Abstract
Key message
The regulator MdERF1B in the apple (Malus × domestica) ethylene pathway mainly acts on MdMYB9 and MdMYB11 to regulate anthocyanin and proanthocyanidin accumulation.
Abstract
Dietary anthocyanins and proanthocyanidins (PAs) have health benefits for humans, and are associated with decreased risks of coronary heart disease and cancer. Ethylene can enhance reddening of apple (Malus × domestica), but the regulatory mechanism is poorly understood. In this study, an ethylene response factor (ERF), MdERF1B, was identified and functionally characterized. ‘Orin’ calli overexpressing MdERF1B were generated and then analyzed by quantitative reverse transcription-PCR. Compared with the control calli, the MdERF1B-overexpressing calli showed increased expression levels of MdACO1, MdERF1, and MdERF3 in the ethylene pathway and MdCHS, MdCHI, MdF3H, MdDFR, MdANS, MdLAR, MdANR, MdMYB9 and MdMYB11 in the flavonoid pathway. As a result, the levels of anthocyanins and PAs were significantly increased in the MdERF1B-overexpressing calli. MdERF1B interacted with MdMYB9, MdMYB1, and MdMYB11 proteins in yeast two-hybrid, pull-down, and bimolecular fluorescence complementation assays. Furthermore, in yeast one-hybrid and electrophoretic mobility shift assays, MdERF1B also bound to the promoters of MdMYB9, MdMYB1, and MdMYB11. In a luciferase reporter assay, MdERF1B mainly activated proMdMYB9 and proMdMYB11, promoting their expression levels. This was in agreement with MdERF1B’s overexpression in calli, which barely affected MdMYB1 expression. Taken together, our findings provide an insight into the regulatory mechanisms in the ethylene pathway that increase anthocyanin and PA accumulation in apple.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
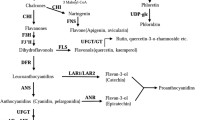
Flavonoids are important secondary metabolites in plants, and include anthocyanins, flavonols, and proanthocyanidins (PAs) (Winkel-Shirley 2001). Flavonoids have important biological functions, including flower pigmentation for pollinator attraction, protection of plants from ultraviolet-B damage; and defense against herbivores and pathogens. As part of the human diet, they eliminate free radicals and help to prevent coronary heart disease and cancer (Dixon and Paiva 1995; Harborne and Williams 2000; Ross and Kasum 2002; Wang et al. 2009). The key enzymes in the flavonoid synthesis pathway are phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and glycosyltransferase (UFGT). Anthocyanins and PAs share the same early synthesis pathway (Honda et al. 2002; Lister et al. 2015), but in the late stage of PA synthesis, leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) are required to produce catechin and epicatechin, respectively (Xie et al. 2003).
The transcription factors (TFs) involved in flavonoid synthesis have been extensively studied, especially the MYB–bHLH–WD40 (MBW) ternary complex that synergistically regulates the synthesis of anthocyanins and PAs (Baudry et al. 2004; Quattrocchio et al. 2006; An et al. 2012; Xie et al. 2012). In Arabidopsis thaliana, the R2R3 MYB TF TT2 regulates the accumulation of PAs (Nesi et al. 2001). In grapes, VvMYBPA1 specifically activates PA biosynthesis (Bogs et al. 2007). In apple, the MYB TFs that regulate the flavonoid pathway include MdMYB1 and MdMYBA, which are involved in anthocyanin synthesis in apple fruit skin (Takos et al. 2006; Ban et al. 2007), and MdMYB10, which is responsible for anthocyanin synthesis in type 1 red-fleshed apple (Espley et al. 2007). In addition, a 23-bp repeat motif in MdMYB10 strongly promotes anthocyanin accumulation (Espley et al. 2009). MdMYB110a_JP is found in type 2 red-fleshed apple, and is responsible for the red flesh but not the skin color (Umemura et al. 2013). MdMYB9 and MdMYB11 activate flavonoid structural genes through the MBW ternary complex, which promotes anthocyanin and PA biosynthesis (An et al. 2015). Recently, Wang et al. (2017) found that MdMYB12 interacts with MdbHLH3 and MdbHLH33 to regulate PA synthesis, and that MdMYB22 directly binds to the promoter of the gene encoding flavonol synthase to promote the flavonol pathway.
Ethylene is an important gaseous phytohormone that is widely involved in plant growth and developmental processes, such as seed germination, seedling growth, fruit maturation, and senescence, as well as in responses to pathogens and wounding (O’Donnell et al. 1996; Penmetsa and Cook 1997; Barry and Giovannoni 2007). In ethylene biosynthesis, S-adenosyl methionine is converted by 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) into ACC, and then ACC is oxidized by ACC oxidase (ACO) to form ethylene (Yang and Hoffman 1984). The rate-limiting enzyme in this process is ACO (Kende 2003). In the ethylene transduction pathway, the ETR membrane receptors recognize ethylene and lose their activity (Liu and Wen 2012). When ethylene is absent, CTR1 negatively regulates the ethylene response by inhibiting the activity of EIN2 (Kieber et al. 1993). In the presence of ethylene, EIN2 activates the downstream positive regulatory factors EIN3 and EIL1, and then activates the expression of the downstream TF ERF1, which activates the expression of ethylene-responsive genes (Solano et al. 1998).
