Abstract
Key message
Flavonoids are important secondary metabolites in plants. Sugar–sugar glycosyltransferases are involved in the final step of flavonoid biosynthesis and contribute to the structural diversity of flavonoids. This manuscript describes the first cloning of a sugar–sugar glucosyltransferase gene in the UGT family that attaches glucose to the 6″-position of sugar bound to a flavonol. The results provide a glimpse on the possible evolution of sugar–sugar glycosyltransferase genes and identify putative amino acids responsible for the recognition of the hydroxyl group of the sugar moiety and specification of sugar. A scheme for the genetic control of flavonol glycoside biosynthesis is proposed.
Abstract
Flavonol glycosides (FGs) are predominant in soybean leaves and they show substantial differences among genotypes. In previous studies, we identified two flavonoid glycoside glycosyltransferase genes that segregated in recombinant inbred lines developed from a cross between cultivars Nezumisaya and Harosoy; one was responsible for the attachment of glucose to the 2″-position of glucose or galactose that is bound to the 3-position of kaempferol and the other was involved in the attachment of glucose to the 6″-position. This study was conducted to clone and characterize the 6″-glucosyltransferase gene. Linkage mapping indicated that the gene was located in the molecular linkage group I (chromosome 20). Based on the genome sequence, we cloned a candidate cDNA, GmF3G6"Gt from Harosoy but the corresponding cDNA could not be amplified by PCR from Nezumisaya. The coding region of GmF3G6″Gt in Harosoy is 1386 bp long encoding 462 amino acids. This gene was not expressed in leaves of Nezumisaya. The GmF3G6″Gt recombinant protein converted UDP-glucose and kaempferol 3-O-glucoside or kaempferol 3-O-galactoside to kaempferol 3-O-glucosyl-(1→6)-glucoside or kaempferol 3-O-glucosyl-(1→6)-galactoside, respectively. These results indicate that GmF3G6″Gt encodes a flavonol 3-O-glucoside/galactoside (1→6) glucosyltransferase and corresponds to the Fg1 gene. GmF3G6″Gt had an amino acid similarity of 82 % with GmF3G6″Rt encoding flavonol 3-O-glucoside/galactoside (1→6) rhamnosyltransferase, suggesting a recent evolutionary divergence of the two genes. This may be the first cloning of a sugar-sugar glucosyltransferase gene in the UGT family that attaches glucose to the 6″-position of sugar bound to a flavonol. A scheme for the control of FG biosynthesis is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flavonoids are important secondary metabolites of plants. They act as attractants to pollinators and symbionts, as sunscreens to protect against UV irradiation, as allelochemicals, and as antimicrobial or antiherbivory factors (Dixon and Pasinetti 2010; Harborne and Williams 2000). Glycosylation is often the final step in flavonoid biosynthesis and is catalyzed by the family 1 glycosyltransferases referred to as UDP glycosyltransferases (UGTs) that transfer a glycosyl moiety from UDP sugars to a wide range of acceptors including flavonoids (Yonekura-Sakakibara and Hanada 2011). Flavonoid glycoside glycosyltransferases (FGGs) attach additional sugars to an existing sugar moiety of flavonol glycosides (FGs), resulting in a wide variety of binding positions and sugar combinations.
Leaves of soybean (Glycine max (L.) Merr.) contain various FGs. Derivatives of quercetin with 3′,4′-dihydroxyl groups in the B-ring, and derivatives of kaempferol with 4′-hydroxyl group are predominant (Buttery and Buzzell 1973). Four flavonol glycoside genes, Fg1 (β(1–6)-glucoside present), Fg2 (α(1–6)-rhamnoside present), Fg3 (β(1–2)-glucoside present) and Fg4 (α(1–2)-rhamnoside present) have been proposed in the biosynthesis of FGs (Buzzell and Buttery 1974). These alleles are defined by the ability to bind glucose or rhamnose at either 2″ or 6″-position of glucose that is bound to the 3-position of flavonols. Further, a new allele of the Fg2 locus was reported, resulting in a series of alleles, viz., Fg2-a, Fg2-b and fg2 (Buzzell and Buttery 1992). Fg3 and Fg4 are linked with a recombination frequency of 12 % in the molecular linkage group (MLG) C2 (chromosome 6) (Buzzell 1974). Soybean plants with the Fg1Fg3 alleles have a lower rate of photosynthesis, lower concentration of leaf chlorophyll, lower leaf weight, and lower seed yield (Buttery and Buzzell 1976). Further, Fg1 and Fg3 control waviness of leaf margins in soybean (Buzzell and Buttery 1998; Rode and Bernard 1975). A scheme for the genetic control of FG biosynthesis was proposed in which glucose or rhamnose is attached to either 2″ or 6″-position of glucose that is bound to the 3-position of flavonol (Buttery and Buzzell 1975). Actually, either glucose or galactose was attached to the 3-position of flavonol (Rojas Rodas et al. 2014). Furthermore, FGs having rhamnose at the 4″-position of 3-O-galactose were identified in soybean leaves (Di et al. 2015; Murai et al. 2013; Rojas Rodas et al. 2014), suggesting the existence of flavonol 3-O-glycoside (1→4) rhamnosyltransferase. Accordingly, the schematic diagram for this biosynthetic pathway needed to be revised (Rojas Rodas et al. 2014).
