Abstract
Aim
Plant-associated bacteria can improve phytoextraction by increasing plant growth and/or metal uptake. This study aimed to characterise the culturable rhizobacterial community associated with two Ni-hyperaccumulators and to obtain a collection of isolates for application in Ni phytomining.
Methods
Non-vegetated and rhizosphere soil samples were collected from the Ni-hyperaccumulator Alyssum serpyllifolium ssp. lusitanicum (three populations) and Alyssum serpyllifolium ssp. malacitanum (one population), as well as from non-hyperaccumulating plants (Dactylis glomerata, Santolina semidentata and Alyssum serpyllifolium ssp. serpyllifolium). Rhizobacteria were isolated and characterised genotypically (BOX-PCR, 16S rDNA sequencing) and phenotypically (Ni tolerance, plant growth promoting (PGP) traits, biosurfactant production).
Results
Hyperaccumulating Alyssum subspecies hosted higher densities of bacteria compared to either non-hyperaccumulators or non-vegetated soil. In some cases hyperaccumulators showed selective enrichment of Ni-tolerant bacteria. Most bacterial strains belonged to the Actinobacteria phylum and presented Ni resistance. Phosphorus-solubilisers were mostly associated with the hyperaccumulators, siderophore-producers with D. glomerata, and IAA-producers with both these species.
Conclusion
Taxonomic diversity and phenotypic characteristics were soil-, plant species- and plant population-specific. Moreover, differences were observed between the two Ni-hyperaccumulating subspecies and amongst plant populations. Several strains presented PGP characteristics which could be useful when selecting microorganisms for bioaugmentation trials.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Serpentine soils are derived from ultramafic rocks, where the term ultramafic refers to igneous or metamorphic rocks containing more than 70 % of ferromagnesium minerals and a low content in silicon (<45 % SiO2) (Brooks 1987). Ultramafic rocks are patchily distributed throughout the world (occupying approximately 1 % of the earth’s surface area; (Proctor 1999)) and are well-known for their physical and chemical anomalies that present a hostile environment for plant growth. Some common traits include an elevated concentration of Mg and Fe, a low availability of Ca relative to Mg (unfavourable for Ca absorption), a deficiency in essential nutrients such as N, P and K, and high concentrations of potentially phytotoxic trace metals such as Ni, Co and Cr. As a result the plant communities in these areas often present a high number of endemic species (serpentinophytes), and have evolved both morphological and physiological adaptations differentiating them from the flora of adjacent geological substrates (Brooks 1987). Serpentine flora includes an unusual plant group, the so-called hyperaccumulators, which are able to accumulate extremely high concentrations of Ni in their aerial biomass (>1000 mg kg−1 dry weight (DW) matter) (Baker and Brooks 1989; Chaney et al. 2007). The Iberian Peninsula hosts two subspecies of Alyssum serpyllifolium Desf. which are both serpentine-endemic and hyperaccumulators of Ni: Alyssum serpyllifolium ssp. lusitanicum from Galicia (NW Spain) and Trás-os-Montes (NE Portugal), and Alyssum serpyllifolium ssp. malacitanum from Andalucía (S Spain).
Nickel-hyperaccumulating plants are considered ideal candidates for application in phytomining, a non-destructive approach for the recovery of high value metals (e.g. Ni) from metal-enriched soils and ores. Plants are cultivated to accumulate trace metals from soils and transport them to the shoots which can then be harvested (Chaney et al. 2007). Bioaugmentation using bacteria associated with hyperaccumulators can improve the plant’s capacity to phytoextract metals from soils (Becerra-Castro et al. 2013; Mengoni et al. 2010; Sessitsch et al. 2013). Plant-associated bacteria can enhance plant growth, reduce stress and/or modify soil metal bioavailability (Becerra-Castro et al. 2013; Cabello-Conejo et al. 2014; Glick 2014; Kidd et al. 2009; Lebeau et al. 2008; Sessitsch et al. 2013). Serpentine soils are a potential source of metal-tolerant (Co, Cr and Ni) bacteria (Pal et al. 2005). Schlegel et al. (1991) found bacterial strains isolated from serpentine soils tolerated up to 10–20 mM Ni (in the culture medium), while strains from other soil types tolerated only 1 mM Ni. Pal et al. (2007) found that bacterial strains isolated from the rhizosphere of the Ni-hyperaccumulators Rinorea bengalensis and Dichapetalum gelonoides tolerated up to 28.9 mM of Ni in the culture medium, while Turgay et al. (2012) found that, bacterial strains isolated from Turkish serpentine soils, tolerated up to 34 mM Ni in the growth medium. Furthermore, the rhizosphere bacterial communities associated with Ni-hyperaccumulating plants have been shown to differ from those of non-accumulating plants growing at the same site or of non-vegetated soil, and are also characterised by a higher number of Ni-tolerant bacteria (Abou-Shanab et al. 2003; Idris et al. 2004; Mengoni et al. 2001; Schlegel et al. 1991). This selective enrichment in Ni-tolerant bacteria in the rhizosphere has been correlated with an increase in soil Ni availability (Becerra-Castro et al. 2009). Amongst the culturable Ni-tolerant bacterial strains isolated from serpentine soils and/or associated with Ni-hyperaccumulating plants, members of the Actinobacteria, Acidobacteria, Chlorobi, Firmicutes, Verrucomicrobia and Proteobacteria have been described (Mengoni et al. 2001; Oline 2006; Pal et al. 2007; Turgay et al. 2012).
In this study, the culturable bacterial community associated with different populations of the two Ni-hyperaccumulating subspecies of Alyssum serpyllifolium (subsp. lusitanicum and subsp. malacitanum) of the Iberian Peninsula were characterised. The rhizosphere bacterial communities associated with these plants were compared with those of non-hyperaccumulating plant species (the Ni-excluder Dactylis glomerata and the facultative serpentinophyte Santolina semidentata) growing at the same sites, as well as with the non-hyperaccumulating Alyssum serpyllifolium subsp. serpyllifolium growing in calcareous soils (developed over limestone and dolomite) in Sierra Nevada (S Spain). A collection of rhizobacterial isolates was obtained and the strains were characterised both genotypically (BOX-PCR and 16S rDNA partial sequencing) and phenotypically (for their resistance to Ni and plant growth promoting (PGP) traits). We focused on the culturable bacterial community since the global aim is to obtain potentially useful isolates which can be used to improve the Ni phytoextraction capacity of Ni-(hyper)accumulating plant species.
Materials and methods
Study areas and collection of samples
The study was carried out in five areas of the Iberian Peninsula in which three subspecies of Alyssum serpyllifolium Desf. (Brassicaceae) are found growing. Four of these areas are serpentine outcrops: two sites in Trás-os-Montes (Samil (S; 41°46′48″N; 6°44′47″W) and Morais (M; 41°31′21″N; 06°49′20″W), NW Portugal), one site in Barazón (L) (42°51′09″N; 08°01′15″W, NW Spain) and one in Sierra Bermeja (SB) (36°28′48″N; 05°11′52″W, S Spain). These serpentinitic areas host the two Ni-hyperaccumulating subspecies of A. serpyllifolium which are endemic to the Iberian Peninsula: A. serpyllifolium subsp. lusitanicum Dudley and P. Silva (hereafter referred to as A. pintodasilvae) in S, M and L, and A. serpyllifolium ssp. malacitanum Rivas Goday (hereafter referred to as A. malacitanum) in SB. The fifth sampling site was the calcareous dolomitic area of Sierra Nevada (SN) (37°7′21″N; 03°26′53″W, S Spain) where the non-hyperaccumulator A. serpyllifolium subsp. serpyllifolium Desfontaines grows (hereafter referred to as A. serpyllifolium). Both Ni-hyperaccumulating subspecies presented leaf Ni concentrations above 10 g kg−1 and the highest Ni accumulation was found in the Spanish population of L (15.5 g kg−1; Table 1). For comparative purposes non-hyperaccumulating plants were also collected in some study sites: Dactylis glomerata L. was sampled at sites M, S and L, Santolina semidentata Hoffmanns. & Link. was sampled at sites M and S.
Trás-os-Montes has a Mediterranean climate, with a mean annual temperature of 12.4 °C and annual precipitation of 720 mm (Carballeira et al. 1983; Menezes de Sequeira and Pinto da Silva 1992). Barazón has a European humid-temperate climate with a mean annual temperature of 12.9 °C and mean annual precipitation of 1381 mm (Carballeira et al. 1983). Sierra Bermeja and Sierra Nevada have a Mediterranean oceanic climate (Rivas-Martínez and Rivas-Saenz 1996–2009): Sierra Bermeja has a mean annual precipitation between 800 and 1600 mm and mean annual temperature between 14 and 16 °C (Gómez-Zotano et al. 2014) and Sierra Nevada has a mean temperature of 12 °C and mean annual precipitation between 450 and 1000 mm (Castillo-Martín 2000).
At each site, 5 to 7 samples of non-vegetated soil (0–15 cm) or of rhizosphere soil were collected for the isolation of bacterial strains. The whole plant and root system (including root-adhering soil) of each species were collected at late flowering stage, and after gently crushing the root ball the tightly held soil (<3 mm from the root surface) was considered as rhizosphere soil. Samples were sieved (<4 mm) and kept at 4 °C until processing. Soil samples are named as follow: L, M, S, SB and SN according to the sampling site (Barazón, Morais, Samil, Sierra Bermeja and Sierra Nevada, respectively); and as A, G and S according to the plant species (Alyssum, Dactylis and Santolina, respectively). Rhizosphere soil is indicated by an R and the non-vegetated soil by NV.
