Abstract
Key message
Contextualization of specific transcriptional responses of Arabidopsis within the stress–tissue–time perspective provides a simplified representation of the cellular transcriptional response pathways to abiotic stress, while reducing the dimensions in gene-oriented response description.
Abstract
Crops resistant to abiotic stresses are a long-term goal of many research programs, thus understanding the progression of stress responses is of great interest. We reanalyzed the AtGenExpress transcription dataset to go beyond gene-level characterization, and to contextualize the discrete information into (1) a process-level signature of stress-specific, time-specific, and tissue-specific responses and (2) identify patterns of response progression across a time axis. To gain a functional perspective, ∼1000 pathways associated with the differentially-expressed genes were characterized across all experiments. We find that the global response of pathways to stress is multi-dimensional and does not obviously cluster according to stress, time or tissue. The early response to abiotic stress typically involves induction of genes involved in transcription, hormone synthesis and signaling modules; a later response typically involves metabolism of amino acids and secondary metabolites. By linking specific primary and secondary response pathways, we outline possible stress-associated routes of response progression. The contextualization of specific processes within stress–tissue–time perspective provides a simplified representation of cellular response while reducing the dimensions in gene-oriented response description. Such simplified representation allows finding stress-specific markers based on process-combinations pointing whether a stress-specific response was invoked as well as provide a reference point for the conductance of comparative inter-plant study of stress response, bypassing the need in detailed orthologous mapping.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most crop plants grow in suboptimal environments. Unfavorable conditions often induce stress responses involving physiological and morphological adaptations (Cramer et al. 2011). Often, stress-induced modifications prevent plants from attaining their full genetic potential (Buchanan et al. 2000), affect production (Shao et al. 2008) and lead to reduced yield (Wang et al. 2003). At the cellular level, resources, otherwise invested in growth, are transformed into the production of stress-induced metabolites (Herms and Mattson 1992; Le Bot et al. 2009). Adjustments of the cellular metabolic activity is achieved through cross-talks between different regulatory levels (Verslues et al. 2006). For example, metabolism of secondary metabolites such as phytohormones have been implicated in different stress responses (Cutler et al. 2010; Hare et al. 1997). Phytohormones further regulate the production of many other metabolites, including primary metabolites such as amino acids, carbohydrates, lipids, and a large variety of secondary metabolites (Akula and Ravishankar 2011; Goossens et al. 2003; Less and Galili 2009; Prescott 1999; Rolland et al. 2002; Rosa et al. 2009; Sasaki-Sekimoto et al. 2005).
Overall, cellular responses to abiotic stress are complex and vary over space, time and stress-type. In the coming decades, changes in global temperatures are predicted to include enhanced peaks of drought, flood, and heat (Mittler 2006), significantly affecting global agricultural systems (Parry 2007). The growing awareness of environmental costs in agriculture leads to extensive studies of plant responses to abiotic stresses. Transcript profiling has been widely employed in studying plant responses to abiotic stress conditions (Ahuja et al. 2010; Bechtold et al. 2016; Grativol et al. 2012; Nakashima et al. 2009). AtGenExpress is one of the most comprehensive and systematic resources providing a four dimensional gene expression profile dataset for the study of abiotic stress transcriptional response in a plant (Kilian et al. 2007), with the dimensions as environmental conditions, time course, tissues and full gene-sets (as covered by the Arabidopsis Affymetrix DNA microarray). Expression profiles were generated for eight different abiotic stresses (osmotic, salt, drought, genotoxic, wound, cold, heat, UV-b), at six time points (0.5, 1, 3, 6, 12, and 24 h of treatment) for two tissues (root and shoot). The published analysis of this data primarily focused on differential analysis of gene expression in response to three of the stress types (UV-B light, drought and cold stress) (Kilian et al. 2007), as well as the characterization of the response in the treated versus untreated tissue (Hahn et al. 2013). Yet much of these data remains under-analyzed.
The integration of gene-level data into higher level structures such as pathways and networks provides a key step in the analyses of big data (Ferrier et al. 2011; Heyndrickx and Vandepoele 2012; Toubiana et al. 2013). Here, we aimed to use available data to delineate the pathway-level response to different stresses. To this end, we made use of this experimental platform and carried an enrichment analysis to elucidate the pathway profiling of all 96 experiments available. We further looked for both common and stress-specific response patterns across each of the experimental dimensions, characterized primary versus secondary response patterns, and delineate links between pathway categories across response progression.