In Arabidopsis, AtERF1 integrates ethylene, jasmonate, and abscisic acid signals during responses to biotic and abiotic stresses (Lorenzo et al. 2003; Cheng et al. 2013). In pear, the AP2/ERF TF PyERF3 interacts with both PyMYB114 and PybHLH3 to enhance anthocyanin biosynthesis (Yao et al. 2017). Exogenous ethylene promotes anthocyanin accumulation in grape berries by stimulating the transcription of CHS, F3H, LDOX, and UFGT (El-Kereamy et al. 2003). Ethylene can also improve flesh reddening in plum (Manganaris et al. 2008). In apple, MdERF1 is prominently expressed in the ripening fruit, while MdERF2 suppresses the expression of MdACS1 to inhibit ethylene biosynthesis during fruit ripening (Wang et al. 2007; Li et al. 2016). Ethylene also promotes apple reddening (Wang and Dilley 2001). However, the specific regulatory mechanisms underlying the effect of ethylene to promote anthocyanin and PA accumulation in apple are poorly understood.
In this study, we identified an AP2/ERF TF, MdERF1B, which belongs to the ERF subfamily. Transcriptional analyses showed that its expression levels corresponded to the degree of apple fruit reddening. Subsequently, we found that MdERF1B interacted with MdMYB9, MdMYB1, and MdMYB11 proteins. In addition, MdERF1B bound to the promoters of MdMYB9, MdMYB1, and MdMYB11, and promoted the expression of MdMYB9 and MdMYB11 but not MdMYB1. Transgenic construct analyses and luciferase (LUC) reporter assays confirmed that MdERF1B promotes anthocyanin and PA accumulation mainly by acting on MdMYB9 and MdMYB11.
Materials and methods
Plant materials
The trees of the early maturing apple ‘Taishanzaoxia’ were located in Liaocheng, Shandong, China. Apple fruits were picked at 57, 63, 69, and 75 days after full bloom (DAFB), their developmental stage was assessed, and then they were quickly transported to the laboratory for further analyses.
The calli of ‘Orin’ were cultured on Murashige and Skoog (MS) medium containing 0.5 mg L−1 6-benzylaminopurine (6-BA) and 1 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) in the dark at 24 °C and subcultured every 14 days.
Determination of ethylene production rate
Three apples were enclosed in a 1 L-container for 2 h, then 1 mL gas was sampled from each container using a syringe. The ethylene concentration was determined using a gas chromatograph (Shimadzu, Kyoto, Japan) equipped with a hydrogen flame ionization detector.
Measurement of anthocyanin concentration
Anthocyanins were measured as described by Giusti and Wrolstad (2001). Briefly, 0.5 g apple peel was ground in liquid nitrogen and then immersed in 10 mL cold 1% HCl-methanol at 4 °C for 24 h. After adding 1 mL supernatant to 4 mL KCl buffer (pH = 1.0) and 1 mL supernatant to 4 mL NaAc buffer (pH = 4.5), the samples were incubated for 15 min in the dark. Then, the absorbance was measured at 510 and 700 nm using a UV–Vis spectrophotometer (UV-2450; Shimadzu, https://www.ssi.shimadzu.com). The anthocyanin content was quantified using the formula: ΔA × 5 × 0.005 × 1000 × 449.2/(26,900 × 0.5), where ΔA = (OD510 − OD700 at pH = 1.0) − (OD510 − OD700 at pH = 4.5).
RNA extractions and quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, http://www.tiangen.com/en/). The synthesis of cDNA was performed using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, http://www.transbionovo.com/). The qRT-PCR reactions were performed with SYBR® Green PCR Master Mix (TransGen) according to the manufacturer’s instructions. The primers used for qRT-PCR analysis are listed in Supplementary Table S1. Transcript data were analyzed using the (Ct) 2− ΔΔCt method (Livak and Schmittgen 2001).
Cloning of MdERF1B and amino acid sequence analysis
MdERF1B (XM_008342898.2) was amplified using Phusion polymerase (Thermo Scientific, http://www.thermofisher.com/). The primers used for cloning CDSs and promoters are listed in Supplementary Table S2. The phylogenetic tree was constructed using MEGA 5.1 software. In total, 36 related ERF protein sequences from different species were aligned using ClustalW. Among them, 11 ERF protein sequences were aligned using DNAMAN software.
Yeast two-hybrid assays (Y2H)
The full-length coding DNA sequences (CDSs) of MdMYB9, MdMYB1, and MdMYB11 were inserted into the vector pGADT7 (Clontech, http://www.clontech.com), and the full-length CDS of MdERF1B was inserted into the vector pGBKT7 (Clontech). MdERF1B had strong self-activation activity; therefore, we screened for the fragment MdERF1B19–197aa with a deleted autoactivation domain. The two recombinant plasmids were co-transformed into the Y2H Gold yeast strain. The cells were cultured on SD/–Leu/–Trp medium initially, and then transferred to SD/–Leu/–Trp/–His/–Ade medium to test for interactions. The empty pGADT7 vector was used as the control.
Pull-down assays
The full-length CDS of MdERF1B was inserted into the pET-32a (+) vector, and the full-length CDSs of MdMYB9, MdMYB1, and MdMYB11 were each inserted into the pGEX-4T-1 vector. The recombinant plasmids were individually transformed into Escherichia coli BL21 (DE3) to form the fusion proteins MdERF1B-histidine (His), MdMYB9-glutathione S-transferase (GST), MdMYB1-GST, and MdMYB11-GST. Two proteins with different tags were mixed for purification with the His-Tagged Protein Purification Kit (CWBIO, http://www.cwbiotech.com/). The proteins were eluted and then tested by western blotting using anti-His and anti-GST antibodies (Abmart, http://www.ab-mart.com.cn/).
Bimolecular fluorescence complementation assays (BiFC)
The full-length CDS of MdERF1B was inserted into the pSPYNE-35S vector, and the full-length CDSs of MdMYB9, MdMYB1, and MdMYB11 were each inserted into the pSPYCE-35S vector. The recombinant plasmids were transformed into Agrobacterium tumefaciens LBA4404 and then cultured in YEP culture medium until an OD600 of 0.6 was reached. Then, 15 mL pSPYNE-35S and 15 mL pSPYCE-35S recombinant plasmids were mixed and used to infect onion epidermal cells by culturing at 28 °C in the dark for 24–48 h. Finally, YFP fluorescence was examined under a confocal laser scanning microscope.
