Abstract
NAC transcription factors (TFs) are plant-specific and play important roles in development, responses to biotic and abiotic cues and hormone signaling. So far, only a few NAC genes have been reported to regulate cell death. In this study, we identified and characterized a NAC55 gene isolated from oilseed rape (Brassica napus L.). BnaNAC55 responds to multiple stresses, including cold, heat, abscisic acid (ABA), jasmonic acid (JA) and a necrotrophic fungal pathogen Sclerotinia sclerotiorum. BnaNAC55 has transactivation activity and is located in the nucleus. BnaNAC55 is able to form homodimers in planta. Unlike ANAC055, full-length BnaNAC55, but not either the N-terminal NAC domain or C-terminal regulatory domain, induces ROS accumulation and hypersensitive response (HR)-like cell death when expressed both in oilseed rape protoplasts and Nicotiana benthamiana. Furthermore, BnaNAC55 expression causes obvious nuclear DNA fragmentation. Moreover, quantitative reverse transcription PCR (qRT-PCR) analysis identified that the expression levels of multiple genes regulating ROS production and scavenging, defense response as well as senescence are significantly induced. Using a dual luciferase reporter assay, we further confirm that BnaNAC55 could activate the expression of a few ROS and defense-related gene expression. Taken together, our work has identified a novel NAC TF from oilseed rape that modulates ROS accumulation and cell death.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The NAC [no apical meristem (NAM), Arabidopsis thaliana transcription activation factor (ATAF1/2) and cup-shaped cotyledon (CUC2)] proteins constitute one of the largest transcription factor (TF) families and are plant-specific (Olsen et al. 2005). NAC-type TFs were originally identified from petunia (Petunia hybrida) NAM (Souer et al. 1996), A. thaliana ATAF1, ATAF2 and CUC2 (Aida et al. 1997). NAC TFs are characterized by a well-conserved N-terminal NAC domain and highly divergent C-terminus. Based on its motif distribution, the NAC domain, which comprises nearly 160 amino acid residues, can be divided into five sub domains (A–E) (Puranik et al. 2012).
There are 117 putative NAC genes in Arabidopsis and 151 in rice (Nuruzzaman et al. 2010). The NAC TFs are multifunctional proteins, which have been shown to be involved in a variety of developmental and physiological processes, such as shoot apical meristem development, lateral root formation, secondary cell wall formation, hormone signal pathways and leaf senescence (Olsen et al. 2005; Puranik et al. 2012). Moreover, quite a few members of the NAC TF family also function in abiotic and biotic stress signaling and tolerance (Nakashima et al. 2012; Nuruzzaman et al. 2013). For instance, Arabidopsis ANAC072 (RD26), ANAC019 and ANAC055 regulate ABA-dependent drought signaling (Fujita et al. 2004; Tran et al. 2004). Arabidopsis ATAF1 and -2 are negative regulators of defense responses against bacterial and fungal pathogens (Delessert et al. 2005; Wang et al. 2009) and ATAF1 plays a dual role in mediating Arabidopsis response to both abiotic and biotic stresses (Lu et al. 2007; Wu et al. 2009). Arabidopsis ANAC019 and ANAC055 mediate drought tolerance, but their overexpression also decreases resistance to B. cinerea (Bu et al. 2008). It is thought that such dual modulation of NAC proteins in plants implies their alliance with distinct regulatory complexes. However, the underlying mechanisms remain to be investigated. Recent evidence also indicates that NAC TFs play a role in endoplasmic reticulum (ER) or osmotic stress-induced cell death in Arabidopsis, rice and soybean, possibly through regulating vacuolar processing enzyme (VPE) or caspase-like protein activity (Kaneda et al. 2009; Faria et al. 2011; Mendes et al. 2013; Yang et al. 2014). However, whether there are other NAC TFs regulating cell death is unknown.