Materials and methods
Stress related microarray databases
We used “The AtGenExpress” dataset of abiotic stress experiment from the GEO platforms on the NCBI database. Experiments conducted on the Affymetrix platforms, Arabidopsis ATH1 Genome Array (GPL198), were chosen for this study as they provide extensive gene coverage and are widely used. GEO dataset of abiotic stress experiment series were (1) GSE5620 (Control Plant) (2) GSE5621 (Cold stress) (3) GSE5622 (Osmotic stress) (4) GSE5623 (Salt stress) (5) GSE5624 (Drought stress) (6) GSE5625 (Genotoxic stress) (7) GSE5626 (UV-B stress) (8) GSE5627 (Wounding stress) (9) GSE5628 (Heat stress). The RNA samples from root and shoot were taken in two biological replicas 0 min, 30 min, 1 h, 3 h, 6 h, 12 h and 24 h after the onset of stress treatment. In all experiments plants were handled in exactly the same way with the exception of the exposure to stress conditions (Kilian et al. 2007). Control data were utilized for normalization of experimental data. Overall the data includes 96 experiments (8 stress × 6 time point × 2 tissues).
Identification and functional analysis of differentially expressed genes
The microarray data were adjusted for the background of optical noise the with GC-RMA (Wu et al. 2004) package in MatLab tool and normalized using quantile normalization non probe-level (Wu and Aryee 2010) followed by median-polish summarization. Gene expression values were normalized to the controls, primary normalization was used to generate the principal component analysis and secondary normalization was used to identify up- or downregulated genes. Further t test and false discovery rate (FDR) were applied to detect differentially expressed genes between treatment and control groups (Kim and Park 2004). Genes were considered differentially expressed, if they were regulated in at least a single experiment. A total of 7300 and 4765 up and down regulated genes, respectively, were detected. The breaking of the numbers of up-regulated genes into categories is now provided in Supplementary Table 1.
Functional annotations for Arabidopsis genes were downloaded from the MapMan annotation scheme (Thimm et al. 2004) that is structured in the form of a hierarchical tree and was specifically designed to cover plant-specific pathways and processes. Supplementary Table 2 specifies for each gene its functional annotations and its profile of up-regulation events across experiments.
Profiles of pathway enrichment in each experiment were determined by calculating the cumulative hypergeometric p value for the probability that a group of genes (up-regulated genes in an experiment) is over-represented within a functional bin at a rate higher than chance expectation (Freilich et al. 2015). We chose to focus on upregulated genes to avoid mixed signals that can result from the co-inclusion of down-regulated genes and would likely introduce contradicting signals (Freilich et al. 2015). A separate analysis of the down-regulated is given in Additional file 1. Multiple tests were performed for all gene groups—functional categories at all hierarchical levels. To control for multiple testing the original p values were then converted into FDR overlaps, with FDR < 0.001 considered significant.
Associating pathway categories
For associating pathways, we considered the following combinations: (i) primary pathways (RNA/hormones at time 0.5) versus secondary pathway (amino acids/secondary metabolites at time 6 h); (ii) amino acid and secondary metabolic pathways at 6 h. For each primary category, we iteratively formed pathway combinations considering all pathways assigned to each the key category (e.g., all categories classified under RNA versus all categories classified under amino acids). Each pathway category was described by its enrichment profile across all stresses in a tissue. For each category-combination we calculated an association score by dividing the number of AND events (experiments where pathways form both response types are significantly enriched) by the number of OR events (experiments where at least one of the pathways is significantly enriched). Results were tested under various cutoffs and a threshold of 0.6 were chosen being both robust and comprehensive (Additional file 1). Calculations were carried employing MatLab tool. Network files providing pathway association across all classification level are provided in Supplementary Table 4. Associations were summed to a uniform level for each category: RNA categories are at the third level of classification at the MapMan scheme (specific TFs); hormones and secondary metabolites classification categories at the 2nd level (specific compounds), and amino acids at the fourth level (specific amino acids). A table presenting associations at the selected level is provided as Supplementary Table 5.
Heat map, hierarchical clustering, PCA and network analysis
For the data mining and visualization, heat map and hierarchical clustering were built in R studio and Matlab. PCA was applied using PAST software (Hammer et al. 2001). Cytoscape 3.2.1 (Shannon et al. 2003) was used for visualization of biological network of pathways using the association index values of primary and secondary responses of abiotic stresses. Network layout was carried automatically using the ‘organic’ layout option that visualizes the clusters in the data. Clustering analysis was carried with both ClusterViz and ModuLand plugins, both detecting a single cluster using default and other cutoffs.