Transformation of ‘Orin’ calli with MdERF1B
The full-length CDS of MdERF1B was inserted into the PRI101-AN vector, which has a CaMV 35S promoter and a green fluorescent protein (GFP) tag. The recombinant plasmid was transformed into A. tumefaciens LBA4404 to infect ‘Orin’ calli. First, the infected calli were co-cultured on MS solid medium in the dark at 24 °C for 1–2 days, and then transferred to screening medium containing 50 mg L−1 kanamycin and 250 mg L−1 carbenicillin. The overexpression of MdERF1B was confirmed by PCR amplification and western blotting.
Yeast one-hybrid assays (Y1H)
The full-length CDS of MdERF1B was inserted into the pGADT7 vector (Clontech), and the promoter fragments of MdMYB9, MdMYB1, and MdMYB11 were inserted into the pHIS2 vector (BD Biosciences, http://www.bdbiosciences.com/). The recombinant plasmids containing MdERF1B and promoter fragments were co-transformed into the Y187 strain, and then the interactions were examined on medium lacking Trp, Leu, and His with the optimal 3-amino-1,2,4-triazole (3-AT) screening concentration.
Electrophoretic mobility shift assays (EMSA)
The fusion protein MdERF1B-His was purified using the His-Tagged Protein Purification Kit (CWBIO). The oligonucleotide probes were synthesized and labeled at their 3′ ends with biotin (Sangon Biotech, http://www.sangon.com/). The double-stranded DNA probes were synthesized with 5× annealing buffer for DNA Oligos (Beyotime, http://www.beyotime.com). The EMSA was performed using the LightShift™ EMSA Optimization and Control Kit (Thermo Scientific, https://www.thermofisher.com). First, 2 µL 10× binding buffer and 1 µL hot probe, with or without 16 µL purified protein, were mixed and maintained at room temperature for 15 min. Then, 1 µL cold probe or 1 µL mutant cold probe were added for the 15-min competition reaction.
Luciferase reporter assays
The full-length CDS of MdERF1B was inserted into the pHBT-AvrRpm1 effector, and the promoter fragments of MdMYB9, MdMYB1, and MdMYB11 were inserted into the pFRK1-LUC-nos reporter. Then, 2 µL UBQ-GUS, 6 µL LUC reporter, and 12 µL effector were transformed into 200 µL protoplasts extracted from ‘Orin’ calli, and then expressed at 24 °C for 6 h. Subsequently, protoplasts were suspended in 100 µL cell lysis buffer. Then, 5 µL of cell extract was mixed with 20 µL 1 mM 4-MUG at 37 °C for 1 h, and 100 µL 0.2 M sodium carbonate was added to stop the reaction. The LUC activity was determined using the Dual-Luciferase© Reporter Assay System (Promega, https://www.promega.com). The fluorescence levels of GUS and LUC were determined using a Multimode Plate Reader (Victor X4, PerkinElmer, http://www.perkinelmer.com/).
4-Dimethylaminocinnamaldehyde (DMACA) staining and PA content measurement
The presence of PAs was detected by DMACA staining. The calli were stained with DMACA reagent (0.2% DMACA, w/v, in methanol:6 M HCl, v/v = 1:1) for 30 min and then washed with 70% (v/v) ethanol. The PA content was measured using the method described by Li et al. (1996). The samples were ground in liquid nitrogen, then 0.4 g powder was dissolved in 1 mL 70% (v/v) acetone solution including 0.1% (w/v) ascorbic acid, and then mixed at 4 °C in the dark for 30 min. The above step was repeated three times and the supernatant was collected by centrifuging. The supernatant (4 mL) was mixed with 3 mL ether at − 20 °C. The resulting lower layer contained soluble PAs. Then, 770 µL soluble PAs, 385 µL methanol, and 192 µL 2% (w/v) DMACA were mixed at room temperature for 20 min, and the absorbance at 643 nm was measured using a UV–Vis spectrophotometer (UV-2450; Shimadzu). The PA concentration was calculated from a standard curve prepared using catechin.
Data analysis
Randomly selected fruits of the early maturing apple variety ‘Taishanzaoxia’ were picked at 57, 63, 69 and 75 DAFB. Their developmental stage was quickly assessed and then samples were frozen. Three biological replicates of each sample were analyzed. Data were analyzed using Tukey’s test. Significant differences are indicated by letters, where lowercase letters represent significant differences at P < 0.05, and uppercase letters represent highly significant differences at P < 0.01.
Results
Ethylene production rate is positively correlated with anthocyanin level
During the development of ‘Taishanzaoxia’ apple, the peak in the ethylene production rate coincided with a rapid increase in anthocyanin content (Fig. 1a–c). We analyzed the expression levels of genes encoding enzymes and TFs in the anthocyanin and ethylene pathways by qRT-PCR. As ethylene release peaked, MdMYB9, MdMYB1, and MdMYB11 were up-regulated. In the ethylene pathway, MdERF1B showed greater increases in its transcript levels than did MdERF1 and MdERF3 (Fig. 1d), which are positive regulators in the ethylene pathway. As shown in Fig. 1b–d, the transcript level of MdERF1B was consistent with the production of ethylene and anthocyanin. Therefore, we hypothesized that the TF MdERF1B was related to structural genes and MYB TFs in the flavonoid synthesis pathway.
