Ishiyama K, Kojima S, Takahashi H, Hayakawa T, Yamaya T (2003) Cell type distinct accumulations of mRNA and protein for NADH-dependent glutamate synthase in rice roots in response to the supply of NH4+. Plant Physiol Biochem 41:643–647
Kinoshita T, Shimazaki K (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol 43:1359–1365
Kosola KR, Bloom AJ (1994) Methylammonium as a transport analog for ammonium in tomato (Lycopersicon esculentum L.). Plant Physiol 105:435–442
Lata JC, Durand J, Lensi R, Abbadie L (1999) Stable coexistence of contrasted nitrification statuses in a wet tropical savanna ecosystem. Funct Ecol 13:762–768
Lata JC, Degrange V, Raynaud X, Maron PA, Lensi R, Abbadie L (2004) Grass populations control nitrification in savanna soils. Funct Ecol 18:605–611
Loqué D, von Wirén N (2004) Regulatory levels for the transport of ammonium in plant roots. J Exp Bot 55:1293–1305
Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J Biol Chem 275:17762–17770
Moore DRE, Waid JS (1971) The influence of washings of living roots on nitrification. Soil Biol Biochem 3:69–83
Ninnemann O, Jauniaux JC, Frommer WB (1994) Identification of a high affinity NH4+ transporter from plants. EMBO J 13:3464–3471
Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol 118:551–555
O’Neill SD, Spanswick RM (1984) Effects of vanadate on the plasma membrane ATPase of red beet and corn. Plant Physiol 75:586–591
Palmgren MG (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Biol 52:817–845
Palmgren MG, Harper JF (1999) Pumping with plant P-type ATPases. J Exp Bot 50:883–893
Palmgren M, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266:20470–20475
Portillo F (2000) Regulation of plasma membrane H+-ATPase in fungi and plants. Biochim Biophys Acta 1469:31–42
Schubert S, Yan F (1997) Nitrate and ammonium nutrition of plants: effects on acid/base balance and adaptation of root cell plasmalemma H+-ATPase. Z Panzenphysiol Bodenkd 160:275–281
Shen H, He LF, Sasaki T, Yamamoto Y, Zheng SJ, Ligaba A, Yan XL, Ahn SJ, Yamaguchi M, Sasakawa H (2005) Citrate secretion coupled with the modulation of soybean root tip under aluminum stress. Up-regulation of transcription, translation, and threonine-oriented phosphorylation of plasma membrane H+-ATPase. Plant Physiol 138:287–296
Subbarao GV, Ishikawa T, Ito O, Nakahara K, Wang HY, Berry WL (2006) A bioluminescence assay to detect nitrification inhibitors released from plant roots: a case study with Brachiaria humidicola. Plant Soil 288:101–112
Subbarao GV, Rondon M, Ito O, Ishikawa T, Rao IM, Nakahara K, Lascano C, Berry WL (2007a) Biological nitrification inhibition (BNI)—is it a widespread phenomenon? Plant Soil 294:5–18
Subbarao GV, Wang HY, Ito O, Nakahara K, Berry WL (2007b) NH4 + triggers the synthesis and release of biological nitrification inhibition compounds in Brachiaria humidicola roots. Plant Soil 290:245–257
Subbarao GV, Nakahara K, Hurtado MP, Ono H, Moreta DE, Salcedo AF, Yoshihashi AT, Ishikawa T, Ishitani M, Ohnishi-Kameyama M, Yoshida M, Rondon M, Rao IM, Lascano CE, Berry WL, Ito O (2009) Evidence for biological nitrification inhibition in Brachiaria pastures. Proc Natl Acad Sci U S A 106:17302–17307
Subbarao GV, Sahrawat K, Nakahara K, Ishikawa T, Kishii M, Rao I, Hash C, George T, Srinivasa Rao P, Nardi P (2012a) Biological nitrification inhibition—a novel strategy to regulate nitrification in agricultural systems. Adv Agron 114:249
Subbarao GV, Sahrawat K, Nakahara K, Rao I, Ishitani M, Hash C, Kishii M, Bonnett D, Berry W, Lata J (2012b) A paradigm shift towards low-nitrifying production systems: the role of biological nitrification inhibition (BNI). Ann Bot 112:297–316
Subbarao GV, Nakahara K, Ishikawa T, Ono H, Yoshida M, Yoshihashi T, Zhu Y, Zakir H, Deshpande S, Hash C (2013) Biological nitrification inhibition (BNI) activity in sorghum and its characterization. Plant Soil 366:243–259
