Abstract
Leaf angle is a key factor in plant architecture and crop yield. Brassinosteroids (BRs) regulate many developmental processes, especially the leaf angle in monocots. However, the BR signalling pathway is complex and includes many unknown members. Here, we propose that Oryza sativa BRASSINOSTEROID-RESPONSIVE LEAF ANGLE REGULATOR 1 (OsBLR1) encodes a bHLH transcription factor, and positively regulates BR signalling to increase the leaf angle and grain length in rice (Oryza sativa L.). Lines overexpressing OsBLR1 (blr1-D and BLR1-OE-1/2/3) had similar traits, with increased leaf angle and grain length. Conversely, OsBLR1-knockout mutants (blr1-1/2/3) had erect leaves and shorter grains. Lamina joint inclination, coleoptile elongation, and root elongation assay results indicated that these overexpression lines were more sensitive to BR, while the knockout mutants were less sensitive. There was no significant difference in the endogenous BR contents of blr1-1/2 and wild-type plants. These results suggest that OsBLR1 is involved in BR signal transduction. The blr1-D mutant, with increased cell growth in the lamina joint and smaller leaf midrib, showed significant changes in gene expression related to the cell wall and leaf development compared with wild-type plants; furthermore, the cellulose and protopectin contents in blr1-D were reduced, which resulted in the increased leaf angle and bent leaves. As the potential downstream target gene of OsBLR1, the REGULATOR OF LEAF INCLINATION1 (OsRLI1) gene expression was up-regulated in OsBLR1-overexpression lines and down-regulated in OsBLR1-knockout mutants. Moreover, we screened OsRACK1A as an interaction protein of OsBLR1 using a yeast two-hybrid assay and glutathione-S-transferase pull-down.
Key message
The overexpression of OsBLR1 caused increased leaf angle and grain length, whereas OsBLR1-knockout mutants had the opposite phenotypes. OsBLR1 is a member in BR signalling pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As one of the most important crops worldwide, rice is a model species for studying monocots. Plant architecture is a key factor in rice yield (Khush and Gupta 2013). The leaf angle, which is the angle between the stem and adaxial side of the blade, is a key component of rice leaf architecture. Erect leaves maximise their carbon earnings through more effective absorption of photosynthetic residues (Zhu et al. 2008; Song et al. 2013). In addition, crops with erect leaves can grow normally at increased plant densities with no additional fertiliser, increasing yield and economic benefits (Sinclair and Sheehy 1999). Recent studies have shown that plant hormones such as brassinosteroids (BRs), auxin, and gibberellin are the main factors that regulate leaf angle (Luo et al. 2016).
BRs, as a class of steroid hormones, are widespread, have high physiological activity, and regulate a series of developmental and physiological processes, including cell differentiation, reproductive development, photomorphogenesis, and stress responses (Bishop and Koncz 2002; Fukuda 2004). In rice, BRs positively regulate grain size and leaf angle (Zhang et al. 2014a). BR signal transduction has been studied systematically in Arabidopsis thaliana. BRI1 recognises the BR signal, which causes separation from BKI1 (Nam and Li 2002; Wang and Chory 2006), and then the BRI1 Associated receptor Kinase 1 (BAK1) binds to BRI1, and the phosphorylated BRI1-BAK1 complex activates BR-signalling kinases (BSKs), which causes the activated BSKs to phosphorylate BSU1 (Tang et al. 2008). On dephosphorylation of BSU1, BIN2 loses its phosphorylation activity on the transcription factor BZR1/BES1, which leads to the accumulation of BZR1 and BES1 in the nucleus (Ryu et al. 2007, 2010). These two key transcription factors ultimately regulate the expression of downstream genes and affect plant development (Wang et al. 2002; Yin et al. 2002). In rice, some BR signalling proteins have been studied. The BR signal is activated by the dephosphorylation of GSK3/SHAGGY-like kinase (OsGSK2, a homologue of BIN2 in rice), which releases its suppression on BRASSINAZOLE-RESISTANT1 (OsBZR1) and DWARF AND LOW-TILLERING (OsDLT) and ultimately increases the leaf angle (Bai et al. 2007; Tong and Chu 2009; Tong et al. 2012). However, the BR signalling pathway is complex, and the functions of many BR-related proteins in rice are unclear.
The bHLH transcription factor family proteins have diverse functions in plants and are involved in growth, stress resistance, biosynthesis, and signal transduction (Jiang et al. 2009; Oikawa and Kyozuka 2009; Ogo et al. 2010). The typical bHLH transcription factor has a highly conserved domain, which comprises approximately 60 amino acids with two functionally distinct regions: the DNA binding region (basic region) and the helix1-loop-helix2 region (HLH region) (Atchley et al. 1999). bHLH transcription factors play an important role in regulating BR signalling. In Arabidopsis thaliana, BES1 interacts with BIM1 (bHLH protein) to bind the E-box of target genes and promote target gene expression (Yin et al. 2002). AIF2 (bHLH protein) interacts with BIN2 to participate in the BR signalling pathway and the overexpression of AIF2 represses the expression of growth-promoting genes, which results in growth retardation (Kim et al. 2017). In rice, ILI1 dimerises with IBH1 (bHLH protein) to regulate the leaf angle under BR induction (Zhang et al. 2009). OsBUL1 (HLH protein) interacts with OsBC1 (bHLH protein) and LO9-177 to form a heterotrimer complex, which responds to BR signalling and regulates leaf angle in rice (Jang et al. 2017). There are approximately 167 members of the bHLH transcription factor family in rice (Li et al. 2006), and studies on the relationships between BR and bHLH proteins are insufficient. Research to identify new bHLH proteins in BR signalling is essential for a deep understanding of BR.
The Receptor for Activated C-Kinase 1 (RACK1) protein family contains the tryptophan-aspartate domain WD40 (Guo and Chen 2008). In plants, RACK1 regulates a series of physiological processes, including seed germination, root and leaf growth, and stress responses (Ullah et al. 2008; Guo et al. 2009). RACK1 family members include RACK1A, RACK1B, and RACK1C, of which RACK1A is the most important (Guo and Chen 2008). RACK1A is involved in many hormone signal pathways in Arabidopsis thaliana (Islas-Flores et al. 2015). The RACK1A mutant is insensitive to gibberellin and BR and exhibits a variety of developmental defects affecting seed germination, leaf production, and flowering (Chen et al. 2006). In rice, OsRACK1A (Os01g0686800) regulates seed germination by taking part in abscisic acid and H2O2-mediated signalling (Zhang et al. 2014b). Recently, it was reported that rice OsRACK1A mutants are shorter (Chen et al. 2019). However, the study of the multifunctional protein OsRACK1A is at an early stage and needs more research in rice.