Analysis of spatiotemporal patterns of ethylene and anthocyanin production, and transcriptional patterns of genes in the flavonoid and ethylene pathway. a ‘Taishanzaoxia’ apple fruit picked at 57, 63, 69, and 75 DAFB. Scale bar = 1 cm. b Ethylene production during development of ‘Taishanzaoxia’ apple. c Anthocyanin production during development of ‘Taishanzaoxia’ apple. d Expression levels of related genes in the flavonoid and ethylene pathway as assessed by qRT-PCR. MdActin was used as the control. FW, fresh weight. Error bars represent SEs of means (n = 3). Lowercase letters represent significant differences at P < 0.05; uppercase letters represent highly significant differences at P < 0.01
Phylogenetic tree and sequence analyses
The ERF family can be divided into groups I–X (Nakano et al. 2006). The phylogenetic analysis showed that MdERF1B belongs to group IX and is homologous with AtERF1. MdERF1B grouped with the JA-responsive ERF TFs AtERF1, ORA59, AaERF1, AaERF2, MdERF3, and tomato LePti4, all of which are all in group IX (Fig. 2a). Many TFs in group IX are involved in the JA and ethylene pathways that function in defense responses (Lorenzo et al. 2003; Cheng et al. 2013). MdERF3 in group IX is induced by MeJA, which upregulates MdACS1 to positively regulate ethylene biosynthesis (Li et al. 2017). The protein sequence alignment indicated that MdERF1B contains an AP2/ERF domain, as described by Sakuma et al. (2002), in which the 14th and 19th amino acids are alanine and aspartate, respectively (Fig. 2b).
Phylogenetic tree analysis and protein sequence alignment. a Phylogenetic tree analysis of MdERF1B protein and related AP2/ERF TFs from different species. b Alignment of AP2/ERF domains between MdERF1B and other AP2/ERF TFs from different species. Dots indicate alanine and aspartate at 14th and 19th amino acids of the domain, respectively. NCBI accession numbers of AP2/ERF TFs are as follows: AaERF1 (JN162091.1), LePti4 (NM_001347076.1), MdERF2 (AB288348), AtERF2 (NM_124093.3), MdERF3 (XM_008339725.2), LePti5 (U89256.1), ORA59 (NM_100497.3), AtERF1 (AT3G23240), MdERF1B (XM_008342898.2), AaERF2 (JN162092.1), MdERF5 (LOC103427506), AtERF5 (NM_124094.3), AtERF6 (NM_117854.3), AtRAP2.1 (BT024691.1), AtRAP2.10 (AT4G36900), AtDREB1A (AB013815.1), AtDREB1B (AB013816.1), DREB2A (At5G05410), AtDREB2B (AT3G11020), AtABR1 (AT5G64750), PyERF3 (MF489220), MdERF1 (AB288347), AtRAP2.2 (AT3G14230), AtRAP2.12 (At1G53910), AtERF10 (NM_100259.2), AtERF11 (NM_102603.3), AtERF4 (NM_112384.2), AtERF8 (NM_104196.3), NtERF4 (NM_001325253.1), AtERF3 (NM_103946.3), AtERF7 (NM_112922.3), AtRAV1 (AB013886.1), AtRAV2 (AB013887.1), AtRAP2.7 (NM_001202696.2), AtAP2 (NM_119856.3) and EjAP2-1 (KM506584.1)
MdERF1B interacts with MdMYB9, MdMYB1, and MdMYB11
To analyze whether there is a connection between MdERF1B and MYB TFs, we performed Y2H assays. The full-length MdERF1B protein had strong self-activation activity; therefore, the assays were performed using five different amino acid fragments, I–V, of MdERF1B. Then, we constructed MdERF1B19–197aa-pGBKT7 (V) with a deleted autoactivation domain (Fig. 3a, b). MdERF1B19–197aa-pGBKT7 (V) was co-transformed separately with MdMYB9-pGADT7, MdMYB1-pGADT7, and MdMYB11-pGADT7 recombinant plasmids into the Y2H Gold yeast strain. In the Y2H assays, MdERF1B interacted with MdMYB9, MdMYB1, and MdMYB11 in vivo (Fig. 3c).
Interactions between MdERF1B and MdMYB9, MdMYB1, and MdMYB11. a I–V represent five different amino acid fragments of MdERF1B. b Each of fragments I–V was cloned into pGBKT7 vector to test self-activation activity using yeast two-hybrid assays. c Validation of interaction between MdERF1B and MdMYB9, MdMYB1, and MdMYB11 in yeast two-hybrid assays. V: amino acid residues 19–197 of MdERF1B. MdERF1B19–197aa was cloned into binding domain; MdMYB9, MdMYB1, and MdMYB11 were each independently cloned into activation domain. d Interaction between MdERF1B and MdMYB9, MdMYB1, and MdMYB11 in pull-down assays. Mixtures of MdERF1B-His with MdMYB9-GST, MdMYB1-GST, or MdMYB11-GST fusion proteins were purified using a His-Tagged Protein Purification Kit with empty GST protein as control. e Bimolecular fluorescence complementation assays confirming interaction between MdERF1B and MdMYB9, MdMYB1, and MdMYB11 in vivo. Strong yellow fluorescent signals were detected in nuclei of onion cells transiently expressing MdERF1B-YN and MdMYB9/1/11-YC fusion proteins. Scale bar = 50 µm
Pull-down assays were performed to confirm the previous results. MdERF1B-His pulled down MdMYB9-GST, MdMYB1-GST, and MdMYB11-GST fusion proteins, but not the empty GST vector (Fig. 3d). Next, BiFC assays were performed to verify the interactions between MdERF1B and MdMYB9, MdMYB1, and MdMYB11 in vivo. MdERF1B-YN was co-transformed with MdMYB9-YC, MdMYB1-YC, MdMYB11-YC, or pSPYCE-35S (YC) into onion epidermal cells. Strong yellow fluorescent signals were detected in onion cell nuclei when MdERF1B-YN was transformed with MdMYB9-YC, MdMYB1-YC, or MdMYB11-YC. The empty vector, serving as the control, produced no fluorescent signal (Fig. 3e).