Genetic analysis of FG composition was performed using recombinant inbred lines (RILs) derived from a cross between cultivars Koganejiro and Kitakomachi (Rojas Rodas et al. 2014). FGs of Koganejiro had rhamnose at the 6″-position of glucose or galactose that is bound to the 3-position of kaempferol, whereas FGs of Kitakomachi were devoid of rhamnose. The FG composition was controlled by a single gene. The candidate gene, GmF3G6″Rt encoding 464 amino acids was located in MLG O (chromosome 10). The recombinant GmF3G6″Rt protein converted UDP-rhamnose and kaempferol 3-O-glucoside to kaempferol 3-O-rutinoside, and catalyzed kaempferol 3-O-galactoside. These results prove that GmF3G6″Rt encodes a flavonol 3-O-glucoside/galactoside (1→6) rhamnosyltransferase and corresponds to the Fg2 gene.
Similarly, genetic analysis was performed using RILs derived from a cross between cultivars Nezumisaya and Harosoy (Di et al. 2015). Harosoy had eight primary HPLC peaks corresponding to FGs (F1–F8), whereas Nezumisaya had seven peaks (F2, F5, F6 and F9–F12); F1, kaempferol 3-O-rhamnosyl-(1→4)-[glucosyl-(1→6)-galactoside]; F2, kaempferol 3-O-rhamnosyl-(1→4)-[rhamnosyl-(1→6)-galactoside]; F3, kaempferol 3-O-glucosyl-(1→6)-galactoside; F4: kaempferol 3-O-glucosyl-(1→6)-glucoside; F5, kaempferol 3-O-rhamnosyl-(1→6)-galactoside; F6, kaempferol 3-O-rhamnosyl-(1→6)-glucoside; F7, kaempferol 3-O-glucoside; F8, apigenin 7-O-glucoside; F9, kaempferol 3-O-glucosyl-(1→2)-[rhamnosyl-(1→6)-galactoside]; F10, kaempferol 3-O-glucosyl-(1→2)-[rhamnosyl-(1→6)-glucoside]; F11, kaempferol glycoside; F12, kaempferol 3-O-glucosyl-(1→2)-glucoside. Thus, FGs of Nezumisaya had glucose at the 2″-position of glucose or galactose that is bound to the 3-position of kaempferol, whereas FGs of Harosoy were devoid of the glucose. Conversely, FGs of Harosoy had glucose at the 6″-position, whereas FGs of Nezumisaya were devoid of the glucose. Among the 91 RILs, 21 RILs had peaks of the Harosoy-type and 23 RILs had peaks of the Nezumisaya-type. 17 RILs had a peak distribution designated as type 3 that lacked F1, F3 and F4 from the Harosoy-type. Further, 28 RILs had a peak distribution designated as type 4 having a mixture of peaks from both cultivars in addition to a unique peak (F13). The segregation fitted to a 1:1:1:1 ratio, suggesting that two genes control the FG pattern. One of the genes was postulated to be responsible for the attachment of glucose to the 2″-position and it probably encodes a flavonol 3-O-glucoside (1→2) glucosyltransferase. Nezumisaya had a dominant allele while Harosoy had a recessive allele of the gene. The other gene was presumed to be involved in attachment of glucose to the 6″-position. Harosoy may have a dominant allele whereas Nezumisaya may have a recessive allele of the gene. RILs of type 3, in which FGs with 2″-glucose and 6″-glucose were absent, were postulated to have double-recessive alleles. RILs of type 4, in which FGs with 2″-glucose and 6″-glucose were present, were presumed to have double-dominant alleles. F13 was presumed to be kaempferol 3-O-glycoside having both 2″-glucose and 6″-glucose.
The candidate gene for 2″-glucosyltransferase, GmF3G2″Gt encodes 459 amino acids and it is located in the MLG C2. The GmF3G2″Gt recombinant protein converted UDP-glucose and kaempferol 3-O-glucoside to kaempferol 3-O-sophoroside, and catalyzed kaempferol 3-O-galactoside. These results indicate that GmF3G2″Gt encodes a flavonol 3-O-glucoside/galactoside (1→2) glucosyltransferase and corresponds to the Fg3 gene. This study was conducted to clone and characterize the second gene that is responsible for the attachment of glucose to the 6″-position.
Materials and methods
Plant materials
A total of 91 RILs derived from a cross between Nezumisaya and Harosoy was used. Methods for RIL development, plant cultivation and HPLC analysis were performed as previously described (Di et al. 2015).
Chemical analysis
To identify FG component corresponding to peak F13 (Di et al. 2015), MeOH extracts from RILs of type 4 were subjected to HPLC analysis performed with the Prominence HPLC System (Shimadzu Corporation) using Kinetex C18 column [2.6 μm, I.D. 4.6 × 100 mm (Phenomenex Inc.)] at a flow rate of 0.8 ml/min, detection wavelengths at 190–700 nm and phosphoric acid/acetonitrile/H2O (0.2:12:88) as eluent, along with the authentic sample of kaempferol 3-O-glucosyl-(1→2)-[glucosyl-(1→6)-glucoside] (Iwashina et al. 2013). Furthermore, the samples were subjected to LC-MS analysis performed with the Shimadzu LCMS-2010EV system using L-column2 ODS column [3 μm, I.D. 2.1 × 100 mm (Chemicals Evaluation and Research Institute)] at a flow rate of 0.2 ml/min, detection wavelengths at 190–700 nm and formic acid/acetonitrile/H2O (1:10:89) as eluent.