Reactive oxygen species (ROS), including the superoxide radical (O2 −), hydrogen peroxide (H2O2), the hydroxyl radical (OH) which are chemically reactive, oxygen-containing molecules, can react with proteins, DNA, and membrane lipids to reduce photosynthesis, increase electrolyte leakage, and accelerate senescence and cell death (De Pinto et al. 2012). ROS play important role in many other biological processes such as growth, development, and stress adaptation (Gechev et al. 2006). Under biotic and abiotic stress conditions, plant cells are capable of producing a burst of ROS that is primarily made up of H2O2. At the same time, ROS represent important signaling molecules in the coordination of stress acclimation pathways (Suzuki et al. 2012). Therefore, a tight control is needed to balance these activities and maintain coordination, including activation of signalling pathways by ROS-responsive regulatory genes and buffering of ROS by ROS-scavenging enzymes and antioxidant molecules (Apel and Hirt 2004). Several enzymatic systems may contribute to this overproduction of ROS. Among these, plasma membrane (PM)-associated respiratory burst oxidase homologs (Rboh), which are similar to NADPH oxidase (NOX) present in mammalian neutrophil cells and generate superoxide in the apoplastic side, has been characterized in several plant species as an essential ROS-producing system activated during the early stages of plant–pathogen interaction (Torres and Dangl 2005). Moreover, oxidative stress increases the activity of antioxidant enzymes, such as superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), glutathione peroxidases (GPX) and glutathione S-transferase (GST), which in turn help to adjust ROS levels and maintain ROS homeostasis (Apel and Hirt 2004).
During defense response, one of the most efficient and immediate immune responses is hypersensitive response (HR) cell death, which is characterized by rapid, localized death of plant cells at the site of pathogen infection (Lam 2004). HR cell death is a form of programmed cell death (PCD). Environmental stresses such as nutrient deficiency, high salt, and drought also induce cell death in plants, suggesting that the process may confer resistance to stress. Although HR cell death shows morphological and biochemical traits similar to animal apoptosis, the most characterized form of PCD, several characteristics differ between these two forms of PCD. On one hand, apoptotic bodies are not formed during plant HR cell death, because of the presence of a cell wall. On the other hand, caspase genes and enzymes are key components of animal apoptosis, however, plants lack obvious homologous sequences of caspases in their genomes, although caspase activities are detected in both animal and plant cells (Bonneau et al. 2008). In addition, vacuoles, which are multifunctional plant cell organelles, have an important function in plant PCD, with a key role for VPE (Hara-Nishimura et al. 2005). Although the existence of HR cell death in plants has been recognized for many years (Van Hautegem et al. 2015), our knowledge of the genetic mechanisms that regulate and execute cell death is still very limited.
ROS have been proposed as key inducers of different types of developmental and/or environmental cell death. The most studied ROS signal is H2O2 (Gechev and Hille 2005). H2O2 is perhaps the most prominent signaling molecule characterized by its relative stability and significant mobility. ROS accumulation, or the oxidative burst, its causal relationship with cell death has been an active area of research and, evidence has accumulated that ROS trigger cell death and are not merely cell death by-products (De Pinto et al. 2012). For instance, pharmacological treatments with ROS antagonists block induction of HR-associated cell death and, transgenic plants with lower levels of antioxidative enzymes display aggravated cell death responses (Van Breusegem and Dat 2006). These observations highlight the importance of understanding how the plant interprets ROS signaling to induce cell death. However, our knowledge of transcriptional induction mechanism of HR cell death remains elusive.
Oilseed rape is a very important oil crop worldwide and its yield is frequently limited by environmental stresses including drought, salinity, cold and fungal pathogens. In our previous studies with oilseed rape, we mined the available expressed sequence tags (ESTs) to identify NAC genes in oilseed rape and, cloned the cDNA sequences of over 41 NAC genes from oilseed rape. We further identified three NAC TF genes that modulate ROS accumulation and cell death with in-depth work still underway (Niu et al. 2014; Wang et al. 2015). In the present study, we describe the characterization of one of the BnaNAC genes, BnaNAC55, which shows similarity to Arabidopsis ANAC055 and could induce ROS over accumulation and elicit cell death when transiently expressed in both oilseed rape protoplasts and N. benthamiana leaves, possibly through modulating ROS and defense-related gene expression.
Materials and methods
Plant materials and growth condition
Oilseed rape, N. benthamiana and Arabidopsis plants were grown in Pindstrup soil mix (Denmark) in the greenhouse at 22 °C with 14 h light/10 h dark, and a light intensity of ca. 150 μEm−2 s−1. The relative humidity is 60–70 %.
RNA isolation and RT-PCR
Young leaves of oilseed rape, N. benthamiana or Arabidopsis seedlings were harvested for RNA isolation using the Plant RNA kit (Omega bio-tek, USA). First-strand cDNA synthesis and high-fidelity PCR amplification using PrimeSTAR HS DNA polymerase (TaKaRa, Japan) were performed as previously described (Liang et al. 2013). Primers used are listed in Supplementary Table S1. PCR products were purified and cloned into pJET1.2 vector (Fermentas, USA). At least two clones were sequenced and sequencing results were analyzed by DNASTAR.