Elemental analysis of field-collected soils and plant material
Non-vegetated (NV) and rhizosphere samples were air-dried and sieved through a 2-mm stainless steel sieve. Soil pH was measured in H2O and KCl using a 1:2.5 soil:solution ratio. Total C and N were analysed by combustion with a CHN analyser (Model CHN-1000, LECO Corp., St Joseph, MI). Exchangeable cations (Ca, Mg, Al, Na and K) were extracted with 1 M NH4Cl and determined by inductively coupled plasma optical emission spectrometry (ICP-OES, model Vista-PRO, Varian). Soils were digested in a 3:1 mixture of concentrated HNO3:HCl and the total concentrations of Co, Cr and Ni were analysed by ICP-OES. Soil metal availability was evaluated after extraction with water after 24 h shaking using a 1:2.5 soil:H2O ratio.
Plant material collected in the field was separated in shoots and roots, washed with pressurised tap water followed by deionised water, oven–dried at 45 °C, weighed and ground. Plant shoots (approximately 0.1 g) were digested in a 2:1 concentrated HNO3:HCl mixture on a hot plate at 120 °C, and the concentration of metals was measured by ICP-OES.
Microbiological analyses
Five grams of fresh rhizosphere soil were suspended in 45 mL sterile sodium hexametaphosphate solution (1 %) and shaken for 30 min in an end-over-end shaker. Soil suspensions were diluted in 10-fold series and plated in duplicate onto 284 agar medium (Schlegel et al. 1991) supplemented with 100 μg mL−1 of the fungicide cycloheximide. The 284 medium contains (per litre): 6.06 g Tris–HCl, 4.68 g NaCl, 1.49 g KCl, 1.07 g NH4Cl, 0.43 g Na2SO4, 0.2 g MgCl2.6H2O, 0.03 g CaCl2.2H2O, 0.04 g Na2HPO4.2H2O, 10 mL Fe(III)NH4 citrate solution (containing 48 mg/100 mL) plus oligoelements (1.5 mg FeSO4.7H2O, 0.3 mg H3BO4, 0.19 mg CoCl2.H2O, 0.1 mg MnCl2.4H2O, 0.08 mg ZnSO4.7H2O, 0.02 mg CuSO4.5H2O, 0.036 mg Na2MoO4.2H2O) adjusted to pH 7. The medium was supplemented with a mixture of different carbon sources: lactate (0.7 g L−1), glucose (0.5 g L−1), gluconate (0.7 g L−1), fructose (0.5 g L−1), and succinate (0.8 g L−1). Densities of metal tolerant bacteria were determined in 284 agar media supplemented with an increasing concentration of Ni (0 mM, 0.5 mM, 1.0 mM, 2.0 mM, 3.0 mM; added as NiSO4.6H2O). After 7 days incubation at 28 °C, colony forming units (CFUs) were counted and calculated per gram DW soil. Distinct colony morphotypes from Ni-enriched media associated with each plant species and from each site were sub-cultured at least three times to ensure purity and cryopreserved at −70 °C in culture medium supplemented with 15 % (v/v) glycerol.
Phenotypic characterisation of bacterial isolates
Rhizobacterial strains were screened for Ni resistance, the ability to produce biosurfactants, and for various plant growth promoting characteristics: phosphate solubilisation capacity, siderophore production, organic acid production, and indoleacetic acid (IAA) production. Nickel resistance of the strains was tested using 284 agar medium (see above) supplemented with an increasing concentration of Ni (0 mM, 1.0 mM, 2.5 mM, 5.0 mM, 10.0 mM; added as NiSO4.6H2O) and incubated at 28 °C for 7 days. The Maximal Tolerable Concentration (MTC) of Ni was recorded for each isolate, as the highest Ni concentration tested where the isolate was able to grow. The ability to solubilise inorganic phosphate was assessed in a modified NBRIP agar medium (1.8 %) supplied with 5 g L−1 of hydroxyapatite and incubated at 28 °C for 5 days (10.0 g glucose, 5.0 g MgCl2.6H2O, 0.25 g MgSO4.7H2O, 0.2 g KCl, 0.1 g (NH4)2SO4, 0.1 g yeast extract in 1 L deionized water adjusted to pH 7.0; modified from (Nautiyal 1999)). A clear halo around the bacterial colony indicated solubilisation of mineral phosphate. Yeast extract was added since some strains were unable to grow in yeast-free NBRIP medium. Siderophore production was detected in a modified 284 liquid medium (without Fe) using the Chrome Azurol S (CAS) method described by Schwyn and Neilands (1987). All glassware used in this assay was previously cleaned with 30 % HNO3 followed by washing in distilled water (Cox 1994). Each isolate was screened for acid production. Single colonies were plated on agar medium containing 0.002 % bromocresol purple (per litre medium): 10.0 g glucose, 1.0 g tryptone, 0.5 g yeast extract, 0.5 g NaCl, 0.03 g CaCl2.2H2O. Colonies forming a yellow halo after 1 day of growth at 28 °C indicated a pH change in the medium and were considered acid producers. The ability of isolated strains to produce IAA was evaluated in liquid medium (5.0 g glucose, 1.0 g (NH4)2SO4, 2.0 g K2HPO4, 0.5 g CaCO3, 0.5 g MgSO4.7H2O, 0.1 g NaCl, 0.1 g yeast extract adjusted to pH 7; modified from Sheng et al. (2008) supplemented with 0.5 mg mL−1 of tryptophan). After 5 days incubation at 28 °C, cultures were centrifuged and the supernatant was incubated with Salkowski reagent for 25 min. The production of IAA was recognized by the presence of red colouring and isolates were considered IAA-producers when the concentration of IAA determined was more than 4 mg L−1 culture. Strains were screened for potential biosurfactant production using the qualitative method of Chen et al. (2007). The strains were inoculated in 284 liquid medium and cultured overnight at 28 °C, at 150 rpm on a rotary shaker. A 100 μL sample was taken from the supernatant of each strain and added to a microwell of a 96-microwell plate. The plate was then viewed using a backing sheet of paper with a black and white grid. The optical distortion of the grid provided a qualitative assay for the presence of surfactants.
Isolates were classified in different phenotype groups, according to their metal resistance (low Ni tolerance (LT): ≤2.5 mM Ni MTC, or high Ni tolerance (HT): 5–10 mM Ni MTC) and PGP traits (N: non PGP traits producers; A: organic acid-producers; P: P-solubilisers; S: Siderophore-producers; T: biosurfactants-producers; H: indoleacetic acid-producers; and their possible combinations).
Eighty four out of the 550 rhizobacterial isolates obtained were already phenotypically characterised in a prior study and used to evaluate the potential of cell-free cultures to mobilise soil Ni under in vitro conditions (Becerra-Castro et al. 2011). This previous study targeted only rhizobacterial strains associated with Alyssum sp. and their capacity for soil Ni mobilisation.
Genotypic characterisation of bacterial isolates
BOX-PCR genomic DNA profiling
BOX-PCR fingerprinting was used to genotype and group bacterial strains within each isolate collection (L, S, M, SB, SN). Crude cell lysates (colonies suspended in 100 μL and heated to 100 °C for 5 min) were used as DNA templates for BOX-PCR reactions. Box reactions were performed in a total volume of 20 μL containing: 1x Taq buffer (Invitrogen), 1.5 mM MgCl2, 0.1 mM of each dNTP, 0.5U Taq polymerase (Invitrogen), 2 μM of BOX A1R primer (5′- CTACGGCAAGGCGACGCTGACG-3′) (Versalovic et al. 1994), and 2 μL of cell lysate. Thermocycling conditions were: 1 cycle of 94 °C for 5 min; 35 cycles of 1 min at 94 °C; 1.5 min at 50 °C and 8 min at 68 °C; and 1 cycle of 8 min at 68 °C. The obtained PCR products were separated by gel electrophoresis in 1.8 % agarose run for 3 h at 3.3 V cm−1 gel. Gel images were analysed, using the Pearson correlation coefficient and UPGMA clustering algorithm of the Gel Compar Bionumerics program (Bionumerics Version 6.6, Applied Maths, Belgium). Rhizobacterial isolates were grouped according to their BOX-PCR profiles at a similarity level of 82 %. BOX-PCR fingerprints were analysed for each sampling site (L, S, M, SB and SN) and moreover, bacterial strains associated with the Ni-hyperaccumulator Alyssum subspecies were analysed separately (LA, SA, MA and SBA). Genetic diversity of isolates was assessed for each plant species and population using the Shannon diversity index (H′ = −∑(xi/x0)ln(xi/x0)) where xi is the number of strains per BOX-PCR group for each level of metal tolerance (LT or HT), and x0 is the total number of strains in either LT or HT. Isolates within each BOX-PCR group were also classified according to the phenotypes (PGP traits and Ni resistance) described above.