Linkage mapping and QTL analysis
Methods for genomic DNA extraction and SSR analysis were performed as previously described (Di et al. 2015). For mapping of a gene responsible for attachment of glucose to the 6″-position, RILs having FG composition of the Nezumisaya type and type 3 were considered to have the genotype of Nezumisaya, whereas RILs having FG composition of the Harosoy type and type 4 were considered to have the genotype of Harosoy. A linkage map was constructed using genotypes indicated by 99 SSR markers and FG patterns by the MAPMAKER/EXP. ver. 3.0 (Lander et al. 1987) with the threshold LOD score of 3.0. Designation of linkage groups followed Cregan et al. (1999). QTL analysis for peak areas (F1–F13) in the HPLC chromatogram was performed by composite interval mapping (Zeng 1993) using the QTL Cartographer ver. 2.5 (Wang et al. 2007). The threshold LOD score was determined by permutation test with 1000 repetitions corresponding to a genome-wide 5 % level of significance. Peaks F7 and F8 were too small to accurately quantify area so these peaks were not subjected to QTL analysis.
Molecular cloning
The methods for RNA extraction and cDNA synthesis were described previously (Di et al. 2015). The full-length cDNA was cloned by end-to-end PCR using a pair of PCR primers (Table 1) whose design was based on the genome sequence of US cultivar Williams 82 deposited in the soybean genome database (Phytozome, http://www.phytozome.net/soybean.php). The genomic fragment including the entire coding region was amplified by PCR from Nezumisaya. In addition, the 5′ upstream region of about 1500 bp was amplified by PCR from Harosoy and Nezumisaya. The PCR mixture contained 0.5 µg of cDNA or 50 ng of genomic DNA, 10 pmol of each primer, 5 pmol of nucleotides and 1 unit of ExTaq in 1× ExTaq Buffer supplied by the manufacturer (Takara Bio) in a total volume of 25 µl. A 30 s denaturation at 94 °C was followed by 30 cycles of 30 s denaturation at 94 °C, 1 min annealing at 59 °C and 1 min extension at 72 °C. A final 7 min extension at 72 °C completed the program. The PCR products with expected molecular lengths were cloned into pCR 2.1 vector (Invitrogen) and sequenced.
Sequencing analysis
Nucleotide sequencing, estimation of intron/exon structure and phylogenetic analysis followed a previous report (Di et al. 2015). Gene prediction in the upstream region of Glyma20g33820 and Glyma20g33831 was performed with the GENSCAN software (http://genes.mit.edu/GENSCAN.html). Transcription factor binding site was searched with TFSEARCH ver. 1.3 (http://diyhpl.us/~bryan/irc/protocol-online/protocol-cache/TFSEARCH.html).
dCAPS analysis
dCAPS analysis was performed using the PCR amplicon of the 5′ upstream region. The base substitution in Nezumisaya is expected to abolish EcoNI site in the amplified product to generate a polymorphism. The PCR mixture contained 30 ng of genomic DNA, 5 pmol of each primer, 10 pmol of nucleotides and 1 unit of ExTaq in 1× ExTaq Buffer in a total volume of 25 µl. After an initial 30 s denaturation at 94 °C, there were 30 cycles of 30 s denaturation at 94 °C, 1 min annealing at 65 °C and 1 min extension at 72 °C. A final 7 min extension at 72 °C completed the program. The amplified products were digested with EcoNI, and the digests were separated in an 8 % polyacrylamide gel. After electrophoresis, the gel was stained with ethidium bromide and the DNA fragments were visualized under UV light.
Expression of recombinant GmF3G6″Gt protein
The entire coding region of GmF3G6″Gt was amplified from cDNA of Harosoy by PCR using the KOD-Plus-DNA polymerase (Toyobo) with high PCR fidelity using primers containing restriction enzyme sites of SacI and HindIII (Table 1). PCR conditions were identical to those in a previous report except that MgSO4 was added to a final concentration of 2 mM (Rojas Rodas et al. 2014). The PCR amplicon was digested by SacI and HindIII, and then cloned into the pCold ProS2 vector (Takara Bio). GmF3G6″Gt proteins were expressed and semi-purified as described previously (Yonekura-Sakakibara et al. 2014).
Enzyme assays
Enzyme assays and MS and MS/MS analyses were conducted as described previously (Rojas Rodas et al. 2014).
Quantitative real-time PCR
cDNA was synthesized by reverse transcription of 5 µg of total RNA derived from leaves at the V3 stage (Fehr et al. 1971), flower petals at the R1 stage, and immature cotyledons, developing seed coats, roots and root nodules at the R7 stage in three replications using the Superscript III First-Strand Synthesis System and an oligo (dT) primer. Primer sequences are shown in Table 1. Expression levels of the soybean actin gene (GenBank Accession Number: J01298) (Shah et al. 1983) were used to normalize the target gene expression. Primers for the actin 1 gene and PCR conditions are identical to a previous report (Rojas Rodas et al. 2014). PCR products from Harosoy were cloned into the pCR 2.1 vector. The nucleotide sequence of sixteen clones was determined.