MdERF1B overexpression promotes anthocyanin accumulation in ‘Orin’ calli
To confirm the role of MdERF1B in regulating anthocyanin and PA production, MdERF1B-overexpressing ‘Orin’ calli were generated and verified by PCR amplification and western blot assays (Fig. 4a, b). Under low temperature and constant light conditions, MdERF1B-GFP calli became red, while the control calli transformed with the empty GFP-containing vector (GFP calli) remained yellow (Fig. 4c). Spectrophotometric analyses revealed that the anthocyanin content was significantly higher in the MdERF1B-overexpressing calli than in the control calli (Fig. 4e). The transcript levels of anthocyanin biosynthesis-related genes and TFs in MdERF1B-GFP and GFP calli were analyzed by qRT-PCR. Most of the structural genes and related TFs were up-regulated in MdERF1B-GFP, compared with the GFP control. However, the MdMYB1 transcript levels were the same in both types of calli (Fig. 4f, g). This result indicated that MdERF1B mainly affects MdMYB9 and MdMYB11 to regulate anthocyanin biosynthesis.
Increased anthocyanin accumulation in MdERF1B-overexpressing ‘Orin’ calli. a, b MdERF1B overexpression in ‘Orin’ calli verified by PCR amplification and western blotting. c MdERF1B-overexpressing ‘Orin’ calli (E1B-OE) and empty GFP vector-containing ‘Orin’ calli (GFP) under low temperature and constant light conditions. d Anthocyanin extracts of MdERF1B-overexpressing calli and GFP calli. e Anthocyanin contents of MdERF1B-overexpressing calli and GFP calli. f, g Transcript levels of anthocyanin biosynthesis-related genes and TFs in MdERF1B-overexpressing calli and GFP calli. FW, fresh weight. MdActin was used as an internal control. Error bars show SEs of means (n = 3). Lowercase letters indicate significant differences P < 0.05, and uppercase letters indicate highly significant differences at P < 0.01
MdERF1B overexpression promotes PA accumulation in ‘Orin’ calli
In apple, MdMYB9 and MdMYB11 promote anthocyanin and PA production (An et al. 2015). In this study, the MdMYB11 was significantly expressed in MdERF1B-overexpressing calli (Fig. 4g). When stained with DMACA, the MdERF1B-GFP calli were deeper blue than the GFP calli (Fig. 5a), and the PA content in MdERF1B-GFP calli was more than 50 times that in the GFP calli (Fig. 5c). Analyses of gene transcript levels by qRT-PCR showed that MdANR and MdLAR1 were up-regulated in MdERF1B-GFP calli compared with in GFP calli, while MdFLS was down-regulated in MdERF1B-GFP calli (Fig. 5d).
Increased proanthocyanidin (PA) accumulation in MdERF1B-overexpressing ‘Orin’ calli. a DMACA staining of MdERF1B-overexpressing ‘Orin’ calli (E1B-OE) and empty GFP vector-containing ‘Orin’ calli (GFP). b PA extracts of MdERF1B-overexpressing calli and GFP calli. c PA contents of MdERF1B-overexpressing calli and GFP calli. d Relative transcript levels of PA and flavonol pathway-related genes in MdERF1B-overexpressing calli and GFP calli. e Relative transcript levels of MdACO1 and MdERFs in MdERF1B-overexpressing calli and GFP calli. FW, fresh weight. MdActin was used as an internal control. Error bars represent SEs of means (n = 3). Lowercase letters indicate significant differences at P < 0.05, and uppercase letters indicate highly significant differences at P < 0.01
MdERF1B interacts with promoters of MdMYB9, MdMYB1, and MdMYB11
A previous study showed that two ERF subfamily TFs in Artemisia annua, AaERF1 and AaERF2, bind to CBF2 (GTCGAC) and RAA (CAACA) motifs (Yu et al. 2012). The promoters of MdMYB9, MdMYB1, and MdMYB11 all contain the RAA motif (Supplementary Fig. S1). Yeast one-hybrid assays were conducted to determine whether MdERF1B binds to the promoters of MdMYB9, MdMYB1, and MdMYB11. The optimal 3-AT concentrations to inhibit the expression of HIS3 in the pHIS2 vector on medium lacking Trp and His were determined to be 100 mM 3-AT for proMdMYB9-pHIS2 and proMdMYB1-pHIS2, and 120 mM 3-AT for proMdMYB11-pHIS2 (Fig. 6a). In Y1H assays, MdERF1B interacted with the promoters of MdMYB9, MdMYB1, and MdMYB11 (Fig. 6b). In EMSAs, MdERF1B specifically bound to the RAA motif in proMdMYB9, proMdMYB1, and proMdMYB11. With an increasing concentration of cold probe, the binding weakened, while the addition of a mutant cold probe did not affect binding (Fig. 6c–e).
Binding of MdERF1B to proMdMYB9, proMdMYB1, and proMdMYB11, and activation of proMdMYB9 and proMdMYB11. a proMdMYB9, proMdMYB1, and proMdMYB11 were cloned into pHIS2 vector, and optimal 3-AT concentration that inhibited the expression of the vector itself was determined. b Interactions between MdERF1B and proMdMYB9, proMdMYB1 and proMdMYB11 in yeast one-hybrid assays. c–e Interactions between MdERF1B and RAA motif in proMdMYB9, proMdMYB1, and proMdMYB11 as determined by electrophoretic mobility shift assay. f Luciferase reporter assays showing activation of proMdMYB9 and proMdMYB11 by MdERF1B. Error bars represent SEs of means (n = 3). Lowercase letters indicate significant differences at P < 0.05, and uppercase letters indicate highly significant differences at P < 0.01
To determine how MdERF1B regulates the activity levels of proMdMYB9, proMdMYB1 and proMdMYB11, we performed LUC reporter assays. As shown in Fig. 6f, MdERF1B transactivated proMdMYB9 and proMdMYB11, but barely affected the activity level of proMdMYB1. Thus, MdERF1B was found to mainly activate proMdMYB9 and proMdMYB11 to regulate anthocyanin and PA biosynthesis.