In this study, we revealed that OsBLR1 (Os02g0705500), as a typical bHLH protein (Li et al. 2006), is a positive regulator in BR signalling. The overexpression of OsBLR1 in rice caused an increased leaf angle, grain length, and BR sensitivity, and the opposite phenotypes appeared in OsBLR1-knockout mutants. We performed additional preliminary studies of the mechanism of OsBLR1 in BR signalling.
Materials and methods
Plant materials and growth conditions
The wild-type Dongjin (DJ, Oryza sativa L. ssp. japonica cv.) and T-DNA insertion mutant seeds of OsBLR1 were purchased from the Korean rice mutant library (https://signal.salk.edu/). Homozygous mutants were identified by two rounds of PCR with three primers (T-DNA border primer: RB-R, Gene primers: BLR1-LP-F and BLR1-RP-R). For homozygous mutants, we could clone the T-DNA border sequence, but could not clone the genome sequence by Gene primers. The T-DNA (pGA2772) insertion copy number analysis was carried out for four hypothetical homozygous mutants by Southern blot, and the specific sequence for 512 bp of T-DNA was marked with digoxin for detection (Zoonbio Biotechnology, Nanjing, China). To generate OsBLR1-overexpression plants, the full-length CDS sequence of OsBLR1 was linked to the pTCK303 plasmid to construct the Ubi:OsBLR1 vector. Then, the vector was transformed into Agrobacterium tumefaciens (EHA105) and introduced into Nipponbare (NIP, Oryza sativa L. ssp. japonica cv.) using the callus genetic transformation method. Positive lines were detected by hygromycin screening and reverse transcription-quantitative PCR (RT-qPCR) analysis. To construct the loss-of-function rice plants of OsBLR1 using the CRISPR/Cas9 technology, two protospacer adjacent motif (PAM) site sequences (PAM1: ACCGTCGAGAGCCTCTGCCAGGG, PAM2: AGAGCAACAGCATCTGCAAGAGG) were located in the first and second exons of OsBLR1 were selected. Following, the sgRNAs (sgRNA1: CCGTCGAGAGCCTCTGCCA, sgRNA2: GAGCAACAGCATCTGCAAG) were linked to the pBWA (V) H2S-cas9pl (osu3)i plasmid to construct the Cas9-OsBLR1 vector. Then, the vector was introduced into NIP in the same method. The homozygous mutants were obtained by gene segregation after the seed propagation of T0-T1 generation. The mutation types were analyzed by sequence alignment of OsBLR1 between mutants and wild type.
For disinfection, seeds were soaked in 5% NaClO solution for 15 min. These sterilized seeds were then sown on 1/2 MS agar medium containing 0.8% phytagel (pH 5.7) and germinated in darkness at 30 °C for 2 days. The germinating seeds were transferred to a 28 °C/25 °C (16 h/8 h) light incubator for culture in water or to a greenhouse for soil culture. Tobacco (Nicotiana benthamiana) seeds were provided by our laboratory. Tobacco seeds were sown evenly into culture soil (nutritious soil:vermiculite = 3:1) and cultured in a 26 °C/24 °C (16 h/8 h) light incubator.
Gene expression analysis
The Trizol method was performed to extract total RNA, and using the FastKing RT Kit (with gDNase) (Tiangen, Beijing, China) for reverse transcribed of cDNA from mRNA. The cDNA was used as the template for semi-quantitative reverse transcription PCR (SqRT-PCR) and RT-qPCR. β-ACTIN was used as the reference gene. For RT-qPCR, the SYBR Green Taq Ready Mix (Tiangen, Beijing, China) and the Bio-Rad CFX96 Real-Time System C1000 Thermal Cycler were used for expression analysis. The following data analysis formula referred to \({\text{R}} = {2}^{{ - [\Delta {\text{C}}_{{\text{t}}} \;{\text{sample}} - \Delta {\text{C}}_{{\text{t}}} \;{\text{control}}]}}\), where R refers to the relative expression level, ΔCt sample is the D-value of gene Ct value to internal reference β-ACTIN Ct value in experimental group, and ΔCt control is the D-value of gene Ct value to β-ACTIN Ct value in the control group. The RT-qPCR data represent the mean ± SE (n = 3). The data were compared by Student’s t-test. *p < 0.05, **p < 0.01.
Subcellular localization analysis
The full-length OsBLR1 coding region without a stop codon was linked to pBI221-GFP plasmid to construct pBI221-OsBLR1-GFP vector. pBI221-OsBLR1-GFP and pBI221-GFP plasmids were transformed into EHA105 and spread on YEP medium (10 g/l tryptone, 10 g/l yeast extract, 5 g/l NaCl, 50 μg/ml Ampicillin, 50 μg/ml Rifampin, 15 g/l Agar for solid medium), and incubated at 28 °C for 3 days. Shaking the positive clone bacteria in YEP medium and resuspending it with MMA (10 mM MES, 10 mM MgCl2, 0.2 mM Acetosyringone, pH 5.6) to OD600 = 0.1. The mixture was injected into tobacco for transient expression, and then incubated in darkness at 25 °C for 2.5 days. Observation was performed by LSM 710 Laser Confocal Microscope (Carl Zeiss AG, Germany).
Scanning electron microscopy and paraffin section
The germinated seeds were transplanted into the soil and grew to filling stage at 30 °C. For Scanning electron microscopy, the lamina joint tissue of flag leaf (2 cm of length) was excised for imaging the adaxial surface. Plant tissues were fixed in 2.5% glutaraldehyde solution, and then in a graded ethanol series. After desiccation for 3 h, sprayed with gold powder, and observed by scanning electron microscope. For paraffin section, the flag leaf tissue distance from stem 3 cm (0.5 cm of length) were placed in FAA solution (50% alcohol:formalin (37–40% formaldehyde):glacial acetic acid; 18:1:1). After dehydration in a graded ethanol series (70%, 80%, 90% and 100%), followed by embedding. The samples were dissected and then observed by light microscope. Parts of these experiments were performed by Servicebio company (Servicebio technology CO, Wuhan, China).