Accession number
Sequence data from this article have been deposited with the DDBJ Data Libraries under the accession number LC126028 (Harosoy) and LC126029 (Nezumisaya).
Results
Chemical analysis, linkage mapping and QTL analysis
Based on HPLC comparison with the authentic sample and LC-MS measurement, peak F13 was identified as kaempferol 3-O-glucosyl-(1→2)-[glucosyl-(1→6)-glucoside]. A total of 99 SSR markers were classified into 28 linkage groups spanning 2172 cM (Di et al. 2015). A gene responsible for attachment of glucose to the 6″-position was mapped in MLG I (chromosome 20) between Satt623 and Sat_419 (Fig. 1). QTLs associated with peaks F1–F6 and F9–F13 in the HPLC chromatogram were found in the vicinity of Sat_419 in MLG I and/or in the vicinity of Satt202 in MLG C2 (Table 2). The only exception was peak F2 that had a QTL in the vicinity of Satt156 in MLG L (chromosome 19) in addition to those in MLGs I and C2. The peak of the QTLs in MLG I was close to the position of the gene responsible for attachment of glucose to the 6″-position whereas the peak of QTLs in MLG C2 was close to the position of the gene for attachment of glucose to the 2″-position (Fg3). Nezumisaya-types at the QTL in MLG I displayed smaller peaks for F1, F3, F4 and F13, and larger peaks for F2, F11 and F12. Nezumisaya-types at the QTL in MLG C2 had smaller peaks for F2–F6, and larger peaks for F9–F13.
Linkage mapping of a gene responsible for the attachment of glucose to the 6″-position of glucose or galactose bound to the 3-position of kaempferol using recombinant inbred lines derived from a cross between soybean cultivars Nezumisaya and Harosoy. The name of the linkage group is indicated at the top followed by the chromosome number in parenthesis. Distances (cM) of markers from the top of the linkage group are shown on the left
Molecular cloning
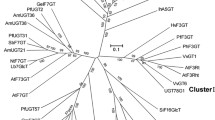
A survey of the genome sequence of Williams 82 suggested that three genes similar to UGT genes, viz., Glyma20g33810 (deduced polypeptide consists of 462 amino acids), Glyma20g33820 (322 amino acids) and Glyma20g33831 (126 amino acids), were aligned in a 15 kb region between Satt623 and Sat_419. Sequence similarity among the deduced amino acids was 52 to 75 %. BLAST analysis and multiple alignment suggested that Glyma20g33820 and Glyma20g33831 retained 3′ ends but lacked 5′ ends. We cloned and sequenced genome fragments of about 500 bp upstream of the presumed coding region from Glyma20g33820 and about 1000 bp upstream from Glyma20g33831. GENSCAN analysis suggested that no coding region existed in the upstream regions of Glyma20g33820 and Glyma20g33831 for both cultivars (data not shown). So we concluded that they might be pseudogenes. The entire coding region of Glyma20g33810 was amplified by RT-PCR from Harosoy and then cloned into plasmid vectors for sequencing. Sequencing analysis revealed that the coding region is 1386 bp long encoding 462 amino acids. We designated the gene as GmF3G6″Gt, and it was designated as UGT79A7 by the UGT Nomenclature Committee (Mackenzie et al. 1997). The GmF3G6″Gt gene had 82 % amino acid similarity with GmF3G6″Rt encoding flavonol 3-O-glucoside/galactoside (1→6) rhamnosyltransferase (Rojas Rodas et al. 2014) (Fig. 2), whereas it had only 35 % similarity with GmF3G2″Gt encoding flavonol 3-O-glucoside/galactoside (1→2) glucosyltransferase. The flavonoid glycosyltransferase phylogenetic tree suggested that GmF3G6″Gt belongs to the FGG gene cluster (Fig. 3).
Unrooted molecular phylogenetic tree of the some flavonoid glycosyltransferases. Bar represents 0.1 amino acid substitutions/site. The GenBank accession numbers for the sequences are shown in parentheses: At3Rt (NM_102790); At3Gt (NM_121711); Vv3Gt (AF000371); Ph3Gt (AB027454); Pf3Gt (AB002818); Hv3Gt (X15694); Zm3Gt (X13501); At5Gt (NM_117485); Pf5Gt (AB013596); Vh5Gt (BAA36423); Ph5Gt (AB027455); Db7Gt (CAB56231); Nt7Gt (AAB36653); Sb7Gt (BAA83484); At7Rt (AY093133); CmF7G2″Rt (AAL06646); CsF7G6″Rt (ABA18631); IpA3G2″Gt (AB192315); PhA3G6″Rt (X71059); BpA3G2″Glt (AB190262); AcA3Ga2″Xt (FG404013); At3G2″Xt (Q9FN26); GmF3G6″Rt (AB828193); GmF3G2″Gt (LC017844). Gt, glucosyltransferase; Rt, rhamnosyltransferase; Xt, xylosyltransferase; Glt, glucuronosyltransferas. Ac, Actinidia chinensis; At, Arabidopsis thaliana; Bp, Bellis perennis; Cm, Citrus maxima; Cs, Citrus sinensis; Db, Dorotheanthus bellidiformis; Gm, Glycine max; Hv, Hordeum vulgare; Ip, Ipomoea purpurea; Nt, Nicotiana tabacum; Pf, Perilla frutescens; Ph, Petunia hybrida; Sb, Scutellaria baicalensis; Vh, Verbena hybrida; Vv, Vitis vinifera; Zm, Zea mays
In contrast to Harosoy, the coding region of GmF3G6″Gt could not be amplified by RT-PCR from Nezumisaya. So we amplified and cloned the genomic fragment containing the entire coding region from Nezumisaya. The fragment corresponding to the coding region is 1386 bp long capable of encoding 462 amino acids similar to Harosoy. There was a single nucleotide polymorphism (SNP) at nucleotide position 1041 between the cultivars, but their amino acid sequences were identical. Comparison between the cDNA sequence and the corresponding genome sequence suggested that GmF3G6″Gt had no intron similar to that of GmF3G6″Rt (Rojas Rodas et al. 2014).