DNA extraction and 16S rRNA gene amplification
For DNA extraction, purified strains were grown in 1/10 strength 869 liquid medium (1.0 g tryptone, 0.5 g yeast extract, 0.5 g NaCl, 0.1 g glucose, 0.035 g CaCl2.2H2O in 1 L deionised water (Mergeay et al. 1985)) and genomic DNA was extracted from bacterial cell pellets. Briefly, the method consists of alkaline cell lysis followed by phenol/chloroform/isopropanol alcohol purification. DNA quality was checked by gel electrophoresis on a 0.8 % agarose gel. PCR amplification targeting the 16S rRNA gene was carried out using the primers 16S-27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16S-1492R (5′-TACGGYTACCTTGTTA CGACTT-3′) (Lane 1991). PCR reactions were performed in a total volume of 50 μL containing: 1x Taq buffer (Invitrogen), 2.5 mM MgCl2, 0.1 mM of each dNTP, 1.75U Taq polymerase (Invitrogen), 0.4 μM of each primer, and 1 μL of extracted DNA. Thermocycling conditions were: 2 min at 94 °C; 30 cycles of 1 min at 94 °C; 1 min at 55 °C and 2 min at 72 °C; and 1 cycle of 10 min at 72 °C. PCR products were partially sequenced (between 750 and 1000 bases) using the primer 16S-27F (Lane 1991). Sequence data were checked using the Chromas v. 1.45 software (Technelysium Pty. Ltd., Australia), uploaded and aligned in the Ribosomal Database Project (RDP; Cole et al. (2009)) and assessed for similarity with sequences in the RDP. The sequences used for identification of the culturable strains are available in the EMBL database (www.ebi.ac.uk) under accession numbers HG941722 - HG942010, HE646570 - HE646571, and FN908759 - FN908797 (the latter group were identified and described for Ni mobilisation capacity by Becerra-Castro et al. 2011).
Genetic diversity of isolates was assessed for each plant species and population using the Shannon diversity index (H′ = −∑(xi/x0)ln(xi/x0)) where xi is the number of strains per genera, and x0 is the total number of strains.
Statistical analyses
Analyses of variance (ANOVA) were used to detect statistically significant differences between bacterial counts. Comparisons of prevalence of PGP phenotypes (Plant Growth Promoting traits) between plant species were achieved through a chi-squared test or Fisher’s exact test when necessary. Statistical analyses were performed using SPSS v.22.0, SPSS Inc., Chicago, IL.
Results
General soil physicochemical properties
Some general physicochemical properties of the soils at each site are given in Table 1, as well as the total concentration of Ni in aboveground shoots of each plant species studied. All four serpentine soils were characterised by pH values close to neutrality, a low C and N content, a predominance of Mg in the exchange complex, and Ca/Mg quotients of <1. In contrast, SN soil presented alkaline pH values and Ca dominating the cation exchange complex, higher organic matter content and C/N ratio. Total Ni concentrations were higher in serpentine soils than the calcareous SN soil. Values were similar in the Portuguese sites (S and M; 3.03 and 2.72 g kg−1, respectively) and lower in the L and SB soils (2.20 and 2.10 g kg−1, respectively). In contrast, total Ni in SN calcareous soil was only 0.03 mg kg−1. In accordance, labile Ni (water-soluble) was highest in the S site. Water-soluble Ni concentrations were higher in the rhizosphere soil of the M population of the Ni-hyperaccumulator compared to the non-hyperaccumulators D. glomerata (1.2-fold lower) and S. semidentata (2.2-fold lower) or the non-vegetated soil (3.1-fold lower). Also the rhizosphere soil of the Ni-hyperaccumulator in SB presented a higher concentration of water-soluble Ni than the non-vegetated soil (2.1-fold lower).
Culturable bacterial densities and the rhizosphere effect
The densities of culturable bacteria in non-vegetated and rhizosphere soils of the different plant species and sites are presented in Table 2. Bacterial densities in non-vegetated serpentine soils did not differ significantly, and ranged from 3.7 × 107 CFU g−1 DW soil in L to 7.9 × 107 CFU g−1 DW soil in SB (Tukey post-hoc test showed no differences between NV serpentine soils; p > 0.05). On the other hand, bacterial densities in non-vegetated soils of SN were at least 10-fold higher than those of the serpentine sites (5.2 × 108 CFU g−1 DW soil).
The effect of the plant on bacterial densities (rhizosphere effect) was assessed using the R / NV ratio (CFUs g−1 rhizosphere soil / CFUs g−1 non-vegetated soil) (Table 2). Bacterial densities were higher (albeit not always significantly) in the rhizosphere of all the plant species compared to non-vegetated soil, with the exception of D. glomerata in M (R / NV of 0.9). The highest densities were always associated with the Alyssum subspecies, and this was the case for all five populations. The increase in bacterial densities in the rhizosphere ranged from 1.3-fold in the S population of D. glomerata to 4.0-fold in the L population of A. pintodasilvae. The most pronounced rhizosphere effect was observed in the L population of A. pintodasilvae (R / NV ratio of 4; p < 0.001), where densities were up to one order of magnitude higher in the rhizosphere soil compared to non-vegetated soil. On the other hand, the biggest differences in rhizosphere bacterial densities between different plant species growing at the same site were observed in M. In this case the bacterial densities associated with A. pintodasilvae were 1.7 × 108 CFU g−1 soil, which was 3.4- and 2.3-fold higher than those found in the rhizosphere soil of D. glomerata (7.5 × 107 CFU g−1 soil; p < 0.05) and S. semidentata (5.0 × 107 CFU g−1 soil; p < 0.001), respectively. In fact, the rhizosphere bacterial densities associated with S. semidentata from this site (5.0 × 107 CFU g−1 soil) were similar to those determined in non-vegetated soil (5.5 × 107 CFU g−1 soil).
Nickel resistance of the culturable bacterial community
Bacterial densities of soils (NV and rhizospheric) plated in culture media supplemented with Ni decreased with an increase in the medium Ni concentration, and this was observed for all plant species and populations (Table 2). Compared to the total bacterial population, densities of Ni-resistant bacteria in serpentine soils were reduced by at most one order of magnitude at the highest metal concentration tested. In contrast, in SN, densities were reduced by up to three orders of magnitude: in non-vegetated soil, densities decreased from 5.2 × 108 CFU g−1 soil (0 mM Ni) to 6.0 × 105 CFU g−1 soil at the highest Ni concentration (3 mM) and in A. serpyllifolium rhizosphere soil they fell from 1.1 × 109 CFU g−1 soil (0 mM Ni) to 1.6 × 106 CFU g−1 soil (3 mM Ni).
As observed for the total culturable bacterial population, the Ni-resistant population (CFU g−1 soil) was highest in the rhizosphere of the Ni-hyperaccumulator compared to non-vegetated soil, these plants also tended to present higher R / NV ratios compared to non-accumulators. A strong rhizosphere effect (R / NV ratios >2) was not observed for any population of S. semidentata or D. glomerata (except for the L population of D. glomerata; Table 2). The M population of A. pintodasilvae hosted up to 6-fold higher densities of Ni-resistant bacteria in culture medium with 2 mM Ni compared to non-vegetated soil (R / NV of 6.1), while the R / NV ratios for the same population of D. glomerata and S. semidentata were at most 1.5. At the same site the abundance of Ni tolerant bacteria was also significantly higher in the rhizosphere of the hyperaccumulator than in the rhizosphere of S.semidentata or D. glomerata. In the SB population of A. malacitanum Ni-resistant bacterial densities were 5-fold higher than those of non-vegetated soil in media supplemented with either 2 or 3 mM Ni (Table 2).
In most cases the presence of the plant did not affect bacterial Ni tolerance. For example, in the case of L no differences were observed between the % Ni-resistant bacteria associated with the different plant species and the non-vegetated soil. In S, at the two lower concentrations of Ni (0.5 and 1 mM) the rhizosphere of all three plant species harboured a lower % of Ni-tolerant bacteria compared to non-vegetated soil. However, in the case of the Ni-hyperaccumulators A. pintodasilvae (M population) and A. malacitanum (SB population), these plants harboured a higher proportion of Ni-tolerant bacteria than their corresponding non-vegetated soils: increasing from 19.6 % in non-vegetated soil to 36.8 % in the rhizosphere of the M population of A. pintodasilvae (at 2 mM Ni) and from 19.5 to 27 % in the SB population of A. malacitanum (at 1 mM Ni) (Table 2).
Phenotypic characteristics of the bacterial isolates
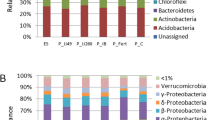
A total of 550 different colony morphotypes (74 associated to LA, 64 to SA, 54 to MA, 95 to SBA, 48 to LG, 41 to SG, 44 to MG, 43 to SS, 45 to MS, and 42 to SNA) were isolated and analysed for their plant growth promoting traits and Ni tolerance (MTC). Figure 1a summarises the frequency of PGP traits found amongst the rhizobacterial isolates. All of the plant species harboured rhizobacteria which were capable of producing organic acids. This was in fact the most common trait amongst the collection of rhizobacteria: 29.3 % of the isolated strains were characterised as organic acid-producers. Depending on the plant species or population between 12 and 61 % of the isolates were organic acid-producers. The highest numbers of organic acid-producers were associated with S. semidentata (51.2 to 62.2 % of the isolates from this plant species showed this trait), followed by the Alyssum subspecies (24.3 to 37.5 %) and finally, D. glomerata (12.2 to 15.9 %). The ability to produce IAA was also present in all plant species and represented 23.1 % of the total of number of isolates. IAA-producers were most common in the L, S and M serpentine sites. Similar proportions of IAA-producers were found for isolates of A. pintodasilvae and D. glomerata in L (31.1–39.6 %) and S (21.9–24.4 %) populations. In M a higher % of IAA-producers were found amongst isolates of A. pintodasilvae (44.4 %), and a similar % was obtained from isolates of either D. glomerata (20.5 %) or S. semidentata (17.8 %). IAA-producers were less frequently associated with either A. malacitanum from SB (9.5 % of isolates) or A. serpyllifolium from SN (14 %). The mean production of IAA (mg L−1) by strains varied from 5.3 in SA to 48.0 in LG, however, a high variability was found between isolates from each site. The maximum IAA production was recorded in the population of D. glomerata, from Barazón, where three isolates showed IAA production higher than 200 mg L−1 (247.9 mg L−1 for strain LG105; 315.9 mg L−1 for strain LG120 and 211.4 mg L−1 for strain LG121). In addition, one strain associated with the grass species in Samil also showed IAA production (up to 118.2 mg L−1; strain SG5), while in the case of the hyperaccumulator, the maximum IAA production was recorded in A. pintodasilvae from Barazón (136.2 mg L−1; strain LA 24).