Measurement of endogenous BRs, cellulose and protopectin contents
blr1-D and wild-type DJ plants (> 50 seeds) grew in a greenhouse for two months. For endogenous BR measurement, the leaf and stem tissue (> 2 g) were mixed. The samples were rapidly frozen and ground to powder and then dissolved in methanol. The endogenous BRs contents were determined by electrospray ionisation/high-performance liquid chromatography/tandem mass spectrometry (ESI-HPLC–MS/MS) (ProNetsBio, Wuhan, China), and the measure method referred to Wu et al. (2013) and Xin et al. (2013). For endogenous BR measurement of wild-type NIP-1/2, blr1-1 and blr1-2, we sampled at 5 cm in the vicinity of the lamina joint and performed measurement in the same method. For cellulose and protopectin measurement, leaf tissue (> 0.3 g) was used for crude extraction and digestion with sulfuric acid, and a determination experiment was performed by Comin Company (Suzhou Comin biotechnology, Suzhou, China).
Exogenous BR treatment analysis
For lamina joint inclination assay, sterilized rice seeds were grown in the dark at 30 °C for 9 days. We obtained equally sized samples by excising approximately 2 cm of tissue near the lamina joint. The samples were floated on distilled water containing various concentrations of 24-Epibrassinolide (eBL, a bioactive form of BRs, dissolved in ethanol) at 30 °C for 30 h in darkness. Leaf angle was measured by Image J software. This experiment was carried out under dim light and repeated three times. For coleoptile elongation assay and root elongation assay, the sterilized seeds grew on 0.4% phytagel medium with different concentrations (0, 1, 10 μM) of eBL at 30 °C for 4.5 days in darkness, and then measured the length of the coleoptile and the primary root. For RNA extraction, 10-day-old seedlings were sprayed with 10 μM eBL, and then the leaves were harvested at various time points. For phenotypic analysis of BR treatment in living plants, different concentrations of eBL solution was applied to specific tissue using transfer liquid gun, and then the plants were cultured in greenhouses at 30 °C.
RNA-Seq analysis
Samples were collected from the leaves of DJ and blr1-D at the tillering stage (2 months), and two biological replicates were carried out. The construction and sequencing of the cDNA library were based on the BGISEQ-500 platform (BGI Genomics, Shenzhen, China). The clean reads obtained by sequencing data filtering were compared with the reference genome IRGSP-1.0. The data of DJ and blr1-D were normalized, and genes whose expression differences were more than two-fold increased were selected and analysed by the differentially expressed genes (DEG)-seq method (Wang et al. 2010). The DEGs were classified according to Gene Ontology (GO) annotation (https://geneontology.org/). The phyper function of R software was used for enrichment analysis and FDR correction of the p-value. FDR ≤ 0.01 was regarded as significant enrichment. The selected differential genes were blasted using the RAP-DB database (https://rapdb.dna.affrc.go.jp/index.html) and the NCBI database (https://www.ncbi.nlm.nih.gov/). The heatmap was plotted by MeV4.9.0 software.
Yeast two-hybrid (Y2H) assay
The full-length CDS sequence of OsBLR1 was linked to the pGBKT7 plasmid to construct the OsBLR1-pGBKT7 vector. OsBLR1-pGBKT7 plasmid was transformed into yeast AH109 for toxicity and auto-activation detection. For yeast two-hybrid library screening assay, OsBLR1-pGBKT7 plasmid was used as bait and mated with a rice seedling yeast two-hybrid library (constructed by Shanghai biogene biotechnology company, Shanghai, China), and then the mated culture was spread on the SD/-Leu/-Trp, SD/-Leu/-Trp/-His and SD/-Leu/-Trp/-His/-Ade plates for positive screening. Finally, the positive clones were detected by Lac Z assay. The selected positive clones were analysed by PCR and sequencing alignment with databases (NCBI and RAP-DB). For the yeast two-hybrid verification assay, the CDS sequence of OsRACK1A was cloned into the pGADT7 plasmid to construct the OsRACK1A-pGADT7 vector, OsBLR1-pGBKT7 and OsRACK1A-pGADT7 plasmids were co-transferred into yeast AH109 and spread on SD/-Leu/-Trp, SD/-Leu/-Trp/-His, SD/-Leu/-Trp/-His/-Ade and then the Lac Z assay was performed. pGBKT7-P53 + pGADT7-largeT was used as a positive control and pGBKT7-Laminc + pGADT7-largeT was used as a negative control.
Glutathione-S-transferase (GST) pull-down assay
The CDS sequence of OsBLR1 was linked to the pGEX-4T1 plasmid to construct the OsBLR1-pGEX-4T1 vector. OsBLR1-GST and GST proteins were expressed in Escherichia coli Rosetta (DE3) after induction by 0.5 mM isopropyl-b-D-thiogalactopyranoside (IPTG) and purified by Glutathione SepHarose™ 4B (GE Healthcare, Boston, USA). The similar method was performed to construct the OsRACK1A-pET-30a vector. OsRACK1A-His protein was expressed in Escherichia coli Rosetta (DE3) after induction by 0.8 mM IPTG and purified by Ni SepHaroseTM 6 Fast Flow (GE Healthcare, Boston, USA). For GST-pulldown assay, glutathione beads containing 0.5 mg OsBLR1-GST or GST purified proteins were incubated with 0.5 mg OsRACK1A-His protein in pull-down buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA and 1 mM DTT) at 4 °C for 5 h. The beads were washed three times with wash buffer (20 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM EDTA, 1 mM DTT and 0.1% (w/v) TritonX-100). The proteins were eluted from the beads using elution buffer (20 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM DTT and 15 mM Glutathione), and then boiled in 2 × sodium dodecyl sulphate (SDS) buffer for 5 min. The mixture was isolated in a 10% SDS–polyacrylamide gel. We analysed the results by Coomassie brilliant blue R-250 staining and Western blot analysis using anti-GST and anti-His antibodies (Abmart, Shanghai, China).