Discussion
The MBW ternary complex is a main regulator of the flavonoid pathway (Baudry et al. 2004; Quattrocchio et al. 2006; An et al. 2012). Some MYB TFs responsible for regulating anthocyanin synthesis in apple have been identified, including MdMYB1, MdMYBA, MdMYB10, MdMYB110a_JP, and MdMYB16 (Takos et al. 2006; Ban et al. 2007; Espley et al. 2007, 2009; Umemura et al. 2013; Xu et al. 2017). In recent studies on PA synthesis, MdMYB9 and MdMYB11 were found to interact with MdbHLH3 to form the MBW ternary complex, and then promote anthocyanin and PA biosynthesis by interacting with the promoters of ANS, ANR, and LAR (An et al. 2015). Subsequently, Wang et al. (2017) found that MdMYB12 and MdMYB22 promote PA and flavonol production, respectively. In addition to MYB TFs, other TFs are involved in flavonoid biosynthesis. In Arabidopsis, TTG2 (WRKY), ANAC078, and COP1 regulate flavonoid biosynthesis (Johnson et al. 2002; Morishita et al. 2009; Maier et al. 2013). In apple, MdHY5 activates proFLS to regulate flavonoid metabolism (Henry-Kirk et al. 2018). The results of the present study showed that MdERF1B regulates anthocyanin and PA biosynthesis in apple mainly by activating MdMYB9 and MdMYB11.
MdERF1B interacts with MdMYB9, MdMYB1, and MdMYB11
Members of the AP2/ERF superfamily participate in various regulatory pathways. In Arabidopsis, AtERF5 and AtERF6 activate the stress-resistance gene AtMYB51 (Dubois et al. 2013). In loquat (Eriobotrya japonica) fruit, the AP2/ERF TF EjAP2-1, which functions as a repressor, interacts with EjMYB1 and EjMYB2 to negatively regulate lignin biosynthesis (Zeng et al. 2015). PyERF3 interacts with PyMYB114 and PybHLH3 to promote anthocyanin biosynthesis in pear (Yao et al. 2017). In this study, MdERF1B was found to directly interact with MdMYB9, MdMYB1, and MdMYB11 in Y2H, pull-down, and BiFC assays (Fig. 3c–e). In Y2H assays, MdERF1B fused with pGBKT7 had stronger self-activation activity, and the N- and C-terminal regions of MdERF1B had self-activation activity (Fig. 3b), like tobacco ERF4 (Ohta and Shinshi 2000). Previous studies have shown that MdMYB1 is a key regulator of anthocyanin biosynthesis in apple, while MdMYB9 and MdMYB11 can interact with MdbHLH3 to promote anthocyanin and PA accumulation in apple (An et al. 2015). Our findings indicated that protein interactions between MdERF1B and MdMYBs may be involved in the regulation of anthocyanin and PA biosynthesis in apple.
MdERF1B participates in the regulation of anthocyanin and PA accumulation
Plant coloration is not only regulated by genetics but is also affected by phytohormones. In Arabidopsis, gibberellic acid inhibits anthocyanin accumulation (Loreti et al. 2008). In apple, high levels of auxin inhibit anthocyanin biosynthesis, cytokinin promotes anthocyanin accumulation (Ji et al. 2015), and MeJA mediates anthocyanin and PA accumulation by degrading MdJAZ2 (An et al. 2015). Ethylene promotes anthocyanin accumulation in apple, grape berries, and plum (Wang and Dilley 2001; El-Kereamy et al. 2003; Manganaris et al. 2008). However, the mechanisms of the ethylene pathway leading to flavonoid production are unclear. In this study, as the release of ethylene peaked, the anthocyanin level also increased (Fig. 1b, c). We investigated the ethylene response factor, MdERF1B, and found that its transcript levels corresponded with those of structural genes and MYB TFs in the flavonoid pathway (Fig. 1d). Thus, we hypothesized that MdERF1B in the ethylene pathway mediates anthocyanin production.
We tested this hypothesis by generating MdERF1B-overexpressing calli. The anthocyanin and PA contents were greater in the MdERF1B-overexpressing calli than in transgenic calli containing the empty vector (Figs. 4e, 5c). The structural genes in the flavonoid biosynthesis pathway can be divided into two groups: early biosynthetic genes such as CHS, CHI and F3H; and late biosynthetic genes such as DFR, ANS, UFGT, FLS, LAR, and ANR (Nesi et al. 2000). In this study, the overexpression of MdERF1B indirectly increased the transcript levels of early and late biosynthetic genes (Figs. 4f, 5d). The only exception was FLS, because in the flavonoid pathway, DFR and FLS have the same substrate and therefore have a competitive relationship. This explains why the increase in DFR expression resulted in a concomitant reduction in FLS expression (Figs. 4f, 5d).
ACS and ACO are essential for ethylene synthesis, and ERFs are the main regulators of the ethylene pathway. Previous studies have shown that in apple, MdERF1 responds to fruit ripening, and MdERF3 promotes the release of ethylene by binding to the MdACS1 promoter (Wang et al. 2007; Li et al. 2016). In our study, MdERF1B significantly stimulated the transcription of MdACO1, MdERF1, and MdERF3. In particular, MdACO1 was significantly upregulated, with its transcript level in MdERF1B-overexpressing calli being more than 34 times that in the control calli (Fig. 5e). These findings indicated that MdERF1B may be a positive regulator in the ethylene pathway, mediating MYBs to regulate anthocyanin and PA production.