Subbarao GV, Yoshihashi T, Worthington M, Nakahara K, Ando Y, Sahrawat KL, Rao IM, Lata J, Kishii M, Braun H (2015) Suppression of soil nitrification by plants. Plant Sci 233:155–164
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tesfamariam T, Yoshinaga H, Deshpande SP, Srinivasa Rao P, Sahrawat KL, Ando Y, Nakahara K, Hash CT, Subbarao GV (2014) Biological nitrification inhibition in sorghum: the role of sorgoleone production. Plant Soil 379:325–335
Ullrich WR, Larsson M, Larsson CM, Lesch S, Novacky A (1984) Ammonium uptake in Lemna gibba G 1, related membrane potential changes, and inhibition of anion uptake. Physiol Plant 61:369–376
Van Beusichem ML, Kirkby EA, Baas R (1988) Influence of nitrate and ammonium nutrition on the uptake, assimilation, and distribution of nutrients in Ricinus communis. Plant Physiol 86:914–921
Walker N, Beilby M, Smith F (1979) Amine uniport at the plasmalemma of charophyte cells: I. Current–voltage curves, saturation kinetics, and effects of unstirred layers. J Membr Biol 49:21–55
Xu GH, Fan XR, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182
Yamashita K, Kasai M, Ezaki B, Shibasaka M, Yamamoto Y, Matsumoto H, Sasakawa H (1995) Stimulation of H+ extrusion and plasma membrane H+-ATPase activity of barley roots by ammonium-treatment. Soil Sci Plant Nutr 41:133–140
Yan F, Feuerle R, Schaffer S, Fortmeier H, Schubert S (1998) Adaptation of active proton pumping and plasmalemma H+-ATPase activity of corn roots to low root medium pH. Plant Physiol 117:311–319
Yan F, Zhu Y, Muller C, Zorb C, Schubert S (2002) Adaptation of H+-pumping and plasma membrane H+ ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol 129:50–63
Yuan L, Loque D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wiren N (2007) The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19:2636–2652
Zakir HAKM, Subbarao GV, Pearse SJ, Gopalakrishnan S, Ito O, Ishikawa T, Kawano N, Nakahara K, Yoshihashi T, Ono H, Yoshida M (2008) Detection, isolation and characterization of a root-exuded compound, methyl 3-(4-hydroxyphenyl) propionate, responsible for biological nitrification inhibition by sorghum (Sorghum bicolor). New Phytol 180:442–451
Zeng HQ, Liu G, Kinoshita T, Zhang RP, Zhu YY, Shen QR, Xu GH (2012) Stimulation of phosphorus uptake by ammonium nutrition involves plasma membrane H+ ATPase in rice roots. Plant Soil 357:205–214
Zeng H, Feng X, Wang B, Zhu Y, Shen Q, Xu G (2013) Citrate exudation induced by aluminum is independent of plasma membrane H+-ATPase activity and coupled with potassium efflux from cluster roots of phosphorus-deficient white lupin. Plant Soil 366:389–400
Zhu YY, Di TJ, Xu GH, Chen X, Zeng HQ, Yan F, Shen QR (2009) Adaptation of plasma membrane H+-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ 32:1428–1440
Zhu YY, Zeng HQ, Shen QR, Ishikawa T, Subbarao GV (2012) Interplay among NH4 + uptake, rhizosphere pH and plasma membrane H+-ATPase determine the release of BNIs in sorghum roots—possible mechanisms and underlying hypothesis. Plant Soil 358:131–141
Acknowledgments
The research presented here is supported through JIRCAS invitation fellowship program to co-authors (Drs. Houqing Zeng, Tingjun Di and Prof. Yiyong Zhu), and is funded by grant-in-Aid for scientific research from Ministry of Agriculture, Forestry and Fisheries of Japan (MAFF) to JIRCAS under BNI project. Funding support also came from Natural Science Foundation of China (NSFC 31172035) and Program of New Century Excellent Talent in Universities (NCET-11-0672).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Ad C. Borstlap.
Rights and permissions
About this article
Cite this article
Zeng, H., Di, T., Zhu, Y. et al. Transcriptional response of plasma membrane H+-ATPase genes to ammonium nutrition and its functional link to the release of biological nitrification inhibitors from sorghum roots. Plant Soil 398, 301–312 (2016). https://doi.org/10.1007/s11104-015-2675-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2675-2