Results
Overexpression of OsBLR1 increased leaf angle and grain length
In the prior researches on the expression profile of gene, we found that OsBLR1 was highly expressed in the leaf and was mainly located in the nucleus (Fig. S1a, b, and c). To explore the function of OsBLR1 in the leaf, we screened T-DNA insertion mutants of OsBLR1, and obtained the homozygous, single-copy T-DNA insertion mutant blr1-D by PCR and Southern blotting (Fig. S2a and b). Sequencing analysis indicated that the reverse insertion site of Ti plasmid containing the 35 s enhancer (Jeong et al. 2006) in blr1-D was 624 bp upstream from the initiation codon of OsBLR1, and the Ti plasmid was located between OsBLR1 and Os02g0705600 on chromosome 2 (Fig. 1a). RT-qPCR data showed that the expression of OsBLR1 was significantly increased, while that of Os02g0705600 was unchanged in blr1-D (Fig. 1b), which demonstrated that blr1-D was an OsBLR1-overexpression line. The phenotype of blr1-D was characterised using T3 generation plants. We found that blr1-D had larger grains than the wild-type DJ (Fig. 1c). Although the grain length of blr1-D increased significantly, there were no significant changes in grain width or thickness (Fig. 1d). blr1-D had a dramatically increased leaf angle during the seedling, tillering, and maturation stages compared with DJ, and had drooping leaves that appeared bent and brittle (Fig. 1e). At the seedling stage (4-week-old rice), the angles of the second and third leaves of DJ were 27° and 32°, respectively, while the corresponding angles of blr1-D were 72° and 67°. At the filling stage, the angles of the top third leaf, the top second leaf and the flag leaf of DJ were 29°, 24°, and 21°, respectively, while the corresponding angles of blr1-D were 70°, 76°, and 87° (Fig. 1f).
The T-DNA insertion mutant of OsBLR1 (blr1-D) had increased leaf angle and grain length. a The reverse insertion site of the Ti plasmid between OsBLR1 and Os02g0705600 was located at 624 bp upstream of OsBLR1. b Expression analysis of OsBLR1 and Os02g0705600 in blr1-D by RT-qPCR. Data represent the mean ± SE (n = 3). **p < 0.01, Student’s t-test. c The grain (T3 generation) photos of DJ and blr1-D. Bar: 2 cm. d Statistical analysis of grain length, grain width and thickness in DJ and blr1-D. Data represent the mean ± SE (n = 15). **p < 0.01, Student’s t-test. e Phenotypic photos of DJ and blr1-D during different development stages. LJ: Lamina joint. Bar: 20 cm. f Statistical analysis of leaf angle in DJ and blr1-D. Data represent the mean ± SE (n = 10). **p < 0.01, Student’s t-test
To verify the function of OsBLR1, we transformed wild-type rice (NIP) with a construct of the Ubi promoter driving the expression of OsBLR1 cDNA and obtained three overexpression lines after hygromycin screening and expression analysis: BLR1-OE-1, BLR1-OE-2, and BLR1-OE-3 (Fig. 2a, b). The T2 generation overexpression lines had larger grains than NIP (Fig. 2c); the grain length of BLR1-OE-1/2/3 was longer, but there were no significant changes in grain width or grain thickness (Fig. 2d). At the tillering stage, BLR1-OE-1/2/3 showed a significant increase in leaf angle compared with NIP (Fig. 2e). The leaf angle of NIP was 28°, while those of BLR1-OE-1, BLR1-OE-2, and BLR1-OE-3 were 62°, 59°, and 68°, respectively (Fig. 2f). In addition, compared with the wild type, the 1000-grain weight, and tiller number of the OsBLR1-overexpression lines were significantly increased (Fig. S3a and b). Considering the consistent phenotypes in blr1-D and BLR1-OE-1/2/3, we inferred that OsBLR1 increased the leaf angle and grain length in rice.
OsBLR1-overexpression lines (BLR1-OE-1/2/3) had increased leaf angle and grain length. a The Ubi:OsBLR1 construct model in pTCK303-OsBLR1 plasmid. b RT-qPCR analysis of the expression of OsBLR1 in BLR1-OE-1/2/3 and NIP. Data represent the mean ± SE (n = 3). **p < 0.01, Student’s t-test. c The grain (T2 generation) photos of BLR1-OE-1/2/3 and NIP. Bar: 2 cm. d Statistical analysis of grain length, grain width and thickness in BLR1-OE-1/2/3 and NIP. Data represent the mean ± SE (n = 15). **p < 0.01, Student’s t-test. e Phenotypic photos of NIP and BLR1-OE-1/2/3 during the tillering stage. LJ lamina joint. Bar: 20 cm. f Statistical analysis of the leaf angle in NIP and BLR1-OE-1/2/3. Data represent the mean ± SE (n = 10). **p < 0.01, Student’s t-test
Knockout of OsBLR1 decreased leaf angle and grain length
To understand the function of OsBLR1, we generated OsBLR1-knockout mutants using the CRISPR/Cas9 technique. The target PAM sequences were located in the first and second exons of OsBLR1. Ultimately, we obtained three mutants with altered OsBLR1amino acid sequences, and named blr1-1, blr1-2, and blr1-3 (Fig. 3a and S4). Interestingly, these mutants had significantly smaller grains than NIP due to the grain length decreased significantly (Fig. 3b); however, there was no significant difference in grain width or thickness (Fig. 3c). blr1-1, blr1-2, and blr1-3 had erect leaves (Fig. 3d). The leaf angle of NIP was 28°, while those of blr1-1, blr1-2, and blr1-3 were 13°, 16°, and 14°, respectively (Fig. 3e). Considering the opposite phenotypes in the overexpression lines and knockout mutants of OsBLR1, we confirmed that OsBLR1 functions to increase leaf angle and grain length. Interestingly, compared with the wild type, the 1000-grain weight, and tiller number of blr1-1, blr1-2, and blr1-3 were decreased (Fig. S3a and b). This confirmed that OsBLR1 functions not only to increase leaf angle and grain length, but also 1000-grain weight, and tiller number.