Results and discussion
Expression pattern of differentially expressed genes in abiotic stresses
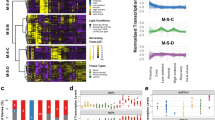
A total of 7300 genes were up-regulated in a total of 49,986 events across the 96 experiments (“Materials and methods” section), within the range of previous reports for the current data set (Kilian et al. 2007). Figure 1a shows a heat map representing the number of up-regulated genes in roots and shoots along a time course following each abiotic stress condition. The results suggest communication between root and shoots tissue. For example, wounding stress was applied only to the aerial parts of the plants; this induced rapid responses in gene expression first in shoot, and a later, milder, response in the roots. The opposite was observed for salt and osmotic stress, where treatment of the roots caused significant alterations first in the root and then in the shoot. Response extent and patterns differ between stress types. A continuous increase over time in the number of induced genes is observed following cold and osmotic exposure (both root and shoot tissues), while the other stresses generally elicited transient alterations in gene expression. Genotoxic stress induced a small number of genes in comparison to the other stresses. Hahn et al. (2013) also observed abiotic stress has capability to generate systemic signals between the shoot and root tissues and suggested it might be essential to regain homeostasis.
Distribution of differentially up-regulated genes or significantly enriched pathways across experiments. a Heat map illustrating the number of upregulated genes. b Heat map illustrating the number of pathways significantly enriched with upregulated genes. c A PCA analysis of the up-regulated gene profiles across the 96 experiments. The data were normalized and auto scaled prior to PCA. Numbers in parenthesis indicate the percentage of variance explained by each PC
This initial analysis confirms and builds on many of the conclusions from Kilian et al. (2007). The majority of these stress-regulated genes are not present in the basal co-regulation network we described previously (Atias et al. 2009). More specifically, while we identified genes as stress-induced, only 1020 of these are found in the basal co-regulation network (Supplementary Table 2), representing 14 % of this network. Thus most stress-induced genes are not co-expressed in the absence of stress conditions.
Identification of pathways enriched in differentially expressed genes across experimental dimensions
To delineate the overall cellular activity during the progression of stress response, genes were assigned to the MapMan hierarchical annotation scheme providing high (general, e.g. amino acid metabolism) and low (specific, e.g. alanine aminotransferase) level descriptions for the functional role of each gene. For each experiment (specific stress, specific time point, specific tissue), we calculated the frequency of genes at each classification level in order to outline the cellular processes that are more significantly associated with the condition. Overall, we identified 1148 significantly enriched pathways at different hierarchical levels (Supplementary Table 3). The number of pathways enriched per experiment (Fig. 1b) is in general agreement with the number of genes upregulated (Fig. 1a).
To identify similarities between experimental dimensions, both the profiles of differentially expressed genes and of enriched pathway subjected to PCA analysis (Fig. 1c; Supplementary Fig. 1, respectively). The analysis shows that the transcriptional responses to osmotic and salt stresses are most similar. The early response to UV, cold and wounding grouped together, while there was no clear separation between other stress responses. In general, this analysis of the pathways was not sufficient to delineate specific cellular processes associated with individual stresses. Similar patterns (or lack thereof) were observed using hierarchical cluster analysis (Supplementary Fig. 2). Hence we looked in detail at the specific pathways involved.
Associating transcriptional response progression during abiotic stress with specific pathway categories
For each top-level category, we counted the total number of enriched pathways in each experiment. Out of 30 top-level categories, we focused on seven key categories, requiring the enrichment of at least ten sub-pathways in at least a single experiment (Fig. 2). This analysis juxtaposed with those in Fig. 1 (and in particular the general pathway analysis in Fig. 1b), giving a more top-down description of the effect of exposure to abiotic stress on cellular activity, and allowing a detailed view of the cellular processes involved in response progression. Though the focusing on seven key categories induces potential bias for more-developed categories, filtering was required for gaining a comprehendible signal, where the full dataset (Supplementary Table 3) allows a more detailed inspection.
Distribution of significantly enriched pathways from key categories across experiments. The heat map illustrates scaled distribution of the number of pathways significantly enriched with up-regulated genes across key MapMan categories. For each stress condition, the data point with the largest number of pathways was defined as 1 (dark red), to which all other points in the same conditions were scaled against (color bar on right)
For example, osmotic and salt response starts early in root tissues followed by a response in the shoot (Fig. 1a, b). Looking at the specific pathways involved, we observe a relatively high number of hormonal pathways enriched during the early response in both tissue types (Fig. 2). The wound response starts in shoot tissues (Fig. 1a, b), also associated with a high number of enriched hormonal pathways. The drought response starts in the root tissue, and after 3 h continues in the shoot (Fig. 1a, b) with up-regulation of genes assigned to hormones pathways being the primary response in the root, and genes assigned to hormones and amino acid pathways being the later response in the shoot (Fig. 2). The transcriptional response to heat stress starts after 1 h in both tissues (Fig. 1a, b), with RNA regulatory pathways as the main contributors (Fig. 2). Cold stress responses are observed in the shoot after 3 h, followed by a response in the root, again here mediated mainly by RNA regulatory pathways (Fig. 2). The main pathway category enriched following genotoxic stress was amino acid metabolism, at the relatively late time of 3 h. Reexamination of the data (Supplementary Table 3) showed that early (30 min) genotoxic exposure initially induced a DNA damage response; however the number of pathways induced was below the threshold for consideration in the analysis shown in Fig. 2 (in correspondence with the small number of genes).