In contrast to the coding region, the 5′-upstream region of GmF3G6″Gt was quite polymorphic between the cultivars; there are eight SNPs, an indel of 52 consecutive nucleotides and a substitution of 30 consecutive nucleotides (Fig. S1). The consecutive substitution may be derived from duplication of 35 nucleotides in Harosoy (Fig. 4a). A nucleotide motif (AACTACCCG) similar to the binding site of myb-homologous P gene responsible for phlobaphene pigmentation (Grotewold et al. 1994) existed in the duplicated region (Fig. 4a). Harosoy had two copies of the motif in tandem because of the fragment duplication, whereas Nezumisaya had one copy in the region.
Partial nucleotide sequences of 5′ upstream region of GmF3G6″Gt gene in soybean cultivars, Harosoy and Nezumisaya, and outline and results of dCAPS analysis. a Nucleotide alignment of 5′ upstream region. Identical nucleotides are shown by asterisks. Dashes represent gaps to improve alignment. Nucleotide fragment duplicated in Harosoy is in white font highlighted in black. A nucleotide motif similar to the binding site of myb-homologous P gene is shown by arrows. b Outline of dCAPS analysis. Mismatched nucleotide (T) introduced in the forward primer is double-underlined. EcoNI site (CCTNNNNNAGG) is shown in white font highlighted in gray. Nucleotide fragments duplicated in Harosoy is in white font highlighted in black. c Results of dCAPS analysis of GmF3G6″Gt gene in soybean cultivars, Harosoy and Nezumisaya, and recombinant inbred lines derived from a cross of the cultivars. PCR amplicon was digested with EcoNI and the digests were separated on an 8 % polyacrylamide gel. ϕ: molecular marker ϕx174/HaeIII; N, Nezumisaya; H, Harosoy. FG pattern of the recombinant inbred lines is exhibited below the gel. N, Nezumisaya-type; H, Harosoy-type; 3, type 3; 4, type 4. The migration of size markers (bp) is shown to the right of the gel
dCAPS analysis
The dCAPS primer generated a band of 169 bp in Harosoy and Nezumisaya. EcoNI digestion of the PCR amplicon generated bands of 152 bp in Nezumisaya, whereas the amplicon of Harosoy was undigested (Fig. 4b). Banding patterns co-segregated with FG patterns; RILs with FGs of the Harosoy type and type 4 had bands of Harosoy type, whereas RILs with FGs of the Nezumisaya type and type 3 had bands of Nezumisaya type (Fig. 4c).
In vitro characterization of recombinant GmF3G6″Gt
The GmF3G6″Gt recombinant protein was expressed in E. coli as a His/ProS2 fusion and then semi-purified. After cleavage of the His/ProS2 tag, the GmF3G6″Gt protein was used for enzymatic assays. GmF3G6″Gt converted kaempferol 3-O-glucoside to kaempferol 3-O-glucosyl-(1→6)-glucoside as confirmed by comparison of retention time, UV spectra and MS/MS ionization with the standard compound (Fig. 5). GmF3G6″Gt showed a broad activity for kaempferol/quercetin 3-O-glucoside/galactoside derivatives (Table 3). GmF3G6″Gt has a higher preference for UDP-glucose than for UDP-galactose, with only 7.5 % activity relative to that for UDP-galactose. No UGT activity was detected for UDP-arabinose and UDP-glucuronic acid. Accordingly, GmF3G6″Gt was defined as a flavonol 3-O-glucoside/galactoside (1→6) glucosyltransferase.
Identification of the reaction product of GmF3G6″Gt protein from soybean cultivar Harosoy. a Elution profiles of the reaction product of GmF3G6″Gt protein (K3Glc + GmF3G6″Gt) and the standards, kaempferol 3-O-glucoside, kaempferol 3-O-glucosyl-(1→6)-glucoside. b UV spectra of the reaction product of GmF3G6″Gt protein and the standard, kaempferol 3-O-glucosyl-(1→6)-glucoside. Mass spectra (c) and MS/MS spectra (d) of the reaction product of GmF3G6″Gt protein and the standard, kaempferol 3-O-glucosyl-(1→6)-glucoside. e The MS/MS fragmentation for kaempferol 3-O-glucosyl-(1→6)-glucoside. K3Glc, kaempferol 3-O-glucoside; K3Glc6″Glc, kaempferol 3-O-glucosyl-(1→6)-glucoside
Gene expression
At the V3 stage, GmF3G6″Gt was expressed in leaves of Harosoy, whereas the gene was not expressed in Nezumisaya in accordance with the results of RT-PCR experiments (Table 4). In Harosoy, the gene was expressed slightly in immature cotyledon and root, expressed strongly in developing seed coat and root nodule and expressed very strongly in flower petals. The PCR products from Harosoy were ascertained to be derived from the expected genomic region by sequencing.