OsBLR1-knockout mutants had erect leaves and shorter grains. a The Cas9-OsBLR1 construct model and mutation types in PAM site sequences of blr1-1/2/3.b The grain (T2 generation) photos of blr1-1/2/3 and NIP. LJ lamina joint. Bar: 20 cm. c Statistical analysis of grain length, grain width and thickness in blr1-1/2/3 and NIP. Data represent the mean ± SE (n = 15). **p < 0.01, Student’s t-test. d Phenotypic photos of NIP and blr1-1/2/3 during the tillering stage. LJ lamina joint. Bar: 20 cm. e Statistical analysis of leaf angle in NIP and blr1-1/2/3. Data represent the mean ± SE (n = 10). **p < 0.01, Student’s t-test
OsBLR1 transduced the BR signalling
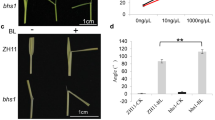
We found that the phenotypes of increased leaf angle and stem elongation in blr1-D seedlings were consistent with wild-type rice seedlings treated with eBL (Fig. S5a–d). Because the transgenic lines of OsBLR1 had these typical phenotypes, especially in the leaf angle, we speculated that OsBLR1 was involved in either the biosynthesis or signal transduction of BR. Lamina joint inclination is one of the most sensitive responses to BR in rice and has been used as a sensitive bioassay for BR (Yamamuro et al. 2000). First, we performed a lamina joint inclination assay using eBL to study the relationship between OsBLR1 and BR. This showed that the leaf angle changed in a dose-dependent manner, and blr1-D reached the maximum bending angle at 1 μM eBL, while the wild-type DJ reached the maximum bending angle at 10 μM (Fig. 4a, b). Similarly, the wild-type NIP reached the maximum bending angle at 10 μM eBL, but the corresponding concentration in BLR1-OE-1/2/3 was 5 μM (Fig. 4c, d). However, in living plants, blr1-1/2/3 mutants did not show significant bending, even at 100 μM eBL (Fig. 4e, f). The BR signalling pathway was blocked in these knockout mutants.
Lamina joint inclination assay in wild type and OsBLR1-transgenic lines. a, c and e The lamina joint inclination assay using different concentration of eBL in blr1-D, BLR1-OE-1/2/3, blr1-1/2/3 and wild-type. b, d and f Statistical analysis of the data in a, c and e. Data represent the mean ± SE (n > 3), *p < 0.05, **p < 0.01, Student’s t-test
Next, we performed coleoptile elongation and root elongation assays. In the wild-type plants, the coleoptile length was elongated in an eBL dose-dependent manner and the root was wavy and its elongation was inhibited by high concentrations of eBL (1 and 10 μM). The coleoptile and root elongation responses to exogenous BR were enhanced in the overexpression lines (blr1-D and BLR1-OE-1) and decreased in the knockout mutants (blr1-1 and blr1-2) (Fig. 5a–c). The above findings suggested that the sensitivity to BR in OsBLR1-knockout mutants was reduced, whereas it was increased in OsBLR1-overexpression lines. Then we measured the contents of endogenous BRs, the brassinolide (BL), castasterone (CS), and 6-deoxocastasterone (6-DeoxoCS) contents in blr1-D were lower than in DJ, whereas there were no significant differences in NIP, blr1-1, and blr1-2 (Fig. 5d). Accordingly, we inferred that OsBLR1 affects the signal transduction but not biosynthesis of BR.
The measurement of sensitivity to eBL (coleoptile elongation and root elongation assays) and contents of endogenous BRs of OsBLR1-transgenic lines. a Seedlings of wild type and transgenic lines of OsBLR1 were photographed after growing in medium in the presence ( +) or absence ( −) of 1 µm BR (eBL) for 4.5 days. Bar: 2 cm. b, c Statistical analysis of coleoptile elongation and root elongation under different concentration of BR treatment. Data represent the mean ± SE (n = 10), *p < 0.05, **p < 0.01, Student’s t-test. d The contents of BL, CS and 6-DeoxoCS in wild type, blr1-D and blr1-1/2 by ESI-HPLC–MS/MS method. In parentheses, the value of the mutant was on the left and the wild type on the right
OsBLR1 regulated leaf development and BR signalling
RNA-Seq analysis was performed to analyse the molecular mechanism of OsBLR1 function in leaf development. DEG-seq analysis showed that approximately 2152 genes were significantly changed in blr1-D compared with DJ (Fig. 6a). The GO annotation classification results of these differentially expressed genes (DEGs) indicated that many genes were associated with cell development, leaf development, and hormone signalling. After the GO enrichment analysis, the ten terms with the smallest Q-values were selected; these terms were closely related to cell development, especially cell wall development (Fig. 6b). Among leaf development genes, the expression of DL was down-regulated, while the expression of OsRLI1, OsGSR1, and OsNCED3 were up-regulated (Fig. 6c). Among cell development genes, the expression of OsEXPs and OsGLPs in blr1-D were up-regulated. However, the expression of the pectin methylesterases OsPME3 and OsPME17, and the cellulose synthases OsBC1, OsBC1L4, and OsBC7 were significantly down-regulated (Fig. 6d).