To delineate a generic trend of transcriptional response progression, we summed together the scaled scores given for each stress (Fig. 2), and looked at the level of representation of each functional category across the stress responses in different time points for each tissue (Fig. 3a). This analysis reveals that the responses can be grossly divided into three steps (Fig. 3b). The early primary transcriptional responses to abiotic stresses (0.5–1 h) include the induction of pathways related to RNA regulation (e.g. transcription factors) and hormone metabolism. This is followed by a late primary response (1–3 h) characterized by the up-regulation of various signaling modules, such as MAP kinases, Ca2+ and receptor kinase pathways. The later secondary responses include up-regulation of genes controlling amino acid, lipid and secondary metabolism pathways (typically 3–6 h). Notably, the current analysis focuses on the transcriptional response whereas an earlier cascade of information transduction is transmitted through a diverse array of molecule types including hormones, siRNA, possibly ssRNA molecules and electrical signals is beyond the detection scope here (Hahn et al. 2013).
Cross-stress response patterns of pathways. a Heat maps illustrating scaled distribution of pathways across multiple stresses. Values in each cell represent the summing together of scaled values across all stress conditions (as in Fig. 2). b Diagram of pathways response progression during abiotic stress
Characterization of stress-specific versus stress-generic pathways
The generic trend of response progression (Fig. 3b), allows narrowing the big picture by focusing on four dominant pathway categories, each category at a single time point: RNA and hormones at time point 0.5; amino acids and secondary metabolites at time point 6 h. For each pathway category, we surveyed enriched pathways at all classification levels.
Notably, across all stresses, we observe that the number of enriched hormonal categories was higher in the tissue directly exposed to the stress (shoot versus root), with the exception of heat stress where the number of hormonal pathways is relatively low. Across all stress types, the most common hormonal pathways are ethylene-associated functions, in accordance with previous reports supporting its key role in stress response regulation (Beguerisse-Dıaz et al. 2012; Kushwah et al. 2011; Zhai et al. 2013). The only experiments where ethylene response was not detected are in tissues not directly exposed to stress (e.g., ethylene response was detected in the shoot but not in root following wound stress) (Fig. 4a). Similarly, gibberellin response was detected in all stress-exposed tissues with the exception of heat (Fig. 4a). Jasomonate response was not detected in cold, heat and UVb stresses. Abscisic response was not detected in genotoxic, heat and UVb stresses (Fig. 4a). The most specific hormonal pathways in this experimental system were auxin (wound, cold) and salicylic acid (UVb) (Fig. 4a).
Enriched pathways from selected key categories in specific stresses within each panel, each color is corresponding to the top-category of the pathways indicated on the right side of the graph. a, b Enriched pathways at 0.5 h following stress classified to a the hormonal category, or b the RNA category. c, d Enriched pathways at 6 h following stress classified to the c amino acids category, or d secondary metabolites category
Within the RNA category, the most common RNA pathways across all stress types are C2C2 (Zn) F-zinc finger family, MYB and WRKY transcription factor functions (Fig. 4b). In support of our results, the dominant involvement of most of these TF families in the regulation of stress response was supported by previous studies. C2H2-type zinc finger proteins were demonstrated to play a crucial role in plant stress responses following exposure to various stress types including low temperature, salt, drought, osmotic stress and oxidative stress (Kiełbowicz-Matuk 2012; Wang et al. 2014). MYB proteins were shown to be regulators of the response to salinity, drought and cold in Arabidopsis (Lippold et al. 2009). WRKYs TFs are up-regulated following exposure to dehydration, cold, heat, UV irradiation, drought, salt and ABA (Qiu and Yu 2009). More stress-specific enrichment patterns were detected for ABI3/VP1-related B3-domain transcription category (shoot tissues following genotoxic and UV stresses) (Fig. 4b); GRF zinc finger family (genotoxic and drought shoot tissues); and bZIP related family (root tissues following osmotic, salt and heat stresses) (Fig. 4b).