a Frequency of plant growth promoting (PGP) traits presented by isolates obtained from the different populations of each plant species (presented as a % of the total number of isolates per group). b Maximal Tolerable Concentration (MTC) of bacteria isolated from the different rhizosphere soils (presented as a percentage of the total number of isolates in each group). The metal resistance was tested using 284 agar medium supplemented with 1, 2.5, 5 or 10 mM Ni added as NiSO4 and incubated at 28 °C for 7 days. LA (A. pintodasilvae from Barazon), LG (D. glomerata from Barazon), SA (A. pintodasilvae from Samil), SG (D. glomerata from Samil), SS (S. semidentata from Samil), MA (A. pintodasilvae from Morais), MG (D. glomerata from Morais), MS (S. semidentata from Morais), SBA (A. malacitanum from S. Bermeja) and SNA (A. serpyllifolium from S. Nevada)
Siderophore-producing strains were found in the rhizosphere of all the Alyssum populations studied except in A. pintodasilvae from M. The frequency of isolates showing this characteristic varied between 6.8 % in L and 33.3 % in SN. The siderophore-producing ability was abundant in isolates from the rhizosphere of D. glomerata collected in M (51.2 %) and S (21.7 %). In contrast, in the rhizosphere of S. semidentata these were much less frequent (only 2.3 % of isolates in S).
The abilities to solubilise mineral phosphate or to produce biosurfactants were relatively rare traits amongst the bacterial isolates, representing 4.5 and 0.5 % of the total number of the isolates, respectively. P solubilisers were not found in isolates from D. glomerata but were isolated from the rhizosphere of the Alyssum and S. semidentata populations (except in L). In Alyssum between 4.8 and 11.6 % of the isolates were able to solubilise P while in S. semidentata this characteristic represented 2.2 and 4.6 % of the isolates. Biosurfactant production was only detected amongst 3 rhizobacteria isolated from A. malacitanum (SB), S. semidentata (of M) and D. glomerata (of S), and the percentage was never greater than 3 %.
Figure 1b shows the frequency of isolates (as a % of the total number of isolates in each group) for each MTC. All of the serpentine plant species harboured rhizobacteria capable of growing in all the tested Ni concentrations. However, the level of Ni tolerance depended on both the soil type and the plant species. About 90 % of rhizobacterial isolates associated with the non-hyperaccumulator (SN) were only able to grow in medium without Ni (MTC < 1 mM), while for the plant species growing in the serpentine sites from 4.2 to 18.5 % of rhizobacteria presented a MTC of 10 mM. Of the rhizobacterial isolates associated with non-hyperaccumulators species growing on serpentine sites the majority presented MTC values < than 1 mM Ni. For instance, most of the isolates associated with the Ni-excluder D. glomerata (66–73 %) did not tolerate Ni concentrations ≥2.5 mM, while in the rhizosphere of the Ni-hyperaccumulators this percentage ranged from 36 to 51 %.
Within the Ni-hyperaccumulators, only 9.4 % of strains associated with the S population of A. pintodasilvae were unable to grow in the presence of Ni in the growth medium. The same population presented the highest percentage (45.3 %) of associated microorganisms with a MTC of 2.5 mM Ni. While the L population presented the highest number of isolates with an MTC of 5 mM (30 %) and the M population of 10 mM (18.5 %). Similarly, it was the Ni-hyperaccumulators which presented the highest numbers of associated bacteria with an MTC of 10 mM Ni (6.8, 10.9 and 18.5 % in the L, S and M sites, respectively). Nonetheless, a high percentage (at times similar or even higher than the Ni-hyperaccumulators) of isolates associated with the non-hyperaccumulator S. semidentata also presented elevated Ni MTC values (up to 5 or 10 mM).
Online resource 1 summarises the frequency of each phenotype within the collection of isolates. The most frequent phenotype was LT-N (47 % in L, 25 % in M, 28 % in S, 31 % in SB and 38 % in SN), which represents low Ni tolerance (up to 2.5 mM Ni MTC) and no PGP trait. This was followed in the serpentine sites, by HT-N (high Ni tolerance but no PGP trait) (18 % in L, 16 % in M, 12 % in S, 23 % in SB) and by LT-A in the SN site (17 %). Phenotypes A and H (which correspond with organic acid- and IAA-producers respectively), were also frequently found in LT strains (up to 18 % in L) but were less often represented by HT strains (up to 10 % in S). Phenotypes presenting three PGP traits (phenotypes APH, ASH and PSH) were rarely found (at most 2 % of strains). In general, all phenotypes were represented in both ranges of metal tolerance (LT and HT), except phenotypes T and APH which were exclusive to HT isolates and phenotypes AP, PH, ST and PSH which were only present in LT isolates. Strains isolated from L showed a maximum of two PGP traits (HT-SH, siderophore- and IAA-producers; Online resource 1). Whereas in S, M and SB several strains with more than two PGP traits were found (phenotypes APH, ASH and PSH); and these were mainly associated with the Ni-hyperaccumulator. Rhizobacterial strains associated with Alyssum ssp. generally presented a higher diversity in phenotypes than the other plant species. For example, in the L population A. pintodasilvae strains were allocated into 12 different phenotypes, while only five phenotypes were found in rhizobacteria from the gramineae growing in the same site, and none of these were specific to the grass species. In the Portuguese (M and S) populations, phenotype distribution depended on the plant species. In M, phenotypes LT-AP and HT-APH were associated with A. pintodasilvae, while HT-S, LT-AS and LT-SH (all of which include siderophore-producers) with D. glomerata, and finally, HT-T with S. semidentata. In S, the 18 phenotypes were distributed among the plant species as follows: A. pintodasilvae (15 groups) > D. glomerata (10) > S. semidentata (9); four of the groups were specific to the hyperaccumulator (LT-PH, LT-PSH, HT-AS, and HT-ASH), two to D. glomerata (LT-ST and LT-ASH) and only one (HT-APH) was specific to S. semidentata. In SB and SN, strains associated with Alyssum sp. were represented by ASH and PH phenotypes, respectively.
Identification and genotypic diversity of culturable rhizobacteria
BOX-PCR profiles
A BOX-PCR profile was obtained for 538 out of the 550 isolated bacterial strains. Profiles were analysed on a site-by-site basis (for each plant species present) and for all the Ni-hyperaccumulating Alyssum species together (Online Resource 2- Online Resource 6 and Online Resource 7). Table 3 presents a summary of the total number of BOX-PCR groups obtained in each of these analyses and the distribution of bacterial isolates amongst these groups for each site (Table 3a) and plant species (Table 3b). In the case of L the BOX-PCR profiles of isolates were distributed amongst 19 groups, strains associated with the Ni-hyperaccumulator A. pintodasilvae were distributed amongst 13 groups and those of D. glomerata in 13 groups. Twelve of these BOX-PCR groups are represented exclusively by strains of either A. pindoasilvae or D. glomerata (groups L01-L02, L04, L07-L08, L11-L12, L14 and L16-L19), while seven groups are made-up of isolates from both species (Online Resource 2). In the case of S, the BOX-PCR profiles of isolates from A. pintodasilvae and S. semidentata were distributed amongst 19 and 9 groups each, while for D. glomerata isolates were distributed amongst only 4 groups. Twelve groups were exclusively made up of isolates associated with A. pintodasilvae (S01, S02, S07, S09, S11, S12, S15, S16, S17, S18, S20 and S21), three groups with isolates of S. semidentata (S10, S19 and S22), and some groups (S04, S08 and S13) were composed of isolates from both the Ni-hyperaccumulator and S. semidentata. There were no groups solely represented by isolates of D. glomerata (Online Resource 3). In the case of plant populations of M, isolates were distributed in 24 BOX-PCR groups; A. pintodasilvae isolates were distributed amongst 17 groups, S. semidentata amongst 17 groups and D. glomerata amongst 8 groups. The groups are generally represented by isolates from all three plant species, with some exceptions being group M01 (solely represented by isolates of D. glomerata), M02, M10, M19, M23-M24 (each represented by a single isolate of A. pintodasilvae), M03, M14, M16-18 and M22 (isolates of S. semidentata) (Online Resource 4). Rhizobacterial strains isolated from A. malacitanum (from SB) and from A. serpyllifolium (from SN) were allocated into 18 and 10 BOX-PCR groups, respectively (Online resource 5 and 6).
BOX-PCR profiles of strains associated with the two Alyssum subspecies (from L, M, S, SB) were allocated into 40 groups (Online resource 7). Approximately half of these (19 groups) were made up of strains from various populations. On the other hand, some groups were predominantly represented by isolates from a single population. This was the case for groups A36 or A37 which were principally composed of strains isolated from the SB population of A. malacitanum, or for groups A08, A14, A20, A23 and A38 which were made up of strains almost exclusively isolated from the Portuguese serpentine populations (M or S) of A. pintodasilvae. BOX-PCR group A18 represents isolates associated with A. pintodasilvae from both the Spanish population (L) and Portuguese populations (M and S) (Online Resource 7). In general no correlation was found between BOX- and phenotypic-groups, however some phenotypes were shown to be specific to a particular BOX-group. Phenotype SH was specific to the L05 BOX-group in Barazón (Online Resource 2), phenotype APH was specific to group M08 in Morais and to S06 in Samil (Online Resource 4 and 3), and finally, phenotype ASH was specific to group S05 in Samil (Online Resource 3). In SB, phenotype PSH was associated with SB09 and in SN phenotype ASH was specific to SN07 (Online Resource 5 and 6, respectively). Regarding the Ni-hyperaccumulators, only phenotype ASH (organic acid-, siderophore- and IAA-producer) was found exclusively in the BOX group A36 (Online Resource 7).