Discussion
Soybean cultivars, Harosoy and Nezumisaya have gray pubescence and deposit predominantly kaempferol derivatives in leaves, but their FG components were distinctly different (Di et al. 2015). Genetic analysis suggested that two genes control the FG pattern, one responsible for the attachment of glucose to the 2″-position and another responsible for the attachment of glucose to the 6″-position. The former gene was cloned from the MLG C2 region (Di et al. 2015) in accordance with a previous study (Buzzell 1974).
In this study, we mapped a gene responsible for the attachment of glucose to the 6″-position in the MLG I. A survey of the genome sequence of Williams 82 suggested that three genes similar to GT genes, Glyma20g33810, Glyma20g33820 and Glyma20g33831 were found in this region. Multiple alignment and sequencing of the upstream regions suggested that the last two genes might be truncated pseudogenes. We cloned a cDNA sequence corresponding to Glyma20g33810 from Harosoy and designated it as GmF3G6″Gt. GmF3G6″Gt is 1386 bp long encoding 462 amino acids. Recombinant GmF3G6″Gt protein of Harosoy converted UDP-glucose and kaempferol 3-O-glucoside or kaempferol 3-O-galacoside to kaempferol 3-O-glucosyl-(1→6)-glucoside or kaempferol 3-O-glucosyl-(1→6)-galactoside, respectively. These results proved that GmF3G6″Gt encodes flavonol 3-O-glucoside/galactoside (1→6) glucosyltransferase. dCAPS analysis confirmed the association between the nucleotide polymorphism and FG pattern. We confirmed that peak F13 corresponds to kaempferol 3-O-glucosyl-(1→2)-[glucosyl-(1→6)-glucoside], supporting the hypothesis that the peak was generated by the double-dominant alleles of the Fg1 and Fg3 loci.
The GmF3G6″Gt had no intron similar to GmF3G6″Rt, consistent with most UGT genes that have either none or one intron (Paquette et al. 2003). The GmF3G6″Gt gene was expressed in various tissues including leaves, developing seed coats and root nodules, but it was expressed most strongly in flower petals. This is consistent with the results of a previous study in which kaempferol 3-O-gentiobioside was predominant in flower petals and accounted for more than 70 % of total FG contents (Iwashina et al. 2007). To our knowledge, this is the first cloning of a sugar-sugar glucosyltransferase gene in the UGT family that attaches glucose to the 6″-position of sugar bound to flavonol.
The GmF3G6″Gt gene had 82 % amino acid similarity with GmF3G6″Rt of soybean encoding flavonol 3-O-glucoside/galactoside (1→6) rhamnosyltransferase. In contrast, GmF3G6″Gt had only 35 % of amino acid similarity with GmF3G2″Gt encoding flavonol 3-O-glucoside/galactoside (1→2) glucosyltransferase. This is consistent with the notion that FGG genes established recognition mechanisms for the hydroxyl group of the sugar moiety (2″-position or 6″-position) before developing the ability to specify the nature of the sugar (glucose or rhamnose) (Yonekura-Sakakibara et al. 2012). GmF3G6″Gt was closer to GmF3G6″Rt according to phylogenetic analysis and had higher amino acid similarity compared with flavonoid glucoside (1→6)-rhamnosyltransferases of other plant species; it had 56 % similarity with flavanone 7-O-glucoside (1→6)-rhamnosyltransferase of Citrus sinensis (Frydman et al. 2013) and 53 % similarity with anthocyanin 3-O-glucoside (1→6)-rhamnosyltransferase of petunia (Kroon et al. 1994). These results suggest that soybean FG6″Gs established substrate specificity for nature of sugar after speciation of plants.
Multiple alignment of FGGs (four FG6″Gs and six FG2″Gs) suggested 31 amino acids specific to FG6″Gs, and 3 amino acids specific to FG2″Gs (Fig. S2). In addition, three amino acids associated with sugar donor (glucose or rhamnose) were found; amino acid positions 26 (Ala or Gly for glucose/Pro for rhamnose), 136 (Ser or Thr for glucose/Val or Pro for rhamnose) and 218 (Ser or Arg for glucose/Lys for rhamnose). The second amino acid corresponded to that responsible for sugar donor specificity in an FGG gene involved in saponin biosynthesis (Ser for xylose/Gly for glucose) (Sayama et al. 2012). Site-directed mutagenesis may reveal which amino acids are responsible for glycosylation of specific positions of the sugar moiety or specification of the sugar donor.
In contrast to Harosoy, the coding region of GmF3G6″Gt could not be amplified by RT-PCR from Nezumisaya. Quantitative real-time PCR analysis suggested that GmF3G6″Gt of Nezumisaya may not be expressed in leaves. Thus, the intensity of gene expression is associated with the dominance relationships of this gene. The 5′ upstream region was quite polymorphic between the cultivars; there were eight SNPs, an indel of 52 nucleotides and a substitution of 30 consecutive nucleotides in Harosoy. Further, a nucleotide motif similar to the binding site of myb-homologous P gene responsible for phlobaphene pigmentation existed in duplicate in Harosoy, but Nezumisaya had only one instance of this motif in the promoter region. The nucleotide polymorphism(s) in this region may be responsible for non-expression of this gene in the leaves of Nezumisaya. Transgenic experiments or transient assays using deletion clones may be necessary to determine if the duplication or other nucleotide polymorphism(s) are critical for gene expression.