Unlike the relatively generalized enrichment pattern of hormonal and RNA activities, the metabolism of amino acids demonstrate higher specificity in response to stress. Most specifically, the synthesis of aspartate and cysteine are observed only following drought stress in the shoot tissue; degradation of asparagine was unique to root tissue following salt stress; and GABA synthesis was detected in root tissue following osmotic stress (Fig. 4c). The most diverse response in regard to the metabolism of amino acids was detected following exposure to drought and osmotic stresses in both root and shoot tissues (Fig. 4c). Shoot versus root responses show similarities between the two stress types. More generally, root versus shoot responses following stress do not typically cluster together suggesting the tissue types differ in their subsequent cellular modifications. Finally, we see similar number of enriched pathways involved in degradation and in synthesis (Fig. 4c). Notably, amino acids act as common precursors of secondary metabolites (Akula and Ravishankar 2011) whose production was in many cases induced following abiotic stress (Nakabayashi and Saito 2015). Our survey points at glucosinolates, flavonoids, isoprenoids and phenylpropanoids as the main secondary metabolites for which we detect enrichment in upregulated genes (Fig. 4d).
Network analysis linking primary and secondary pathway responses
Highlighting the key regulatory (hormonal, RNA) and metabolic processes (amino acids, secondary metabolites) allows further delineating associations between pathways, based on co-occurrence patterns. We calculated a similarity coefficient between pathways classified under the four categories (RNA, hormones, amino acids, secondary metabolites) based on the frequency a pathway from category A and a pathway from category B were enriched under the same experimental setting (tissue exposed to stress type; “Materials and methods” section). To this end, we iteratively crossed all primary responses with secondary responses, forming pair-wise combinations of a primary response pathway with a secondary response pathway. Similarly, pathways from the amino acids categories were iteratively crossed with pathways from the secondary metabolism categories. The links aim at revealing both regulatory (primary/regulatory processes with secondary-metabolic processes) and biosynthetic (amino acids with secondary metabolites) associations.
Overall links were formed between 5 hormonal pathways and 11 transcription regulation pathways (primary response), 22 amino acids metabolism pathways (synthesis and degradation) and 7 secondary metabolism pathways (late response) in root and shoot tissues (Fig. 5a). Reassuringly, many of the links are supported by previous studies. For example, associations between transcription factors and secondary metabolites include links between MYB transcription factors and glucosinolate biosynthesis in the root, in accordance with the reported role of MYB proteins as regulators of glucosinolate biosynthesis in Arabidopsis stress response (Hirai et al. 2007). Examples for hormonal and secondary metabolites associations include links between jasmonate pathway and flavonoids and glucosinolate, in accordance with the role of jasmonate as a regulator of the synthesis of these metabolites during UV and wound stresses (Barah et al. 2013; Demkura et al. 2010). Glucosinolates were also connected with abscisic acid and ethylene in Arabidopsis, and in a few other Brassica crop species (Yan and Chen 2007).
Network association of primary and secondary categories at selected time points. Nodes represent pathways in the following categories: Primary response (0.5 h)—Hormones (green) and RNA transcriptome (red); Secondary response (6 h)—amino acid (blue) and secondary metabolite (yellow). The edges were drawn between nodes when their co-appearance score was higher than threshold (“Materials and methods” section). Nodes of hormonal and secondary metabolites categories are shown at the second-level of classification; Nodes of RNA and amino-acids categories are shown at the third and fourth level of classification, respectively. a The full network containing associations from both shoot and root. Subnetworks composed of the direct neighbors of the nodes b for methionine synthesis and degradation and c for asparagine synthesis and degradation
Associations between hormones and amino-acids include links between ethylene and aspartate and ethylene and alanine (shoots and roots, respectively). A link between ethylene and aspartate was supported by Arraes’ model suggesting an ethylene-mediated aspartate biosynthesis in drought stress (Arraes et al. 2015). The link between ethylene and alanine can possibly reflect the role of β-alanine as an ethylene precursor (Stinson and Spencer 1969). Aromatic amino acids are linked with jasmonic acid and abscisic acid (in root) and auxin (in shoot). Such links can be related to either biochemical or regulatory associations. Phenylalanine, tyrosine and tryptophan serve as precursors for auxin (Tzin and Galili 2010). Jasomonate and abscisic acid induced signaling regulates tryptophan and tyrosine biosynthesis, respectively (Dombrecht et al. 2007).