The Shannon diversity index (H′) is presented in Online resource 8. Diversity indices were calculated separately for isolates presenting low (LT) and high (HT) Ni tolerance. No correlation was found between plant species and bacterial diversity of isolates with low Ni tolerance (LT), the highest H′ values were recorded in the bacterial community associated with the M population of S. semidentata. (H′ = 1.04) and the S population of A. pintodasilvae (H′ = 0.95). However, bacterial diversity of isolates with high Ni tolerance (HT), tended to be higher in the rhizosphere of the Ni-hyperaccumulating species than the non-accumulators. The highest index was recorded in the bacterial population of A. pintodasilvae growing in Morais (H′ = 0.96) while that of the excluder D. glomerata growing at the same site presented the lowest bacterial diversity (H′ = 0.58).
16S rDNA identification
A total of 329 isolates were identified through comparative sequence analysis of 16S rDNA sequences, and isolates were affiliated with a total of 24 different genera belonging to four phyla. The Shannon diversity (H′) index was also calculated on the basis of the 16S rDNA sequencing. The highest value was found in the SN population of A. serpyllifolium (1.9), while for the serpentine populations, the highest diversity was associated with the Ni-hyperaccumulators, and particularly the S population of A.pintodasilvae (1.7) (Online resource 8).
Figure 2 presents the taxonomic breakdown for each plant species and population. Four different phyla were represented in the bacterial collection, two phyla correspond with Gram-positive bacteria (Actinobacteria and Firmicutes) and two with Gram-negative (Bacteroidetes and Proteobacteria). The distribution of isolates within these phyla differed according to the soil type, 94 % of the isolates obtained from serpentine soils (L, S, M, SB) were Gram-positive bacteria and 6 % were Gram-negative (data not shown). In contrast, in the calcareous soil (SN) Gram-positive bacteria represented 77 % of the total community and Gram-negative 23 %. Proteobacteria or Actinobacteria were the most diverse taxonomic classes (including 9 and 11 different genera, respectively).
Taxonomic breakdown of 16 s rDNA sequences a 35 sequences from Alyssum pintodasilvae (Barazón), b 25 from Dactylis glomerata (Barazón), c 35 from Alyssum pintodasilvae (Morais), d 26 from Dactylis glomerata (Morais), e 28 from Santolina semidentata (Morais), f 50 from Alyssum pintodasilvae (Samil), g 30 from Dactylis glomerata (Samil), h 30 from Santolina semidentata (Samil), i 42 from Alyssum malacitanum (S. Bermeja), j 26 from Alyssum serpyllifolium (S. Nevada). The central pie shows percentages by phyla; each outer ring progressively breaks these down to finer taxonomic levels (phyla, class, family, genera)
Differences in bacterial diversity were observed regarding the distribution of phyla between plant species and populations. For the M population of S. semidentata (MS) and the S population of D. glomerata (SG) 100 % of the isolates were affiliated with the phylum Actinobacteria (Fig. 2e and g). For the Ni-hyperaccumulators, the proportion of isolates belonging to the Actinobacteria decreased in the order SB (95 %) (Fig. 2i), S (90 %) (Fig. 2f), L (88.6 %) (Fig. 2a) and M (74.3 %) (Fig. 2c). Actinobacteria was least represented in the SN population of A. serpyllifolium, although 63 % of isolates were still affiliated with this phylum (Fig. 2j). The second most important phylum was Proteobacteria (Alpha- and Gamma-proteobacteria), particularly in the SN population of A. serpyllifolium (25.9 % of isolates belonged to this phylum) (Fig. 2j). The phylum Firmicutes was primarily associated with the Alyssum subspecies (11.4 % of isolates in M, 10.0 % in S, 5 % in SB and 11.1 % in SN) (Fig. 2c, f, i and j), while isolates belonging to the Bacteroidetes were only found in the L population of A. pintodasilvae (5.7 % of isolates) (Fig. 2a).
Regarding the distribution of genera amongst the bacterial isolates associated with each plant species (considering all their populations together), the Ni-hyperaccumulators showed a higher diversity and hosted members of 19 different genera, while only 9 genera were associated with the non-hyperaccumulating A. serpyllifolium. The rhizosphere community of D. glomerata and S. semidentata were represented by members of 10 and 6 genera, respectively (data not shown). Some bacterial genera, such as Arthrobacter, Streptomyces and Rhodococcus (all members of the Actinobacteria), were found in the rhizosphere of all the plant species (Fig. 2a to j). In contrast, some genera were only isolated in specific plant species: members of the genera Stenotrophomonas (Gammaproteobacteria), Methylobacterium (Alphaproteobacteria), Staphylococcus (Firmicutes), Amycolatopsis and Mycobacterium (both Actinobacteria) and the phylum Bacteroidetes (exclusively represented by the genera Olivibacter and Chitinophaga) were only found in association with the Ni-hyperaccumulators (either A. pintodasilvae or A. malacitanum) (Fig. 2a, c, f and i). On the other hand, members of the genera Janibacter (Actinobacteria) and Mesorhizobium and Aminobacter (Alphaproteobacteria) were exclusively isolated from the rhizosphere of D. glomerata (specifically its L population, Fig. 2b), and Enterobacter (Gammaproteobacteria) from S. semidentata (S population, Fig. 2h). Finally, members of two genera, Pseudomonas and Rhizobium (Gamma- and Alpha-proteobacteria, respectively), were only found in the rhizosphere of the non-hyperaccumulator A. serpyllifolium (Fig. 2j).
Amongst the Ni hyperaccumulators the predominant genera represented by isolates associated with the L population of A. pintodasivae (Fig. 2a) were Arthrobacter (48.6 %) and Amycolatopsis (25.7 %). While the main genera associated with the two Portuguese populations of A. pintodasilvae were (Fig. 2c and f): Arthrobacter (37.1 % in M and 20 % in S) and Streptomyces (37.1 % in M and 32.0 % in S). Similarly, the rhizosphere community of A. malacitanum (SB) (Fig. 2i) was predominantly represented by Arthrobacter (31.0 %) and Streptomyces (40.5 %). The latter two genera were also important components of the rhizobacterial community of the non-hyperaccumulator A. serpyllifolium, but in this case 18.5 % of isolates were identified as Pseudomonas. As observed for the hyperaccumulators, members of the genera Arthrobacter and Streptomyces were predominant in the rhizosphere of both D. glomerata and S. semidentata: 60 % of isolates of the L population of D. glomerata were affiliated with Arthrobacter, while 53 % and 75 % of isolates from S and M populations of D. glomerata and S. semidentata, respectively, were affiliated with Streptomyces. Members of the genera Nocardia were generally found in association with D. glomerata from Portuguese sites (M and S), representing 19.2 % and 16.7 % of isolates, respectively.
Discussion
This study describes the culturable rhizosphere bacterial community associated with different populations of the only two Ni-hyperaccumulating subspecies of Alyssum serpyllifolium in the Iberian Peninsula. The Ni tolerance, PGP traits and diversity of these communities were compared with those of non-hyperaccumulating plants growing at the same sites and with a close relative and non-hyperaccumulator, A. serpyllifolium, growing in calcareous soils. Trace metals are well known to affect the soil bacterial density and activity, as well as the community structure and diversity (Bååth 1989). A high soil metal content has been suggested to explain the lower bacterial densities observed in serpentine soils compared to nearby non-serpentine soils (Amir and Pineau 1998; Pal et al. 2005). Higher bacterial densities in the non-serpentine (SN) soil were therefore to be expected. Toxic concentrations of trace metals were not detected in this calcareous soil and the higher organic matter content should favour microbial development (Acea and Carballas 1986). In accordance, bacteria from the serpentine soils (L, M, S, SB) showed a higher resistance to Ni than those from the calcareous (SN) soil. An increase in the Ni concentration of the culture medium led to a significant decrease in bacterial densities of all soils. However, this decrease was less pronounced in the serpentine soils, indicating their adaptation to the phytotoxic levels of Ni to which they are normally exposed. Microorganisms from serpentine soils have previously been shown to present a high level of tolerance to trace metals, such as Ni, Co and Cr (Becerra-Castro et al. 2009; Pal et al. 2005; Schlegel et al. 1991; Turgay et al. 2012).