QTLs for the area of the peaks in the HPLC chromatogram were found close to the position of the Fg1 and/or Fg3 genes except for peak F2 which had an additional minor QTL in MLG L. A gene with small effects on FG composition may exist in MLG L. Thus, results of QTL analysis are consistent with those of genetic analysis and linkage mapping. The QTLs in MLGs C2 and the QTLs in MLG I may correspond to the Fg3 and Fg1 genes, respectively. The scheme for genetic control of FG biosynthesis is shown in Fig. 6. The dominant allele of the Fg1 gene increased the amount of F1, F3 and F4 that had glucose at the 6″-position. The dominant allele of the Fg3 gene increased the amount of F9, F10 and F12 that had glucose at the 2″-position. The same allele decreased the amount of F2, F5 and F6. This is probably because F5 and F6 are substrates of the Fg3 protein as well as substrates of flavonol 3-O-glycoside (1→4) rhamnosyltransferase. The Fg3 protein may have catalyzed F5 and F6 in competition with the flavonol 3-O-glycoside (1→4) rhamnosyltransferase. The amounts of F3, F4 and F12 were controlled by both Fg1 and Fg3 genes, probably because F7 is a substrate of both Fg1 and Fg3 proteins.
Schematic presentation of genetic control of flavonol glycoside (FG) biosynthesis in soybean leaves based on FG composition of cultivars Harosoy and Nezumisaya, and recombinant inbred lines developed from a cross of the cultivars. K3G, kaempferol 3-O-glucoside/galactoside. K3G is followed by a suffix indicating the position (2″, 4″ or 6″-position) and nature of sugar (Rh, rhamnose, Gl glucose) that is attached to the glucose/galactose. 4″Rt: flavonol 3-O-glycoside (1→4) rhamnosyltransferase. Corresponding peaks in HPLC chromatogram are indicated in parenthesis
We cloned Fg1 encoding flavonol 3-O-glucoside/galactoside (1→6) glucosyltransferase, Fg2 encoding flavonol 3-O-glucoside/galactoside (1→6) rhamnosyltransferase and Fg3 encoding flavonol 3-O-glucoside/galactoside (1→2) glucosyltransferase, but Fg4 probably encoding flavonol 3-O-glycoside (1→2) rhamnosyltransferase remains to be cloned (Buzzell and Buttery 1974). In addition, FGs having rhamnose at the 4″-position of 3-O-galactose (kaempferol 3-O-rhamnosyl-(1→4)-[glucosyl-(1→6)-galactoside] and kaempferol 3-O-rhamnosyl-(1→4)-[rhamnosyl-(1→6)-galactoside]) have been identified in soybean leaves (Di et al. 2015; Murai et al. 2013; Rojas Rodas et al. 2014), suggesting the existence of a gene that attaches rhamnose to the 4″-position. Soybean FGs having rhamnose at the 4″-position of flavonol are galactosides having glucose or rhamnose at the 6″-position. Flavonol 3-O-glycoside (1→4) rhamnosyltransferase of soybean possibly has a narrow substrate specificity in contrast to Fg1–Fg3 proteins that have wide substrate specificity. Cloning of the 4″-rhamnosyltransferase gene is intriguing because FGG genes responsible for attachment of sugar to the 4″-position have not been cloned from any plant species as of this writing. Overall, the biosynthesis of FGs in soybean leaves may be as follows; glucose or galactose is attached to the 3-position of kaempferol or quercetin. Subsequently, either glucose or rhamnose is attached to 2″-, 4″- or 6″-positions of glucose or galactose, resulting in a wide variety of FGs (Rojas Rodas et al. 2014). Cloning and analysis of Fg4 gene and a gene responsible for attachment of rhamnose to the 4″-position may help further understand the evolution of FGG genes, the recognition mechanism of specific positions of the sugar moiety and ability to specify nature of sugar to be attached to an existing sugar moiety.