Amino-acids serve as precursors for the synthesis of secondary metabolites. In accordance with the biochemical pathway, phenylpropanoids and flavonoids are linked with aromatic amino acids (Casati and Walbot 2005); glucosinolates are linked with methionine, branched-chain and aromatic amino-acids (Halkier and Gershenzon 2006). Cysteine, also linked with glucosinolates, acts as a donor for all reduced sulfur in plant cells and involved in biosynthesis and metabolism of glucosinolates in Arabidopsis (Takahashi et al. 2011). Tryptophan and glutamate, linked with indole-glucosinolates, are known to be catabolized into many indole-containing secondary metabolites (Hildebrandt et al. 2015). The most highly connected categories are glucosinolate: indolic glucosinolate in root and aliphatic glucosinolate in shoot (Supplementary Table 5). This further supports the centrality of glucosinolates metabolism in the response to environmental stress in Arabidopsis.
Beyond specific connections, the network perspective contextualizes individual links and allows identifying multiple pathway associations at several functional levels. Attempts to take a “guilt by association” approach (Oliver 2000) were hampered by the low modularity of the network, where pathways could not be clustered into meaningful groups, in accordance with the typical pleiotropic nature of stress response. Taking an alternative approach, we focused on the links formed between amino-acids—the most stress-specialized category (Fig. 4), and other pathway categories. In Fig. 5b, c we show sub-networks containing methionine and asparagine respectively—the two amino acids for which both synthesis and degradation categories were included in the network, and their direct neighbors. Notably, in both networks we observe that the regulatory elements are typically exclusively associated with either synthesis or degradation processes. In the methionine network, degradation is associated with PHOR1, C3H zinc finger and GRAS transcription factor families; synthesis is associated with C2H2 zinc finger family. Similarly, B3 transcription factor family is associated with asparagine degradation only (Fig. 5c). The glucosinolate category provides a bridge between asparagine synthesis and degradation. Looking at the lower level of glucosinolate pathways (Supplementary Table 5) we observe that glucosinolate synthesis is directly linked with asparagine synthesis; glucosinolate degradation is indirectly linked with asparagine degradation, via its association with B3 transcription factor family. All these three pathways—degradation of asparagine and glucosinolate and B3 transcription factor—are enriched in the root tissue following exposure to salt stress conditions (Fig. 4), pointing at a possibly higher level coordinated process. All pathways associated with asparagine synthesis are co-connected between themselves. That is, both hormonal categories associated with asparagine synthesis and both secondary metabolites categories associated with asparagine synthesis are co-associated. Hence, taking this networks approach we predict a number of potential pathway interactions not previously identified, as well as possible higher-level, multi-pathways, functional modules.
Conclusion
Numerous research programs have studied the transcriptional response to various abiotic stresses. However, much of the data remains under-analyzed. Here we aimed at further delineating the molecular responses arising from abiotic stress conditions and understanding the transcriptional response progression by gaining a pathway perspective of the functions carried out by up-regulated genes. The current analysis further allows mapping more than 1000 stress-associated processes into stress type–tissue–time coordinates. This pathway representation provides a simplified representation of cellular response while reducing the dimensions in gene-oriented response description, and ultimately allows for the identification of novel pathway dependencies. Notably, we expect different plants to differ at their response progression patterns. Hence, such simplified representation allows the conductance of comparative inter-plant studies, bypassing the need in detailed orthologous mapping and providing a constructed reference perspective.
Beyond comparative analyses, such mapping allows finding marker processes pointing whether a stress response was invoked. As many of the transcription factor and hormones are common to multiple stress type, stress-specific markers can be based on more complex combinations. Our reanalysis of the publically available data and particularly the predictions for higher-level functional associations arising from the network analysis contributes an additional layer of knowledge. While many of the pathways detected in the analysis were previously reported to take part in the cellular response to stress, novel connections are predicted. Moreover, supporting evidences are anecdotal and link-specific, each representing a piece in the puzzle. Their association within a framework of a network attempts to capture the bigger picture and provide a step forwards towards a system-level understanding of the progression of stress specific responses. Delineating the metabolic activity following exposure to stress can shed light on the cost of specific responses to plant productivity and broaden our understanding of the trade-off between stress response and plant’s primary metabolism. Hence, it can promote the design of metabolic engineering strategies, aiming, for example at the enhanced production of selected amino acids. The links formed between processes, though not necessarily representing casualty, further provide a source for predictions associating pathways and allow better understanding of the regulatory and metabolic processes behind the transcriptional response to abiotic stress. The association of stress types with specific cellular processes lays a foundation for deciphering the roll of different response routes in term of agricultural productivity.