Phylogenetic tree reconstruction and bioinformatics
The predicted amino acid sequences of NACs of oilseed rape and other species were aligned using ClustalX1.83 and then a phylogenetic tree was reconstructed using the maximum parsimony (MP) algorithm implemented in MEGA6.06 (release 6,140,226). Motif analysis of BnaNACs was determined by using Prosite program (http://prosite.expasy.org/prosite.html). The respective domains of NAC proteins were aligned using ClutsalX1.83 and illustrated by Boxshade (http://www.ch.embnet.org/software/BOX_form.html).
Transcriptional activity assay in yeasts
The coding regions of TFs was cloned into the pGBKT7 (BD) vector (Clontech, USA), using the primers listed in Supplementary Table S1. The assay in yeast was performed as described previously (Wang et al. 2015).
Transactivation assay in plants
The coding regions of NAC genes were amplified by high-fidelity PCR with primers included in Supplementary Table S1. The PCR products were digested and then cloned into pYJHA plasmid. Reporter plasmid 3xNACRS::LUC contains triple tandem repeats of NACRS element and the minimal TATA region of 35S promoter of cauliflower mosaic virus (CaMV), which are located upstream of the firefly luciferase (LUC) gene in the pGreenII0800-LUC vector (Hellens et al. 2005). Internal control is Renilla (REN) LUC driven by 35S promoter. The control plasmid is a binary vector pYJGFP, in which expression of green fluorescent protein (GFP) gene is controlled by CaMV35S promoter. All plasmids were introduced into agrobacterium strain GV3101 individually through a freeze-thaw method and agrobacterial cultures transformed with the effector plasmid and reporter plasmid (9:1, v/v) were co-infiltrated into the lower epidermal side of 30 d old N.benthamiana leaves. At 2 and 3 dpi, leaf discs with a diameter of 1 cm were harvested and ground in liquid nitrogen and extracted in 300 μl of lysis buffer, with supernatant used to assay LUC and REN activity by dual LUC assay kit according to the manufacturer’s instructions (Promega) on a GloMax™ 20–20 luminometer (Promega). The binding ability of NAC TFs to the NACRS element was indicated by the ratio of LUC to REN. Triplicate measurements were performed.
Quantitative RT-PCR (qRT-PCR) assay
Oilseed rape tissues of three independent biological replicates were prepared as described previously (Niu et al. 2014). Total RNA samples were isolated and the first-strand cDNAs were synthesized from 2.5 μg of total RNA as described previously (Liang et al. 2013). qRT-PCR was performed using tenfold diluted cDNA samples and SYBR Premix Ex Taq (TaKaRa, Japan) on the CFX96 real-time thermocycler (Bio-Rad, USA). Primers used for qRT-PCR were designed using PrimerSelect (DNASTAR Inc. USA), which targeted mainly 3′UTRs, with an amplicon size of 75–300 bp (Supplementary Table S1). The specificity and amplification efficiency of each pair of primers were examined through both BLASTn searches in the NCBI database and by running standard curves with melting curves. Isolation RNA from N. benthamiana leaf discs was performed as described above, again from three biological replicates. The NbPP2A, NbL23 and NbF-box were used as the reference genes (Liu et al. 2012). Two technical replicates for each biological replicate were run and the significance was determined using a t test in SPSS statistical software (p ≤ 0.05).
Subcellular localization and bimolecular fluorescence complementation (BiFC) assays
To examine the localization of BnaNAC55 in planta, the coding region was amplified using Pfu polymerase (Bioer, China) with primers listed in the Supplementary Table S1. After purification, PCR products were restricted and then fused upstream of GFP gene in the pYJGFP vector. Transformed Agrobacterium tumefaciens GV3101 culture was resuspended in infiltration media before infiltration into 32 d old leaves of N. benthamiana (Liang et al. 2013). 2 days later, leaf discs were observed of GFP under a confocal microscope (FV1000MPE, Olympus, Japan).
For BiFC, the coding region of BnaNAC55 was subcloned into both 35S-SPYNE(R)173 and 35S-SPYCE(M) vectors (Waadt et al. 2008) before being introduced into A. tumefaciens strain GV3101 and coinfiltrated into 32 d old N. benthamiana leaves. The fluorescence of yellow fluorescence protein (YFP) in the lower epidermal cells of leaves was examined 3 d after infiltration on a Nikon A1 confocal microscope (Nikon, Japan).