Plant root exudates, secretions and lysates contain labile C sources and growth factors which are well known to stimulate microbial growth and metabolic activity (Delorme et al. 2001; Grayston et al. 1998). Bacterial densities were therefore unsurprisingly higher in the rhizosphere soils of all the plant species studied compared to corresponding non-vegetated soils (R / NV ratios > 1). This rhizosphere effect was most pronounced in the case of Alyssum (in both the Ni-hyperaccumulators and non-hyperaccumulator). The Ni-hyperaccumulating A. pintodasilvae and A. malacitanum also hosted a higher density of Ni-resistant bacteria in their rhizosphere than either the non-accumulating plants from serpentine sites or non-vegetated soil. This effect was not observed in the case of the non-hyperaccumulator A. serpyllifolium (SN). Population-specific differences were observed in the case of A. pintodasilvae, and the highest densities of Ni-resistant bacteria were associated with the M population. In addition, both the M population of A. pintodasilvae and the SB population of A. malacitanum showed a selective enrichment of Ni-resistant bacteria in the rhizosphere. This has been observed for other metal-hyperaccumulating plants, such as Alyssum bertolonii, Noccaea goesingense and Sebertia acuminata (Lodewyckx et al. 2002; Mengoni et al. 2001, 2010; Schlegel et al. 1991). Likewise, Zn tolerance has often been described in bacterial communities exposed to this metal: Delorme et al. (2001) found higher densities of Zn-tolerant bacteria in the rhizosphere of the Zn-hyperaccumulator N. caerulescens compared to the non-accumulator Trifolium pratense or to non-vegetated soil. Moreover, bacteria isolated from serpentine soils have been shown to present co-resistance to various metals, including Cr, Co and Zn (Mengoni et al. 2010). It would also have been interesting to study root architecture of the different plant species and in particular to compare those of the different populations of Alyssum. Root architecture and proliferation would influence exudation and therefore also the rhizosphere effect.
Trace metal uptake by hyperaccumulators has been found to be associated with partial depletion of labile, easily bioaccessible trace metal pools in the rhizosphere (e.g. Fitz et al. 2003; Puschenreiter et al. 2005; Whiting et al. 2001) and active root proliferation towards soil patches rich in metals (Schwartz et al. 1999). Such depletion of labile metal pools has often been associated with sustained or even enhanced metal solubility (i.e. soil solution concentration) due to a more intense weathering of Ni-bearing silicates in the rhizosphere (Kidd et al. 2009; Puschenreiter et al. 2005). In agreement, water-soluble Ni concentrations in the rhizosphere soils of the Ni-hyperaccumulating Alyssum from M and SB were significantly higher than those detected in the rhizosphere soil of non-accumulators (e.g. D. glomerata or S. semidentata) or of non-vegetated soil. In addition, previous studies have also shown a higher labile Ni fraction (assessed via sequential extraction procedures or Sr(NO3)2-extractable concentrations) in the rhizosphere soil of these Alyssum populations compared to non-vegetated soils or non-accumulators (Becerra-Castro 2006; Cabello-Conejo 2015). This tendency towards a higher Ni mobility in the rhizosphere of the hyperaccumulator could be attributed to the activity of the Ni-tolerant bacteria and at the same time lead to their further enrichment in the rhizosphere. Of the 508 isolates obtained from serpentine soils the vast majority (65 %) were able to growth with the presence of Ni in the medium (with concentrations > 1 mM Ni). In contrast, 90 % of isolates from the close relative of the Ni-hyperaccumulator, A. serpyllifolium were affected by the presence of the minimal concentration of Ni tested (1 mM) and none of them were able to tolerate more than 2.5 mM. Our results are in agreement with previous studies: Abou-Shanab et al. (2003) found the majority of rhizobacteria isolated from serpentine soils in Oregon (USA) were able to grow in the presence of 8 mM Ni, and Mengoni et al. (2001) found bacterial isolates from serpentine soils in Tuscany (Italy) resisted between 7 and 10 mM Ni in their growth medium. These authors did not study the Ni resistance of bacterial isolates associated with non-accumulators growing at the same site. Here the MTC for isolates associated with the hyperaccumulators was higher than that of the Ni-excluder D. glomerata, which could reinforce the idea that the activity of these hyperaccumulator plants leads to an increase in labile Ni and hence enrichment in Ni-tolerant bacteria or that this plant group selects for Ni-tolerant bacteria which in turn modify soil Ni availability. However, Ni-resistance was an equally important phenotype of the bacterial isolates associated with the serpentinophyte S. semidentata (a similar % of isolates showed Ni MTC of 10 mM Ni), indicating that this may not be a phenomenon specific to the hyperaccumulators.
The potential application of these Ni-tolerant plant-associated bacteria in phytoextraction (or phytomining) processes is based on their ability to improve plant growth and biomass production and/or modify soil metal availability and plant uptake. Bacterial strains for bioaugmentation trials are frequently selected on the presence of PGP traits (such as the production of IAA, ACC deaminase or siderophore production) or their ability to release compounds which could potentially modify metal bioavailability (such as the production of biosurfactants, siderophores or organic acids) (Abou-Shanab et al. 2003; Braud et al. 2006; Cabello-Conejo et al. 2014; Idris et al. 2004). Half of the isolates obtained in this study showed at least one PGP trait (54 % of isolates). The rarest phenotypes were the biosurfactant-producers and the ability to solubilise inorganic phosphate. Biosurfactant-producing strains were only detected amongst isolates from A. malacitanum, D. glomerata or S. semidentata (and were identified as members of the genus Bacillus (Firmicutes) and Arthrobacter (Actinobacteria)). Some authors have shown that microbial produced surfactants can increase the mobility of Cd, Cu, Pb and Zn in soil, and in some cases, increase plant metal uptake (Venkatesh and Vedaraman 2012). It would be interesting to further characterise the biosurfactant-producing bacterial strains identified in this study and particularly their Ni-solubilising capacities. Isolates which were able to solubilise inorganic phosphate were primarily associated with the Ni-hyperaccumulators (at most 12 % isolates). Most of these were Actinobacteria and identified as members of the genera Arthrobacter (strains SA37, SA40, SBA82, SNA110), Streptomyces (strains SA46, SBA57, SBA89) or Rhodococcus (strains SBA86 and SBA70). One P-solubilising isolate from the M population of A. pintodasilvae was identified as Methylobacterium (Alphaproteobacteria). Phosphate-solubilising bacteria are effective in promoting plant growth and biomass by releasing P from inorganic and organic P pools through solubilisation and mineralisation (Rodrı́guez and Fraga 1999). These bacterial associations may play an important role in P mobilisation for hyperaccumulating species of Alyssum, which seems to form non or weak mycorrhizal associations since reports of arbuscular mycorrhizal symbiosis with hyperaccumulating Alyssum sp. cannot be found in the literature.
Jeong et al. (2013) inoculated Brassica juncea with strains of phosphate-solubilising Bacillus sp. According to these authors the release of organic acids and drop in soil pH led to a mobilisation of Cd and consequent increase in Cd uptake as well as an enhanced plant biomass. An increase in plant growth and P uptake were reported by Ma et al. (2010) after inoculating Ricinus communis and Helianthus annuus with the rhizobacterial strain Psychrobacter sp. SRS8. Since metal-enriched soils are characteristically deficient in essential nutrients such as P, the identification of bacterial inoculants which could potentially improve the nutritive state of the phytoextracting plant are of interest. More than 50 % of the hyperacccumulator associated P-solubilisers also showed Ni tolerance (tolerating Ni concentrations in the growth medium of ≥2.5 mM). One of these strains (Arthrobacter nicotinovorans SA40) was shown to significantly improve the biomass production of A. pintodasilvae when grown in contrasting Ni-rich soils (serpentine soils and sewage sludge-amended agricultural soils) (Cabello-Conejo et al. 2014). Strain SA40 was also characterised as a siderophore- and IAA-producer. The fact that this strain has a beneficial effect on plant growth in both naturally-rich and anthropogenic-contaminated soils is noteworthy since it could be applied to a wider range of soils with contrasting edaphic properties.
A substantial number of isolates were able to produce organic acids, siderophores or IAA, although the frequency of these traits varied according to the host plant species or population. Plant-associated bacteria have been shown to modify soil trace metal availability through the release of organic acids, such as citric, oxalic or acetic acid, which in turn can lead to an increase in plant metal uptake (Sessitsch et al. 2013). More than 50 % of organic acid-producers isolated from the hyperaccumulators or S. semidentata tolerated > 2.5 mM Ni and some of them showed additional PGP characteristics. In a previous study, Becerra-Castro et al. (2011) showed that by using the cell-free culture of 13 of the strains associated with the Alyssum sp. (included in this study) the extractable Ni concentration from serpentine soils was increased. However, these authors found no relation between the capacity for Ni mobilisation and the phenotypic traits of the strains. In a more detailed study, strain LA44 (identified as Arthrobacter nitroguajacolicus) was shown to be an efficient mobiliser of Ni from ultramafic rocks under in vitro conditions, and principally liberated Ni associated with Mn oxides through the exudation of oxalate (Becerra-Castro et al. 2013). This strain also shows intense IAA-production, is highly Ni-resistant, and in a previous study was able to increase shoot Ni concentrations in A. pintodasilvae growing in serpentine soil (likely due to an enhanced Ni phytoavailability and hence plant uptake). The strains identified in this study as P-solubilisers or organic acid-producers, and particularly those which show high Ni tolerance, would be good candidates for further studies related to Ni mobilisation.