References
Ahuja I, de Vos RC, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15:664–674
Akula R, Ravishankar GA (2011) Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 6:1720–1731
Arraes FBM et al (2015) Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol 15:1
Atias O, Chor B, Chamovitz DA (2009) Large-scale analysis of Arabidopsis transcription reveals a basal co-regulation network. BMC Syst Biol 3:86
Barah P, Winge P, Kusnierczyk A, Tran DH, Bones AM (2013) Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection PLoS One 8(3):e58987. doi:10.1371/journal.pone.0058987
Bechtold U et al (2016) Time-series transcriptomics reveals that AGAMOUS-LIKE22 links primary metabolism to developmental processes in drought-stressed Arabidopsis. Plant Cell TPC2015–00910
Beguerisse-Dıaz M, Hernández-Gómez MC, Lizzul AM, Barahona M, Desikan R (2012) Compound stress response in stomatal closure: a mathematical model of ABA and ethylene interaction in guard cells. BMC Syst Biol 6:146
Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry & molecular biology of plants, vol 40. American Society of Plant Physiologists: Rockville
Casati P, Walbot V (2005) Differential accumulation of maysin and rhamnosylisoorientin in leaves of high-altitude landraces of maize after UV-B exposure. Plant Cell Environ 28:788–799
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Ann Rev Plant Biol 61:651–679
Demkura PV, Abdala G, Baldwin IT, Ballaré CL (2010) Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiol 152:1084–1095
Dombrecht B et al (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19(7):2225–2245.
Ferrier T, Matus JT, Jin J, Riechmann JL (2011) Arabidopsis paves the way: genomic and network analyses in crops. Curr Opin Biotechnol 22:260–270
Freilich S et al (2015) Systems approach for exploring the intricate associations between sweetness, color and aroma in melon fruits. BMC Plant Biol 15:71
Goossens A et al (2003) A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci 100:8595–8600
Grativol C, Hemerly AS, Ferreira PCG (2012) Genetic and epigenetic regulation of stress responses in natural plant populations. Biochim Biophys Acta 1819:176–185
Hahn A et al (2013) Plant core environmental stress response genes are systemically coordinated during abiotic stresses. Int J Mol Sci 14:7617–7641
Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57:303–333
Hammer Ø, Harper D, Ryan P (2001) PAST: Paleontological Statistics Software Package for education and data analysis. Palaeontolia Electronica 4
Hare P, Cress W, Van Staden J (1997) The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regul 23:79–103
Herms DA, Mattson WJ (1992) The dilemma of plants: to grow or defend. Q Rev Biol 283–335
Heyndrickx KS, Vandepoele K (2012) Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol 159:884–901
Hildebrandt TM, Nesi AN, Araújo WL, Braun H-P (2015) Amino acid catabolism in plants. Mol plant 8:1563–1579
Hirai MY et al (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci 104:6478–6483
Kiełbowicz-Matuk A (2012) Involvement of plant C 2 H 2-type zinc finger transcription factors in stress responses. Plant Sci 185:78–85
Kilian J et al (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kim RD, Park PJ (2004) Improving identification of differentially expressed genes in microarray studies using information from public databases. Genome Biol 5:R70
Kushwah S, Jones AM, Laxmi A (2011) Cytokinin interplay with ethylene, auxin and glucose signaling controls Arabidopsis seedling root directional growth. Plant Physiol. doi:10.1104/pp.111.175794
Le Bot J, Bénard C, Robin C, Bourgaud F, Adamowicz S (2009) The ‘trade-off’ between synthesis of primary and secondary compounds in young tomato leaves is altered by nitrate nutrition: experimental evidence and model consistency J Exp Bot 60(15):4301–4314
Less H, Galili G (2009) Coordinations between gene modules control the operation of plant amino acid metabolic networks. BMC Syst Biol 3:14
Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible W-R, Hincha DK, Udvardi MK (2009) AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol 149(4):1761–1772
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19
Nakabayashi R, Saito K (2015) Integrated metabolomics for abiotic stress responses in plants. Curr Opin Plant Biol 24:10–16
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Oliver S (2000) Proteomics: guilt-by-association goes global. Nature 403:601–603
Parry ML (2007) Climate change 2007: contribution of working group II to the fourth assessment report of the intergovernmental panel on climate change, 2007. Climate change 2007: Working Group II: impacts, adaptation and vulnerability
Prescott SM (1999) A thematic series on kinases and phosphatases that regulate lipid signaling. J Biol Chem 274:8345–8346
Qiu Y, Yu D (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65(1):35–47
Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14:S185–S205
Rosa M, Prado C, Podazza G, Interdonato R, González JA, Hilal M, Prado FE (2009) Soluble sugars—metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal Behav 4:388–393
Sasaki-Sekimoto Y et al (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44(4):653–668
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shao HB, Chu LY, Jaleel CA, Zhao CX (2008) Water-deficit stress-induced anatomical changes in higher plants. C R Biol 331:215–225. doi:10.1016/j.crvi.2008.01.002