OsBLR1 regulated leaf development and BR signalling. a Genes regulated by OsBLR1 using RNA-Seq. b Analysis of the top 10 GO terms in the smallest Q value using GO enrichment of DEGs in blr1-D. c Heatmap showing the fold changes in gene expression involved in leaf development in blr1-D. d Heatmap showing the fold changes in gene expression involved in cell wall development in blr1-D. Log2 (fold changes) are represented by a colour scale from green (down-regulated) to red (up-regulated). e The contents of protopectin and cellulose in the leaves of blr1-D and DJ. Data represent the mean ± SE (n = 3). **p < 0.01, Student’s t-test. f The increased cell growth on the adaxial side of the lamina joint in blr1-D shown by scanning electron microscopy. Bar: 200 μm. g Histological section analysis of the cross section of blade in blr1-D and DJ. Bar: 200 μm. LV: large vascular, SV: small vascular. h The expression of OsBLR1 at different points in time under the induction of eBL (10 μM). i The expression s of OsRLI1 and OsBZR1 in the OsBLR1-transgenic lines. Data represent the mean ± SE (n = 3). *p < 0.05, **p < 0.01, Student’s t-test
Additionally, blr1-D showed increased cell growth on the adaxial side of the lamina joint and had a smaller leaf midrib on scanning electron microscopy and paraffin section observations (Fig. 6e, f). The cellulose and protopectin contents in leaves of blr1-D were reduced by 13.2% and 27.2%, respectively, compared with DJ (Fig. 6g). Moreover, the expression of OsBLR1 was down-regulated by 10 µM eBL (Fig. 6h). To explore the function of OsBLR1 in the BR signalling, we analysed the expression of a series of marker genes in the BR signal network in the OsBLR1 transgenic lines. Of these, OsBZR1 and OsRLI1 were up-regulated in the OsBLR1-overexpression lines, whereas OsRLI1 was down-regulated in the OsBLR1-knockout mutants; however, the expression of OsBRI1, OsGSK2, OsDLT, and OsLIC were unchanged in the OsBLR1-transgenic lines and wild type (Figs. 6i and S6). Above results suggested that OsBLR1 regulates leaf development and transduces BR signalling.
OsBLR1 interacted with OsRACK1A
To function, transcription factors usually interact with other proteins. To investigate the OsBLR1 binding proteins, we performed Y2H screening with a rice seedling yeast library using the full-length OsBLR1 protein as bait. The pGBKT7-OsBLR1 bait plasmid was nontoxic to yeast cells, but self-activated, and 20 mM 3-AT completely inhibited the self-activation of OsBLR1 (Fig. 7a). After mating pGBKT7-OsBLR1 with the yeast library, 22 strong interactive proteins were obtained after screening SD-THL, SD-THLA, and LacZ detection (Fig. S7). OsRACK1A was one of the candidate proteins, and has been reported to respond to multiple hormone signals. To verify the interaction between OsBLR1 and OsRACK1A, we co-transformed pGADT7-OsRACK1A with pGBKT7-OsBLR1 into yeast (AH109). The yeast cells grew well in both SD-THL and SD-THLA medium, and LacZ detection showed blue colour (positive results) (Fig. 7b). We further proved the interaction in a GST pull-down experiment, SDS-PAGE and Western blot data showed that GST protein could not pull down OsRACK1A-His protein, but OsBLR1-GST protein could pull down OsRACK1A-His protein (Fig. 7c). We treated rice plants with 10 µM eBL and found that the expression of OsRACK1A was up-regulated by eBL (Fig. 7d); moreover, OsRACK1A was up-regulated in the OsBLR1-overexpression lines (Fig. 7e). Accordingly, we speculated that OsRACK1A interacts with OsBLR1 and is up-regulated by BR.
OsBLR1 interacted with OsRACK1A. a 20 mM 3-AT completely inhibits the self-activation of OsBLR1. b Y2H assay indicates OsBLR1 interacts with OsRACK1A. OsBLR1-pGBKT7 + OsRACK1A-pGADT7 as the experimental group, pGBKT7-P53 + pGADT7-largeT as the positive control and pGBKT7-Laminc + pGADT7-largeT as the negative control. SD-TL: SD/-Leu/-Trp, SD-THL: SD-Leu/-Trp/-His, SD-THLA: SD/-Leu/-Trp/-His/-Ade. c GST pull-down assay provides evidence for the interaction between OsBLR1 and OsRACK1A. M protein marker. d The expression of OsRACK1A in leaves in the induction of eBL. e The expression of OsRACK1A in OsBLR1-transgenic lines. Data represent the mean ± SE (n = 3). *p < 0.05, **p < 0.01, Student’s t-test
Discussion
OsBLR1 is a typical member of the bHLH transcription factor family, but its specific function and mechanism in rice are unknown. In this study, we demonstrated the involvement of OsBLR1 in BR signalling by constructing and identifying transgenic lines of OsBLR1. OsBLR1 transduces the BR signal to increase leaf angle and grain length. OsBLR1 regulated the expression of OsDL, OsEXPs, OsGLPs, OsBC1, OsBCIL4, and OsRLI1, which affected leaf development, cell wall development, and BR signalling, resulting in lower pectin and cellulose contents, a smaller midrib, and decreased vascular bundles in leaves. We also screened OsRACK1A as an OsBLR1-binding protein using Y2H and GST pull-down.
OsBLR1 transduces the BR signal to increase leaf angle and grain length
We studied the function of OsBLR1 using transgenic lines, including overexpression lines (blr1-D and BLR1-OE-1/2/3) and knockout mutants (blr1-1/2/3). The overexpression lines had increased leaf angles and grain lengths, while the knockout mutants had erect leaves and shorter grains. Considering the opposing phenotypes in the overexpression lines and knockout mutants, we inferred OsBLR1 to be a positive regulator of leaf angle and grain length. Similarly, the overexpression lines OsbHLH153, OsbHLH173, and OsbHLH174 has increased leaf angles (Dong et al. 2018). Our data provide new experimental evidence that deepens our understanding of the function of bHLH transcription factors in regulating leaf angle and grain length.
Mutants with overdoses of BRs or enhanced BR signalling exhibit a dramatically increased leaf angle. BR-related mutants, such as d2-1 (loss-of-function of CYP90D2) (Hong et al. 2003), d61-1 (loss-of-function of OsBRI1) (Yamamuro et al. 2000), and brd3-D (loss-of-function of OsCYP734A4) (Qian et al. 2017), have erect leaves, and brd3-D has smaller grains. A significant increase in leaf angle can be induced by promoting rice BR biosynthesis or signal transduction (Zhang et al. 2014a). Given that the OsBLR1 mutants had typical phenotypes similar to those of d2-1, d61-1, and brd3-D, we first performed lamina joint inclination, coleoptile elongation, and root elongation assays to measure the BR sensitivity of OsBLR1-transgenic lines. This showed that blr1-1/2/3 were less sensitive to eBL, whereas blr1-D and BLR1-OE-1/2/3 were hypersensitive to eBL. The phenotypes of the lines sensitive or insensitive to BR are similar to those of the d61, dlt, and m-qgl3 mutants (Yamamuro et al. 2000; Tong et al. 2009; Gao et al. 2019). Next, we measured the BL, CS, and 6-DeoxoCS contents. There was no significant difference between the wild type and OsBLR1 mutants and, surprisingly, we detected BL in rice. The BL contents in NIP and DJ were 0.70, 0.31, and 0.64 ng/g, respectively. This is the first direct evidence of BL in rice. We measured the BL content using ESI-HPLC–MS/MS method, which is more sensitive to BL than the gas chromatography/mass spectrometry method used previously (the original BL data are shown in Supplementary 2). Based on these results, we speculated that OsBLR1 transduces the BR signal to increase leaf angle, but not biosynthesis. The function of OsBLR1 is similar to those of OsILI1 and OsBU1 (OsbHLH172), which have been reported to participate in BR signalling to regulate leaf angle (Tanaka et al. 2009; Zhang et al. 2009).