Zaidi et al. (2006) reported that an IAA-producing Bacillus subtilis strain was able to promote the growth of Brassica juncea and thereby increased Ni extraction. In this study, IAA-producers were particularly associated with the Ni-hyperaccumulator A. pintodasilvae, while the production of siderophores was associated with strains isolated from D. glomerata. The fact that siderophore-producing bacteria were found in association with the grass species is interesting since this type of plant is known to be a highly efficient phytosiderophore producer. The presence of associated siderophore-producing bacteria could be related to competition between the bacteria and the plant for obtaining Fe. Siderophore-producing bacterial strains were mainly affiliated with the genera Streptomyces and Arthrobacter, although one isolate associated with D. glomerata was identified as Microbacterium sp. (strain SG22) and one isolate associated with A. malacitanum was identified as Mycobacterium sp. (strain SBA60). Abou-Shanab et al. (2003) found that inoculating ultramafic soils with the actinobacterial Microbacterium arabinogalactanolyticum AY509224 increased soil Ni extractability and uptake by Alyssum murale. The predominance of siderophore-producers within the isolate collection obtained from the non-hyperaccumulator (A. serpyllifolium) could be associated with the limited availability of Fe in calcareous soils (due to the pH-dependent low solubility of Fe). The appearance of a specific phenotype linked to a particular plant species also suggests a plant-driving effect in microorganism selection, while specific phenotypes associated with sites indicates that the soil conditions are important factors shaping the bacterial community.
Bacterial diversity was assessed using the BOX-PCR technique and 16S rDNA sequencing. Although the majority of BOX-PCR groups were affiliated with isolates from two or three different plant species, or from different populations of the same species, some were specific to a certain plant population or plant species. The phenotypes associated with the different BOX-PCR groups, shows that there is no correlation between BOX-patterns and phenotype, probably because of the high intra-BOX phenotypic variability. These results are in agreements with Mengoni et al. (2001), who found no correlation between OTUs obtained by ARDRA (restriction analysis of 16S) and heavy-metal tolerant phenotype in bacterial strains isolated from the rhizosphere of A. bertolonii.
To our knowledge differences in the bacterial communities associated with different populations of the same Ni-hyperaccumulating species have not previously been shown. The phyla Proteobacteria and, in particular, Actinobacteria, dominated the culturable rhizobacterial community. Isolates were affiliated with members of genera, such as Arthrobacter, Streptomyces, Rhodococcus or Microbacterium, which have been frequently described amongst soil bacteria. Bacteria of the phyla Proteobacteria (particularly Alphaproteobacteria) and Actinobacteria were also the most numerous within the rhizosphere of the Ni-hyperaccumulator Alyssum murale (Abou-Shanab et al. 2010). In the rhizosphere of Alyssum bertolonii the culturable bacterial population was dominated by Ni-resistant Pseudomonas strains (Gammaproteobacteria) (Mengoni et al. 2001). In this study, strains identified as Pseudomonas sp. were exclusively found in the rhizosphere of A. serpyllifolium growing in the calcareous soil. Culturable rhizobacteria associated with the Ni-hyperaccumulator N. goesingense were mainly represented by Methylobacterium spp., an alphaproteobacterial genus, as well as Rhodococcus spp. and Okibacterium spp., belonging to the Actinobacteria (Idris et al. 2004). Gremion et al. (2003) constructed clone libraries based on the 16S rRNA and 16S rDNA and found Actinobacteria to be a dominant part of the metabolically active bacteria in the rhizosphere of the Cd/Zn-hyperaccumulator N. caerulescens. The predominance of actinobacterial strains in serpentine soils has been related to the high adaptability of such gram-positive bacteria to toxic concentrations of trace metals (DeGrood et al. 2005). Metal toxicity or nutrient deficiency has been shown to lower the frequency of r-strategists (bacteria capable of rapid growth and utilization of resources), such as Bacillus or Pseudomonas, as these are more sensitive to toxic substances (Kozdrój 1995; Kunito et al. 2001). This could partly explain the lower frequency of r-strategists in the serpentine soils. On the other hand, despite a reduced growth and metabolic activity, k-strategists (such as the Actinomycetales) present stable populations over a long time period (making them recommended candidates for bioaugmentation purposes) (Lebeau et al. 2008). The major differences in taxonomic diversity were therefore found between serpentine and non-serpentine soils.
Since the effects of the same bacterial inoculum on plant growth and metal bioaccumulation can be both plant- (Becerra-Castro et al. 2012) and soil-specific (Cabello-Conejo et al. 2014), the selection of microorganisms adapted to the soil characteristics and an ability to colonize the rhizosphere will be a pre-requisite for successful bioaugmentation. This study generated a large number of plant-associated rhizobacterial strains with potential application in these techniques. Many were affiliated with genera which have previously been shown to be beneficial for soil metal removal processes. Although plant population-specific differences in the diversity of associated rhizobacteria were not large and the Ni-hyperaccumulators did not seem to harbour a characteristic bacterial taxonomic diversity; a small number of strains were only found in association with the Alyssum hyperaccumulators and would be interesting candidates for further studies related to the application of Ni hyperaccumulating species in phytomining. For example, strains MA98 and MA106 both identified as Stenotrophomonas sp. and IAA- and organic acid-producers, respectively; MA91 which is a P-solubiliser (identified as Methylobacterium sp.); LA24, LA22, and LA23, all of them IAA-producers (identified as Amycolatpsis sp.) or SBA60 identified as Mycobacterium sp. and siderophore-producers. Moreover, the beneficial effects of stains could also be more pronounced when these are used as a mixed inoculum. These strains will be used in future bioaugmentation trials (individually or as mixed inoculum) to evaluate their effects on the Ni phytoextraction capacity of Ni-hyperaccumulating plant species.
References
Abou-Shanab RI, Delorme TA, Angle JS, Chaney RL, Ghanem K, Moawad H, Ghozlan HA (2003) Phenotypic characterization of microbes in the rhizosphere of Alyssum murale. Int J Phytorem 5:367–379
Abou-Shanab RAI, van Berkum P, Angle JS, Delorme TA, Chaney RL, Ghozlan HA, Ghanem K, Moawad H (2010) Characterization of Ni-resistant bacteria in the rhizosphere of the hyperaccumulator Alyssum murale by 16S rRNA gene sequence analysis. World J Microbiol Biotechnol 26:101–108. doi:10.1007/s11274-009-0148-6
Acea M, Carballas T (1986) Estudio de la población microbiana de diversos tipos de suelos de zona húmeda (N.O. de España). Ann Edafol Agrobiol 45:381–398
Amir H, Pineau R (1998) Effects of metals on the germination and growth of fungal isolates from New Caledonian ultramafic soils. Soil Biol Biochem 30:2043–2054. doi:10.1016/S0038-0717(98)00079-0
Bååth E (1989) Effects of heavy metals in soil on microbial processes and populations (a review). Water Air Soil Pollut 47:335–379. doi:10.1007/BF00279331
Baker A, Brooks R (1989) Terrestrial higher plants which hyperaccumulate metallic elements. A review of their distribution, ecology and phytochemistry. Biorecovery 1:81–126
Becerra-Castro C (2006) Relacións planta – solo – microorganismo nunha especie hiperacumuladora de níquel endémica de solos serpentiníticos. B. Sc. Dissertation, Universidade de Santiago de Compostela (Spain)
Becerra-Castro C, Monterroso C, García-Lestón M, Prieto-Fernández A, Acea MJ, Kidd PS (2009) Rhizosphere microbial densities and trace metal tolerance of the nickel hyperaccumulator Alyssum serpyllifolium subsp. lusitanicum. Int J Phytorem 11:525–541. doi:10.1080/15226510902717549
Becerra-Castro C, Prieto-Fernández Á, Álvarez-López V, Monterroso C, Cabello-Conejo MI, Acea MJ, Kidd PS (2011) Nickel solubilizing capacity and characterization of rhizobacteria isolated from hyperaccumulating and non-hyperaccumulating subspecies of Alyssum serpyllifolium. Int J Phytorem 13(Suppl 1):229–244. doi:10.1080/15226514.2011.568545
Becerra-Castro C, Monterroso C, Prieto-Fernández A, Rodríguez-Lamas L, Loureiro-Viñas M, Acea MJ, Kidd PS (2012) Pseudometallophytes colonising Pb/Zn mine tailings: a description of the plant-microorganism-rhizosphere soil system and isolation of metal-tolerant bacteria. J Hazard Mater 217–218:350–359. doi:10.1016/j.jhazmat.2012.03.039
Becerra-Castro C, Kidd P, Kuffner M, Prieto-Fernández A, Hann S, Monterroso C, Sessitsch A, Wenzel W, Puschenreiter M (2013) Bacterially induced weathering of ultramafic rock and its implications for phytoextraction. Appl Environ Microbiol 79:5094–5103
Braud A, Jézéquel K, Vieille E, Tritter A, Lebeau T (2006) Changes in extractability of Cr and Pb in a polycontaminated soil after bioaugmentation with microbial producers of biosurfactants, organic acids and siderophores. Water Air Soil Pollut Focus 6:261–279
Brooks RR (1987) Serpentine and its vegetation: a multidisciplinary approach. Croom Helm, Dioscorides Press