Stinson RA, Spencer M (1969) β-Alanine as an ethylene precursor. Investigations towards preparation, and properties, of a soluble enzyme system from a subcellular particulate fraction of bean cotyledons. Plant Physiol 44:1217–1226
Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62:157–184
Thimm O et al (2004) Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Toubiana D, Fernie AR, Nikoloski Z, Fait A (2013) Network analysis: tackling complex data to study plant metabolism. Trends Biotechnol 31:29–36
Tzin V, Galili G (2010) The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. Arabidopsis Book 8:e0132
Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523–539
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14. doi:10.1007/s00425-003-1105-5
Wang S, Wei X, Cheng L, Tong Z (2014) Identification of a C2H2-type zinc finger gene family from Eucalyptus grandis and its response to various abiotic stresses. Biol Plant 58:385–390
Wu Z, Aryee MJ (2010) Subset quantile normalization using negative control features. J Comput Biol 17:1385–1395
Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F (2004) A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99:909–917
Yan X, Chen S (2007) Regulation of plant glucosinolate metabolism. Planta 226:1343–1352
Zhai Y et al (2013) Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 513:174–183
Acknowledgments
We thank Profs. Georg Jander, Doron Holland and Gad Galili for helpful comments on the manuscript. We also thank Eyal Zinger and Dr. Efrat Oron for technical support related to R and MatLab languages. This research was supported by the I-CORE Program of the Planning and Budgeting Committee, The Israel Science Foundation (Grant No. 757/12) and the Chief Scientist of Israel Ministry of Agriculture and Rural Development (Grant No. 277059016). DAC is a member Edmond J. Safra Center for Bioinformatics at Tel-Aviv University. B.Y.S. was supported by a fellowship from the PBC (Planning and Budgeting Committee) Fellowship Program for Outstanding Post-Doctoral Researchers from India.
Author contributions
BSY carried out the analyses and wrote the manuscript; TL aided programming and MatLab analyses; ER aided in gene-enrichment analyses; DAC and SF conceived study, oversaw its execution and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2016_539_MOESM1_ESM.xls
Figure 1. Distribution of differentially up-regulated genes or significantly enriched pathways across experiments. A. Heat map illustrating the number of upregulated genes. B. Heat map illustrating the number of pathways significantly enriched with upregulated genes. C. A PCA analysis of the up-regulated gene profiles across the 96 experiments. The data were normalized and auto scaled prior to PCA. Numbers in parenthesis indicate the percentage of variance explained by each PC. (XLS 29 KB)
11103_2016_539_MOESM2_ESM.xls
Figure 2. Distribution of significantly enriched pathways from key categories across experiments. The heat map illustrates scaled distribution of the number of pathways significantly enriched with up-regulated genes across key MapMan categories. For each stress condition, the data point with the largest number of pathways was defined as 1 (dark red), to which all other points in the same conditions were scaled against (color bar on right). (XLS 4729 KB)
11103_2016_539_MOESM3_ESM.xls
Figure 3. Cross-stress response patterns of pathways. A. Heat maps illustrating scaled distribution of pathways across multiple stresses. Values in each cell represent the summing together of scaled values across all stress conditions (as in Figure 2). B. Diagram of pathways response progression during abiotic stress. (XLS 1061 KB)
11103_2016_539_MOESM4_ESM.xlsx
Figure 4. Enriched pathways from selected key categories in specific stresses within each panel, each color is corresponding to the top-category of the pathways indicated on the right side of the graph. A, B. Enriched pathways at 0.5 hours following stress classified to (A) the hormonal category, or (B) the RNA category. C, D. Enriched pathways at 6 hours following stress classified to the (C) amino acids category, or (D) secondary metabolites category. (XLSX 16 KB)
11103_2016_539_MOESM5_ESM.xls
Figure 5. Network association of primary and secondary categories at selected time points. Nodes represent pathways in the following categories: Primary response (0.5 h) - Hormones (green) and RNA transcriptome (red); Secondary response (6 h) - amino acid (blue) and secondary metabolite (yellow). The edges were drawn between nodes when their co-appearance score was higher than threshold (Methods). Nodes of hormonal and secondary metabolites categories are shown at the second-level of classification; Nodes of RNA and amino-acids categories are shown at the third and fourth level of classification, respectively. (A) The full network containing associations from both shoot and root. Subnetworks composed of the direct neighbors of the nodes (B) for methionine synthesis and degradation and (C) for asparagine synthesis and degradation. (XLS 29 KB)
Rights and permissions
About this article
Cite this article
Yadav, B.S., Lahav, T., Reuveni, E. et al. Multidimensional patterns of metabolic response in abiotic stress-induced growth of Arabidopsis thaliana . Plant Mol Biol 92, 689–699 (2016). https://doi.org/10.1007/s11103-016-0539-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0539-7