MdERF1B stimulates the activity of proMdMYB9 and proMdMYB11
Based on the number of AP2/ERF domains and sequence similarities, the AP2/ERF superfamily can be divided into three groups; the ERF, AP2, and RAV families. The ERF family proteins can be classified into groups I–X (Nakano et al. 2006). Our phylogenetic tree revealed that MdERF1B belongs to group IX, along with AtERF1, ORA59, AaERF1, AaERF2, MdERF3, and LePti4 (Fig. 2a). In Arabidopsis, AtERF1 integrates ethylene, JA, and abscisic acid signals to respond to biotic and abiotic stresses (Lorenzo et al. 2003; Cheng et al. 2013). Additionally, ORA59 is an integrator of the JA and ET pathways and directly binds to the GCC box of PDF1.2 to activate defense responses (Zarei et al. 2011). LePti4 responds to JA-regulated pathogen invasion in tomato. In A. annua, AaERF1 and AaERF2 respond to JA to promote artemisinin biosynthesis (Yu et al. 2012). In apple, MeJA regulates MdERF3 to promote ethylene production (Li et al. 2017). The protein sequences of MdERF1B and AtERF1 shared 60.94% identity (Supplementary Fig. S2), but different from the function of AtERF1, the function of MdERF1B was to promote the accumulation of anthocyanin and PA (Figs. 4c–e, 5a–c).
The ERF family contains ERF and CBF/DREB subfamilies (Nakano et al. 2006). The CBF/DREB subfamily mainly binds to the DRE/CRT (A/GCCGAC) motif in response to abiotic stress (Sakuma et al. 2002), and the ERF subfamily mainly binds to GCC-box (AGCCGCC) elements in response to biotic stress (Ohme-Takagi and Shinshi 1995; Pré et al. 2008; Zarei et al. 2011). In A. annua, both AaERF1 and AaERF2 in the ERF subfamily bind to CBF2 (GTCGAC) and RAA (CAACA) motifs (Yu et al. 2012) to promote artemisinin biosynthesis. In this study, we found RAA motifs in the promoters of MdMYB9, MdMYB1, and MdMYB11 (Supplementary Fig. S1). In a series of analyses, such as Y1H and EMSA, MdERF1B was found to interact with proMdMYB9, proMdMYB1, and proMdMYB11 (Fig. 6b–e). Finally, we performed LUC reporter assays, in which MdERF1B stimulated the activity of proMdMYB9/11, but not proMdMYB1 (Fig. 6f). In the MdERF1B-overexpressing calli, MdERF1B did not affect the expression of MdMYB1 (Fig. 4g). The specific reasons for these results still need to be studied. Previous studies have shown that MdMYB1 is an important factor in anthocyanin synthesis in apple fruit skin (Takos et al. 2006), and that MdMYB9 and MdMYB11 promote anthocyanin and PA expression through the MBW complex (An et al. 2015). The results of the present study showed that MdERF1B regulates the accumulation of anthocyanin and PA mainly by affecting MdMYB9 and MdMYB11.
Conclusion
On the basis of our results, we propose a model for the function of MdERF1B in the ethylene pathway to regulate anthocyanin and PA accumulation in apple (Fig. 7). Our results showed that MdERF1B not only interacts with MdMYB9 and MdMYB11, but also binds to the promoters of MdMYB9 and MdMYB11. Therefore, it promotes anthocyanin and PA accumulation mainly by regulating MdMYB9 and MdMYB11.
Putative regulatory mechanism by which MdERF1B in ethylene pathway promotes anthocyanin and proanthocyanidin (PA) biosynthesis in apple. MdERF1B not only interacts with MdMYB9/11 proteins, but also binds to their promoters through their RAA motifs to activate their expression, which increases anthocyanin and PA production
Abbreviations
- PAs:
-
Proanthocyanidins
- PAL:
-
Phenylalanine ammonia-lyase
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- F3H:
-
Flavanone 3-hydroxylase
- DFR:
-
Dihydroflavonol 4-reductase
- ANS:
-
Anthocyanidin synthase
- UFGT:
-
Glycosyltransferase
- LAR:
-
Leucoanthocyanidin reductase
- ANR:
-
Anthocyanidin reductase
- FLS:
-
Flavonol synthase
- GFP:
-
Green fluorescent protein
- GST:
-
Glutathione S-transferase
- His:
-
Histidine
- Anti:
-
Antibody
- MS:
-
Murashige and Skoog
- YFP:
-
Yellow fluorescent protein
- Trp:
-
Tryptophan
- Leu:
-
Leucine
- Ade:
-
Adenine
- CDS:
-
Coding DNA sequence
- 3-AT:
-
3-Amino-1,2,4-triazole
- BiFC:
-
Bimolecular fluorescence complementation assay
- DAFB:
-
Days after full bloom
- EMSA:
-
Electrophoretic mobility shift assay
- TFs:
-
Transcription factors
- DMACA:
-
4-Dimethylaminocinnamaldehyde
- qRT-PCR:
-
Quantitative reverse transcription-PCR
- LUC:
-
Luciferase
- ET:
-
Ethylene
- 6-BA:
-
6-Benzylaminopurine
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- JA:
-
Jasmonate
- MBW:
-
MYB–bHLH–WD40
- MeJA:
-
Methyl jasmonate
- bHLH:
-
Basic helix–loop–helix
- Y1H:
-
Yeast one-hybrid
- Y2H:
-
Yeast two-hybrid
- 4-MUG:
-
4-Methylumbellifery-β-d-glucuronide hydrate
References
An XH, Tian Y, Chen KQ, Wang XF, Hao YJ (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol 169:710–717
An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56:650–662
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Barry CS, Giovannoni JJ (2007) Ethylene and fruit ripening. J Plant Growth Regul 26:143–159
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097
Dubois M, Skirycz A, Claeys H, Maleux K, Dhondt S, De BS, Vanden BR, De ML, Yoshizumi T, Matsui M, Inzé D (2013) ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol 162:319–332
El-Kereamy A, Chervin C, Roustan JP, Cheynier V, Souquet JM, Moutounet M, Raynal J, Ford C, Latché A, Pech JC, Bouzayen M (2003) Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol Plant 119:175–182
Espley RV, Hellens RP, Jo P, Stevenson DE, Sumathi KA, Allan AC (2007) Red coloration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Giusti M, Wrolstad RE (2001) Characterization and measurement of anthocyanins by UV-visible spectroscopy. In: Current protocols in food analytical chemistry. Wiley, Hoboken, F1.2.1–F1.2.13