Cabello-Conejo MI (2015) Nickel hyperaccumulating plants: strategies to improve phytoextraction and a characterisation of Alyssum endemic to the Iberian Peninsula. PhD Thesis, Universidade de Santiago de Compostela (Spain)
Cabello-Conejo MI, Becerra-Castro C, Prieto-Fernández A, Monterroso C, Saavedra-Ferro A, Mench M, Kidd PS (2014) Rhizobacterial inoculants can improve nickel phytoextraction by the hyperaccumulator Alyssum pintodasilvae. Plant Soil 379:35–50
Carballeira A, Devesa C, Retuerto R, Santillán E, Ucieda F (1983) Bioclimatología de Galicia. Fundación Pedro Barrié de la Maza, Conde de Fenosa, A Coruña
Castillo-Martín A (2000) Parque nacional de Sierra Nevada clima e hidrología. In: Canseco (ed) Parque nacional de Sierra Nevada
Chaney RL, Angle JS, Broadhurst CL, Peters CA, Tappero RV, Sparks DL (2007) Improved understanding of hyperaccumulation yields commercial phytoextraction and phytomining technologies. J Environ Qual 36:1429–1443. doi:10.2134/jeq2006.0514
Chen C-Y, Baker SC, Darton RC (2007) The application of a high throughput analysis method for the screening of potential biosurfactants from natural sources. J Microbiol Methods 70:503–510. doi:10.1016/j.mimet.2007.06.006
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi:10.1093/nar/gkn879
Cox CD (1994) Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol 235:315–329
DeGrood SH, Claassen VP, Scow KM (2005) Microbial community composition on native and drastically disturbed serpentine soils. Soil Biol Biochem 37:1427–1435. doi:10.1016/j.soilbio.2004.12.013
Delorme TA, Gagliardi JV, Angle JS, Chaney RL (2001) Influence of the zinc hyperaccumulator Thlaspi caerulescens J. & C. Presl. and the nonmetal accumulator Trifolium pratense L. on soil microbial populations. Can J Microbiol 47:773–776. doi:10.1139/cjm-47-8-773
Fitz WJ, Wenzel WW, Zhang H, Nurmi J, Štipek K, Fischerova Z, Schweiger P, Köllensperger G, Ma LQ, Stingeder G (2003) Rhizosphere characteristics of the arsenic hyperaccumulator Pteris vittata L. and monitoring of phytoremoval efficiency. Environ Sci Technol 37:5008–5014. doi:10.1021/es0300214
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39
Gómez-Zotano J, Román-Requena F, Hidalgo-Triana N, Pérez-Latorre A (2014) Biodiversity and conservation values of the serpentine ecosystems in Spain: Sierra Bermeja (Málaga province). Bol Asoc Geógr Esp 65:451–456
Grayston SJ, Wang S, Campbell CD, Edwards AC (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378. doi:10.1016/S0038-0717(97)00124-7
Gremion F, Chatzinotas A, Harms H (2003) Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ Microbiol 5:896–907. doi:10.1046/j.1462-2920.2003.00484.x
Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A (2004) Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl Environ Microbiol 70:2667–2677. doi:10.1128/aem.70.5.2667-2677.2004
Jeong S, Moon HS, Shin D, Nam K (2013) Survival of introduced phosphate-solubilizing bacteria (PSB) and their impact on microbial community structure during the phytoextraction of Cd-contaminated soil. J Hazard Mater 263(Part 2):441–449. doi:10.1016/j.jhazmat.2013.09.062
Kidd P, Barceló J, Bernal MP, Navari-Izzo F, Poschenrieder C, Shilev S, Clemente R, Monterroso C (2009) Trace element behaviour at the root–soil interface: implications in phytoremediation. Environ Exp Bot 67:243–259. doi:10.1016/j.envexpbot.2009.06.013
Kozdrój J (1995) Microbial responses to single or successive soil contamination with Cd or Cu. Soil Biol Biochem 27:1459–1465. doi:10.1016/0038-0717(95)00070-U
Kunito T, Saeki K, Nagaoka K, Oyaizu H, Matsumoto S (2001) Characterization of copper-resistant bacterial community in rhizosphere of highly copper-contaminated soil. Eur J Soil Biol 37:95–102. doi:10.1016/S1164-5563(01)01070-6
Lane D (1991) 16s/23s rRNA sequencing. In: Stackerbrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Lebeau T, Braud A, Jezequel K (2008) Performance of bioaugmentation-assisted phytoextraction applied to metal contaminated soils: a review. Environ Pollut 153:497–522. doi:10.1016/j.envpol.2007.09.015
Lodewyckx C, Mergeay M, Vangronsveld J, Clijsters H, Van der Lelie D (2002) Isolation, characterization, and identification of bacteria associated with the zinc hyperaccumulator Thlaspi caerulescens subsp. calaminaria. Int J Phytorem 4:101–115. doi:10.1080/15226510208500076
Ma Y, Rajkumar M, Vicente JAF, Freitas H (2010) Inoculation of Ni-resistant plant growth promoting bacterium Psychrobacter sp. strain SRS8 for the improvement of nickel phytoextraction by energy crops. Int J Phytorem 13:126–139. doi:10.1080/15226511003671403
Menezes de Sequeira E, Pinto da Silva AR (1992) Ecology of serpentinized areas of north-east Portugal. In: Roberts BA, Proctor J (eds) The Ecology of Areas with Serpentinized Rocks, vol 17. Geobotany. Springer Netherlands, pp 169–197. doi:10.1007/978-94-011-3722-5_7
Mengoni A, Barzanti R, Gonnelli C, Gabbrielli R, Bazzicalupo M (2001) Characterization of nickel-resistant bacteria isolated from serpentine soil. Environ Microbiol 3:691–698. doi:10.1046/j.1462-2920.2001.00243.x
Mengoni A, Schat H, Vangronsveld J (2010) Plants as extreme environments? Ni-resistant bacteria and Ni-hyperaccumulators of serpentine flora. Plant Soil 331:5–16
Mergeay M, Nies D, Schlegel H, Gerits J, Charles P, Van Gijsegem F (1985) Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162:328–334
Nautiyal CS (1999) An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett 170:265–270. doi:10.1111/j.1574-6968.1999.tb13383.x
Oline DK (2006) Phylogenetic comparisons of bacterial communities from serpentine and nonserpentine soils. Appl Environ Microbiol 72:6965–6971
Pal A, Dutta S, Mukherjee PK, Paul AK (2005) Occurrence of heavy metal-resistance in microflora from serpentine soil of Andaman. J Basic Microbiol 45:207–218. doi:10.1002/jobm.200410499
Pal A, Wauters G, Paul AK (2007) Nickel tolerance and accumulation by bacteria from rhizosphere of nickel hyperaccumulators in serpentine soil ecosystem of Andaman, India. Plant Soil 293:37–48. doi:10.1007/s11104-007-9195-7
Proctor J (1999) Toxins, nutrient shortages and droughts: the serpentine challenge. Trends Ecol Evol 14:334–335. doi:10.1016/S0169-5347(99)01698-5
Puschenreiter M, Schnepf A, Millán IM, Fitz WJ, Horak O, Klepp J, Schrefl T, Lombi E, Wenzel WW (2005) Changes of Ni biogeochemistry in the rhizosphere of the hyperaccumulator Thlaspi goesingense. Plant Soil 271:205–218. doi:10.1007/s11104-004-2387-5
Rivas-Martinez S, Rivas-Saenz S (1996–2009) Worldwide bioclimatic classification system. Phytosociological Research Center, Spain. http://www.globalbioclimatics.org
Rodrı́guez H, Fraga R (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17:319–339. doi:10.1016/S0734-9750(99)00014-2
Schlegel HG, Cosson JP, Baker AJM (1991) Nickel-hyperaccumulating plants provide a niche for nickel-resistant bacteria. Bot Acta 104:18–25. doi:10.1111/j.1438-8677.1991.tb00189.x
Schwartz C, Morel JL, Saumier S, Whiting SN, Baker AJM (1999) Root development of the zinc-hyperaccumulator plant Thlaspi caerulescens as affected by metal origin, content and localization in soil. Plant Soil 208:103–115
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi:10.1016/0003-2697(87)90612-9
Sessitsch A, Kuffner M, Kidd P, Vangronsveld J, Wenzel WW, Fallmann K, Puschenreiter M (2013) The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol Biochem 60:182–194. doi:10.1016/j.soilbio.2013.01.012
Sheng X-F, Xia J-J, Jiang C-Y, He L-Y, Qian M (2008) Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ Pollut 156:1164–1170. doi:10.1016/j.envpol.2008.04.007
Turgay OC, Gormez A, Bilen S (2012) Isolation and characterization of metal resistant-tolerant rhizosphere bacteria from the serpentine soils in Turkey. Environ Monit Assess 184:515–526. doi:10.1007/s10661-011-1984-z
Venkatesh NM, Vedaraman N (2012) Remediation of soil contaminated with copper using rhamnolipids produced from Pseudomonas aeruginosa MTCC 2297 using waste frying rice bran oil. Ann Microbiol 62:85–91. doi:10.1007/s13213-011-0230-9
Versalovic J, Schneider M, De Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Whiting SN, Leake JR, McGrath SP, Baker AJM (2001) Assessment of Zn mobilization in the rhizosphere of Thlaspi caerulescens by bioassay with non-accumulator plants and soil extraction. Plant Soil 237:147–156
Zaidi S, Usmani S, Singh BR, Musarrat J (2006) Significance of Bacillus subtilis strain SJ-101 as a bioinoculant for concurrent plant growth promotion and nickel accumulation in Brassica juncea. Chemosphere 64:991–997. doi:10.1016/j.chemosphere.2005.12.057
Acknowledgments
This research was supported by the Spanish Ministerio de Economía y Competitividad (CTM2012-39904-C02-01) and FEDER, and by the 7th Framework Program of the European Commission (FP7-KBBE-266124, GREENLAND).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Antony Van der Ent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
(DOCX 29 kb)
Online resource 2
(DOCX 24 kb)
Online resource 3
(DOCX 27 kb)
Online resource 4
(DOCX 28 kb)
Online resource 5
(DOCX 22 kb)
Online resource 6
(DOCX 19 kb)
Online resource 7
(DOCX 48 kb)
Online resource 8
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Álvarez-López, V., Prieto-Fernández, Á., Becerra-Castro, C. et al. Rhizobacterial communities associated with the flora of three serpentine outcrops of the Iberian Peninsula. Plant Soil 403, 233–252 (2016). https://doi.org/10.1007/s11104-015-2632-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2632-0