OsBLR1 regulates leaf development and BR signalling
Many reports have shown that BRs regulate cell elongation to promote lamina joint bending, such as BR functional enhancement lines: ili1-D (ILI1 high expression mutant), OsBUL1D (OsBUL1 mutant), and RLI1-OE-3 (OsRLI1 high expression line) (Zhang et al. 2009; Jang et al. 2017; Ruan et al. 2018). We observed increased cell growth on the adaxial side of the lamina joint by scanning electron microscopy, which led to the lamina joint bending in blr1-D. We also found that the leaf midrib had developmental defects in blr1-D, which is similar to DL mutants that form an abnormal midrib and have drooping leaves (Nagasawa et al. 2003). We performed RNA-Seq to study the molecular mechanism of OsBLR1. The results showed that DL, as a positive regulatory factor of midrib development, was significantly down-regulated in blr1-D, which may result in the smaller midrib of blr1-D. We also found that the development of vascular bundles was impaired in the midrib compared with DJ. In addition, the transcription of OsEXPs and OsGLPs was significantly up-regulated in blr1-D. OsEXPs regulate stem and leaf development by facilitating cell wall extension (Li et al. 2002). OsGLPs promote cell wall modification and reconstruction by regulating the H2O2 content (Bolwell et al. 1995). These factors promote cell development in blr1-D. However, the cellulose and protopectin contents in the leaves of blr1-D were significantly decreased. RNA-Seq data showed that the expression of pectin methylesterase and cellulose synthetase genes, such as OsBC1 and OsBC1L4, was significantly decreased (Dai et al. 2011; Liu et al. 2013). We speculate that these changes led to lower pectin and cellulose contents and abnormal cell walls, which may ultimately result in the reduced vascular bundle tissue in the leaves of blr1-D. These factors are important for the increased leaf angle and drooping leaves in blr1-D.
In addition, RNA-Seq and RT-qPCR data showed that the expression of OsRLI1 was up-regulated in OsBLR1-overexpression lines, but down-regulated in OsBLR1-knockout mutants. We know that OsRLI1 positively regulates leaf inclination by regulating lamina joint cell elongation. The rli1 mutants have reduced leaf angles and RLI1 overexpression results in increased leaf angles (Ruan et al. 2018); the phenotypes of the OsRLI1 mutants are similar and consistent with the OsBLR1 mutants. We propose that OsRLI1 is a downstream gene of OsBLR1 in BR signalling, but this hypothesis needs further study.
OsBLR1 combines with OsRACK1A
In this study, we demonstrate an interaction between OsBLR1 and OsRACK1A using Y2H and GST pull-down assays. Recently, some studies have shown that the MYB-bHLH-WD40 complex plays a key role in BR signalling (Cheng et al. 2014). In Arabidopsis thaliana, the sensitivity of the RACK1A mutant to exogenous BR is decreased (Chen et al. 2006); this is consistent with the reduced sensitivity to BR in blr1-1/2/3. In this study, RT-qPCR showed that the expression of OsRACK1A was up-regulated by exogenous BR. The function of OsRACK1A is complicated, and the function of the interaction between OsBLR1 and OsRACK1A in BR signalling needs further study.
References
Atchley WR, Terhalle W, Dress A (1999) Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol 48:501–516. https://doi.org/10.1007/pl00006494
Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14–3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104:13839–13844. https://doi.org/10.1073/pnas.0706386104
Bishop GJ, Koncz C (2002) Brassinosteroids and plant steroid hormone signaling. Plant Cell 14(Suppl):S97–110. https://doi.org/10.1105/tpc.001461
Bolwell GP, Butt VS, Davies DR, Zimmerlin A (1995) The origin of the oxidative burst in plants. Free Radic Res 23:517–532. https://doi.org/10.3109/10715769509065273
Chen JG et al (2006) RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot 57:2697–2708. https://doi.org/10.1093/jxb/erl035
Chen K et al (2019) Translational regulation of plant response to high temperature by a dual function tRNAHis guanylyltransferase in rice. Mol Plant. https://doi.org/10.1016/j.molp.2019.04.012
Cheng Y, Zhu W, Chen Y, Ito S, Asami T, Wang X (2014) Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife. https://doi.org/10.7554/eLife.02525
Dai X, You C, Chen G, Li X, Zhang Q, Wu C (2011) OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol Biol 75:333–345. https://doi.org/10.1007/s11103-011-9730-z
Dong H et al (2018) Genome-wide association studies reveal that members of bHLH subfamily 16 share a conserved function in regulating flag leaf angle in rice (Oryza sativa). PLoS Genet. 14:e1007323. https://doi.org/10.1371/journal.pgen.1007323
Fukuda H (2004) Signals that control plant vascular cell differentiation. Nat Rev Mol Cell Biol 5:379–391. https://doi.org/10.1038/nrm1364
Gao X et al (2019) Rice qGL3/OsPPKL1 Functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31(5):1077–1093. https://doi.org/10.1105/tpc.18.00836
Guo J, Chen JG (2008) RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol 8:108. https://doi.org/10.1186/1471-2229-8-108
Guo J, Wang J, Xi L, Huang WD, Liang J, Chen JG (2009) RACK1 is a negative regulator of ABA responses in Arabidopsis. J Exp Bot 60:3819–3833. https://doi.org/10.1093/jxb/erp221
Hong Z et al (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15:2900–2910. https://doi.org/10.1105/tpc.014712
Islas-Flores T, Rahman A, Ullah H, Villanueva MA (2015) The receptor for activated C kinase in plant signaling: tale of a promiscuous little molecule. Front Plant Sci 6:1090. https://doi.org/10.3389/fpls.2015.01090
Jang S, An G, Li HY (2017) Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol 173:688–702. https://doi.org/10.1104/pp.16.01653
Jeong D-H, An S, Park S, Kang H-G, Park G-G, Kim S-R, Sim J, Kim Y-O, Kim M-K, Kim S-R, Kim J, Shin M, Jung M, An G (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45:123–132. https://doi.org/10.1111/j.1365-313X.2005.02610.x