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Henry-Kirk RA, Plunkett B, Hall M, Mcghie T, Allan AC, Wargent JJ, Espley RV (2018) Solar UV light regulates flavonoid metabolism in apple (Malus × domestica). Plant Cell Environ 41:675–688
Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem 40:955–962
Ji XH, Wang YT, Zhang R, Wu SJ, An MM, Li M, Wang CZ, Chen XL, Zhang YM, Chen XS (2015) Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f.niedzwetzkyana). Plant Cell Tissue Organ Cult 120:325–337
Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14:1359–1375
Kende H (2003) Ethylene biosynthesis. Annu Rev Plant Biol 44:283–307
Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72:427–441
Li YG, Tanner G, Larkin P (1996) The DMACA–HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70:89–101
Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A (2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88:735–748
Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A (2017) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29:1316–1334
Lister CE, Lancaster JE, Walker JRL (2015) Developmental changes in enzymes of flavonoid biosynthesis in the skins of red and green apple cultivars. J Sci Food Agric 71:313–320
Liu Q, Wen CK (2012) Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol 158:1193–1207
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Lorenzo O, Piqueras R, Sánchezserrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15:165–178
Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P (2008) Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol 179:1004–1016
Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hülskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74:638–651
Manganaris GA, Vicente AR, Crisosto CH, Labavitch JM (2008) Effect of delayed storage and continuous ethylene exposure on flesh reddening of ‘Royal Diamond’ plums. J Sci Food Agric 88:2180–2185
Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S (2009) Arabidopsis NAC transcription factor, ANAC078, regulates flavonoids biosynthesis under high. Plant Cell Physiol 50:2210–2222
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12:1863–1878
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2144
O’Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ (1996) Ethylene as a signal mediating the wound response of tomato plants. Science 274:1914–1917
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182
Ohta MTM, Shinshi H (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22:29–38
Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275:527–530
Pré M, Atallah M, Champion A, De VM, Pieterse CM, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147:1347–1357
Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R (2006) PH4 of Petunia Is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18:1274–1291
Ross JA, Kasum CM (2002) DIETARY FLAVONOIDS: bioavailability, metabolic effects, and safety. Annu Rev Nutr 22:19–34
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12:3703–3714
Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Umemura H, Otagaki S, Wada M, Kondo S, Matsumoto S (2013) Expression and functional analysis of a novel MYB gene, MdMYB110a_JP, responsible for red flesh, not skin color in apple fruit. Planta 238:65–76
Wang Z, Dilley DR (2001) Aminoethoxyvinylglycine, combined with ethephon, can enhance red color development without over-ripening apples. HortScience 36:328–331
Wang A, Tan D, Takahashi A, Zhong Li T, Harada T (2007) MdERFs, two ethylene-response factors involved in apple fruit ripening. J Exp Bot 58:3743–3748
Wang L, Lee IM, Zhang SM, Blumberg JB, Buring JE, Sesso HD (2009) Dietary intake of selected flavonols, flavones, and flavonoid-rich foods and risk of cancer in middle-aged and older women. Am J Clin Nutr 89:905–912
Wang N, Xu H, Jiang S, Zhang Z, Lu N, Qiu H, Qu C, Wang Y, Wu S, Chen X (2017) MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f.niedzwetzkyana). Plant J 90:276–292
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299:396–399
Xie XB, Shen LI, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35:1884–1897
Xu H, Wang N, Liu J, Qu C, Wang Y, Jiang S, Lu N, Wang D, Zhang Z, Chen X (2017) The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol Biol 94:149–165
Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35:155–189
Yao G, Ming M, Allan AC, Gu C, Li L, Wu X, Wang R, Chang Y, Qi K, Zhang S, Wu J (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J 92:437–451
Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5:353–365
Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol 75:321–331
Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, Chen KS (2015) EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol J 13:1325–1334
Acknowledgements
This work was supported by the National Key Research Project (2016YFC0501505) and the National Natural Science Foundation of China (31572091, 31730080). We thank Shujing Wu Laboratories for providing the vectors. We thank Lesley Benyon, PhD, and Jennifer Smith, PhD, from Liwen Bianji, Edanz Group China (http://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
XC and JZ: conceived and designed the experiments. JZ and HX: analyzed the data. NW, SJ, HF, ZZ, GY, YW, MS and LX: contributed reagents/materials/analysis tools. JZ: wrote the paper.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2018_770_MOESM3_ESM.tif
Fig. S1 RAA motifs in proMdMYB1, proMdMYB9 and proMdMYB11. Transcriptional start sites (ATG) are indicated with +1. (TIF 133 KB)
Rights and permissions
About this article
Cite this article
Zhang, J., Xu, H., Wang, N. et al. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol Biol 98, 205–218 (2018). https://doi.org/10.1007/s11103-018-0770-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0770-5