References
Buttery BR, Buzzell RI (1973) Varietal differences in leaf flavonoids of soybeans. Crop Sci 13:103–106
Buttery BR, Buzzell RI (1975) Soybean flavonol glycosides: identification and biochemical genetics. Can J Bot 53:219–224
Buttery BR, Buzzell RI (1976) Flavonol glycoside genes and photosynthesis in soybeans. Crop Sci 16:547–550
Buzzell RI (1974) Soybean linkage tests. Soyb Genet Newsl 1:11–14
Buzzell RI, Buttery BR (1974) Flavonol glycoside genes in soybeans. Can J Genet Cytol 16:897–899
Buzzell RI, Buttery BR (1992) Inheritance of an anomalous flavonol glycoside gene in soybean. Genome 35:636–638
Buzzell RI, Buttery BR (1998) Genetics of leaf waviness in soybean. Soyb Genet Newsl 25:23–24
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung L, Specht JE (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Di S, Yan F, Rojas Rodas F, Rodriguez TO, Murai Y, Iwashina T, Sugawara S, Mori T, Nakabayashi R, Yonekura-Sakakibara K, Saito K, Takahashi R (2015) Linkage mapping, molecular cloning and functional analysis of soybean gene Fg3 encoding flavonol 3-O-glucoside/galactoside (1→2) glucosyltransferase. BMC Plant Biol 15:126
Dixon RA, Pasinetti GM (2010) Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol 154:453–457
Fehr WR, Caviness CE, Burmood DT, Pennington JS (1971) Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci 11:929–931
Frydman A, Liberman R, Huhman DV, Carmeli-Weissberg M, Sapir-Mir M, Ophir R, Sumner LW, Eyal Y (2013) The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyl-transferases under domestication. Plant J 73:166–178
Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls plobaphene pigmentation in mize foral ogans by drectly ativating a favonoid biosynthetic gene subset. Cell 76:543–553
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Iwashina T, Githiri SM, Benitez ER, Takemura T, Kitajima J, Takahashi R (2007) Analysis of flavonoids in flower petals of soybean near-isogenic lines for flower and pubescence color genes. J Hered 98:250–257
Iwashina T, Tamura MN, Murai Y, Kitajima J (2013) New flavonol glycosides from the leaves of Triantha japonica and Tofieldia nuda. Natl Prod Commun 8:1251–1254
Kroon J, Souer E, Degraaff A, Xue YB, Mol J, Koes R (1994) Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J 5:69–80
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A, FournelGigleux S, Green M, Hum DW, Iyanagi T, Lancet D, Louisot P, Magdalou J, Chowdhury JR, Ritter JK, Schachter H, Tephly TR, Tipton KF, Nebert DW (1997) The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7:255–269
Murai Y, Takahashi R, Rojas Rodas F, Kitajima J, Iwashina T (2013) New flavonol triglycosides from the leaves of soybean cultivars. Natl Prod Commun 8:453–456
Paquette S, Moller BL, Bak S (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62:399–413
Rode WM, Bernard RL (1975) Inheritance of wavy leaf. Soyb Genet Newsl 2:42–44
Rojas Rodas F, Rodriguez TO, Murai Y, Iwashina T, Sugawara S, Suzuki M, Nakabayashi R, Yonekura-Sakakibara K, Saito K, Kitajima J, Toda K, Takahashi R (2014) Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1→6) rhamnosyltransferase. Plant Mol Biol 84:287–300
Sayama T, Ono E, Takagi K, Takada Y, Horikawa M, Nakamoto Y, Hirose A, Sasama H, Ohashi M, Hasegawa H, Terakawa T, Kikuchi A, Kato S, Tatsuzaki N, Tsukamoto C, Ishimoto M (2012) The Sg-1 glycosyltransferase locus regulates structural diversity of triterpenoid saponins of soybean. Plant Cell 24:2123–2138
Shah DM, Hightower RC, Meagher RB (1983) Genes encoding actin in higher plants: intron positions are highly conserved but the coding sequences are not. J Mol Appl Genet 2:111–126
Wang SC, Basten J, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Yonekura-Sakakibara K, Hanada K (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J 66:182–193
Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, Wangwattana B, Yamazaki M, Saito K (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69:154–167
Yonekura-Sakakibara K, Nakabayashi R, Sugawara S, Tohge T, Ito T, Koyanagi M, Kitajima M, Takayama H, Saito K (2014) A flavonoid 3-O-glucoside: 2"-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant J 79:769–782
Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90:10972–10976
Acknowledgments
The authors are grateful to Dr. Joseph G. Dubouzet (New Zealand) for critical reading of the manuscript. This study was partially supported by the Japanese Government (MEXT) Scholarship to F.R.R. a scholarship grant from the China Scholarship Council to S.D. and the JSPS KAKENHI Grant Number 25440148 to KY-S.
Author Contributions
R.T. designed experiments and together with F.R.R. and S.D. carried out genetic analysis, linkage mapping, molecular cloning and gene expression assays. Y.M. and T.I. carried out chemical analysis. S.S., T.M., R.N., K.Y.-S. and K.S. performed functional analysis.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2016_523_MOESM1_ESM.doc
Figure S1 Alignment of the 5′ upstream region of GmF3G6″Gt gene in soybean cultivars Harosoy and Nezumisaya. Polymorphic nucleotides are shown in red font. The coding sequence is underlined. (DOC 32 KB)
11103_2016_523_MOESM2_ESM.pdf
Figure S2 Multiple alignment of flavonoid glycoside glycosyltransferases (FGGs). The PSPG-box is underlined. Amino acid residues conserved in flavonoid FGGs are in white font highlighted in black. Amino acid residues conserved in flavonoid FG6″Gs and FG2″Gs are in white font highlighted in red and blue, respectively. Amino acid residues specific to glucosyltransferases and rhamnosyltransferases are in white font highlighted in orange and green, respectively. The position of amino acid responsible for sugar specificity in an FGG gene involved in saponin biosynthesis is shown by white triangle. (PDF 42 KB)
Rights and permissions
About this article
Cite this article
Rojas Rodas, F., Di, S., Murai, Y. et al. Cloning and characterization of soybean gene Fg1 encoding flavonol 3-O-glucoside/galactoside (1→6) glucosyltransferase. Plant Mol Biol 92, 445–456 (2016). https://doi.org/10.1007/s11103-016-0523-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0523-2