Jiang Y, Yang B, Deyholos MK (2009) Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol Genet Genom 282:503–516. https://doi.org/10.1007/s00438-009-0481-3
Khush GS, Gupta P (2013) Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breed 132:433–436. https://doi.org/10.1111/pbr.1991
Kim Y, Song JH, Park SU, Jeong YS, Kim SH (2017) Brassinosteroid-induced transcriptional repression and dephosphorylation-dependent protein degradation negatively regulate BIN2-interacting AIF2 (a BR signaling-negative regulator) bHLH transcription factor. Plant Cell Physiol 58:227–239. https://doi.org/10.1093/pcp/pcw223
Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128:854–864. https://doi.org/10.1104/pp.010658
Li X et al (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184. https://doi.org/10.1104/pp.106.080580
Liu L et al (2013) Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9:e1003704. https://doi.org/10.1371/journal.pgen.1003704
Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X (2016) Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep 35:2423–2433. https://doi.org/10.1007/s00299-016-2052-5
Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development (Cambridge, England) 130:705–718. https://doi.org/10.1242/dev.00294
Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110:203–212. https://doi.org/10.1016/s0092-8674(02)00814-0
Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawa NK (2010) The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J 51:366–377. https://doi.org/10.1111/j.1365-313X.2007.03149.x
Oikawa T, Kyozuka J (2009) Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21:1095–1108. https://doi.org/10.1105/tpc.108.065425
Qian W, Wu C, Fu Y, Hu G, He Z, Liu W (2017) Novel rice mutants overexpressing the brassinosteroid catabolic gene CYP734A4. Plant Mol Biol 93:197–208. https://doi.org/10.1007/s11103-016-0558-4
Ruan W, Guo M, Xu L, Wang X, Zhao H, Wang J, Yi K (2018) An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 30:853–870. https://doi.org/10.1105/tpc.17.00738
Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19:2749–2762. https://doi.org/10.1105/tpc.107.053728
Ryu H, Kim K, Cho H, Hwang I (2010) Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells 29:291–296. https://doi.org/10.1007/s10059-010-0034-y
Sinclair TR, Sheehy JE (1999) Erect leaves and photosynthesis in rice. Science 283:1455. https://doi.org/10.1126/science.283.5407.1455c
Song Q, Zhang G, Zhu XG (2013) Optimal crop canopy architecture to maximise canopy photosynthetic CO2 uptake under elevated CO2—a theoretical study using a mechanistic model of canopy photosynthesis. Funct Plant Biol 40:109–124. https://doi.org/10.1071/FP12056
Tanaka A et al (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151:669–680. https://doi.org/10.1104/pp.109.140806
Tang W et al (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science (New York, NY) 321:557–560. https://doi.org/10.1126/science.1156973
Tong H, Chu C (2009) Roles of DLT in fine modulation on brassinosteroid response in rice. Plant Signal Behav 4:438–439. https://doi.org/10.1111/j.1365-313X.2009.03825.x
Tong H et al (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803–816. https://doi.org/10.1111/j.1365-313X.2009.03825.x
Tong H et al (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24:2562–2577. https://doi.org/10.1105/tpc.112.097394
Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC (2008) Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci 17:1771–1780. https://doi.org/10.1110/ps.035121.108
Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science (New York, NY) 313:1118–1122. https://doi.org/10.1126/science.1127593
Wang ZY et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2:505–513. https://doi.org/10.1016/s1534-5807(02)00153-3
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics (Oxford, England) 26:136–138. https://doi.org/10.1093/bioinformatics/btp612
Wu Q, Wu D, Shen Z, Duan C, Guan Y (2013) Quantification of endogenous brassinosteroids in plant by on-line two-dimensional microscale solid phase extraction-on column derivatization coupled with high performance liquid chromatography–tandem mass spectrometry. J Chromatogr A 1297:56–63. https://doi.org/10.1016/j.chroma.2013.04.043
Xin P, Yan J, Fan J, Chu J, Yan C (2013) An improved simplified high-sensitivity quantification method for determining brassinosteroids in different tissues of rice and Arabidopsis. Plant Physiol 162:2056–2066. https://doi.org/10.1104/pp.113.221952
Yamamuro C et al (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1606. https://doi.org/10.1105/tpc.12.9.1591
Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191. https://doi.org/10.1016/s0092-8674(02)00721-3
Zhang D et al (2014) OsRACK1 is involved in abscisic acid- and H2O2-mediated signaling to regulate seed germination in rice (Oryza sativa, L.). PLoS ONE 9:e97120. https://doi.org/10.1371/journal.pone.0097120
Zhang L et al (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21:3767–3780. https://doi.org/10.1105/tpc.109.070441
Zhang C, Bai MY, Chong K (2014) Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep 33:683–696. https://doi.org/10.1007/s00299-014-1578-7
Zhu XG, Long SP, Ort DR (2008) What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr Opin Biotechnol 19:153–159. https://doi.org/10.1016/j.copbio.2008.02.004
Author information
Authors and Affiliations
Contributions
W.Z. and Q.Z. provided funds; W.Z. and K.W. conceived and designed the experiments; K.W. performed the major experiments and data analysis; W.Z. and K.W. discussed the results and drafted the manuscript; M.L. performed the identification of blr1-D; Y.C. performed the coleoptile elongation and root elongation assays; B.Z. assisted the Y2H screening.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, K., Li, Mq., Chang, Yp. et al. The basic helix-loop-helix transcription factor OsBLR1 regulates leaf angle in rice via brassinosteroid signalling. Plant Mol Biol 102, 589–602 (2020). https://doi.org/10.1007/s11103-020-00965-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-00965-5