Transient expression and physiological assays
The full-length coding region or different fragments of NAC genes were amplified by high-fidelity Pfu polymerase with primers listed in Supplementary Table S1. After digestion, the PCR products were inserted downstream of a double CaMV 35S promoter in the binary vectors pYJHA and/or pYJGFP, in which a hydrophobic peptide linker of VDGAPGGG exists between target proteins and HA or GFP tag. Coinfiltration of p19 and BnaNAC55 or fragments into the lower epidermal side of 30 day old leaves of N. benthamiana plants were performed as described previously (Sun et al. 2014). The control plasmid pCsGFPBT together with p19 was similarly infiltrated into the other halves of leaves. For each construct, 15 independent leaves of 5 independent plants (3 leaves per plant) were used for each time-point tested. After that, infiltrated plants were kept under normal growth condition. To quantify the degree of cell death, electrolyte leakage was measured according to (Sun et al. 2014). Distribution of H2O2 was detected by 3,3′-diaminobenzidine (DAB) staining according to the previously described protocol (Sun et al. 2014). Total chlorophyll was extracted in absolute ethanol in the dark at 4 °C. Relative chlorophyll levels were determined by fluorescence using a spectrometer (Thermo Scientific, USA). Anthocyanin was extracted in methanol supplemented with 1 % HCl overnight at 4 °C with absorbance at 530 and 657 nm measured using a spectrophotometer (Thermo Scientific, USA). By subtracting the A657 from the A530, the relative amount of anthocyanin per mg of tissue was calculated (Neff and Chory 1998). For malondialdehyde (MDA) content determination, about 100 mg of leaf discs were homogenized in 4 mL of 0.1 % trichloroacetic acid (TCA), with the concentration measured as described previously (Niu et al. 2014). H2O2 was extracted and determined against a standard curve of H2O2 (Chen et al. 2016).
DNA ladder and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assays
DNA ladder assay was performed as described previously (Niu et al. 2014). For TUNEL assay, a GUS gene fragment (2.053 kb) was amplified through high-fidelity Pfu and subcloned into Nco I-Sal I sites of pYJHA plasmid using the same restriction sites, with the resultant transient vector called pYJHA-GUS. An agrobacterial GV3101-mediated infiltration of N.benthamiana leaves was performed as described previously. Leaves from control (GUS) and BnaNAC55 plants were fixed in 3.7 % paraformaldehyde in 1× PBS buffer and treated with DNase-free RNase A (30 μg/ml; Promega) and Proteinase K (20 μg/ml; Sigma-Aldrich) for 5 min before the TUNEL assay according to the manufacturer’s manual (Promega). Tissue was then counterstained with Vectashield and DAPI solution (Vector Labs, USA) for 5–10 min and examined with an FV1000MPE confocal microscopy (Olympus, Japan).
Transient expression assay in oilseed rape protoplasts
The expression cassettes of BnaNAC55 was released through a Sac I–Sph I restriction from the aforementioned pYJHA recombinant plasmid and subcloned into pUC19 plasmid, with the resultant plasmid called pUC19-BnaNAC55. The control plasmid pUC19-GFP was described previously (Wang et al. 2015). Protoplasts were isolated from leaves of 6 week old oilseed rape plants grown in a 12-h light/12-h dark photoperiod with a relative humidity of 60–70 % according to the protocol for Arabidopsis mesophyll protoplasts (Yoo et al. 2007). The protoplast concentration was quantified using a hemocytometer and a light microscope (DM750, Leica, Germany). A PEG4000-mediated transformation of protoplast suspension was performed with 20 μg of the expression plasmids isolated by a Plasmid DNA Maxi kit (CWBIO, China). After incubated for 18 h at 22 °C, GFP signal was observed under a fluorescent microscope (DM5000B, Leica). For cell death detection in protoplasts, transformed protoplasts were first cultivated for 18 h at 22 °C, and then were incubated for 15 min in 0.05 % Evans blue solution (117 mM KCl, 82 mM MgCl2, and 85 mM CaCl2) (Kaneda et al. 2009). Protoplasts were also subjected to fluorescein diacetate (FDA) staining by incubating for 5 min in 0.01 % FDA solution in acetone. After washing, the survival rate of protoplasts was determined by counting multiple individual fields.
Promoter cloning and luciferase reporter assay
Young leaves of N. benthamiana seedlings were harvested for DNA isolation through the 2 % CTAB method (Niu et al. 2014). Primers for amplification of promoter regions were designed through a BLAST search against the SOL Genomics Network database (http://solgenomics.net/tools/blast) based on orthologous published sequences of Nicotiana tabacum and/or Arabidopsis. High-fidelity PCR amplification was performed using PrimeSTAR HS DNA polymerase (TaKaRa, Japan). Primers were designed to amplify the 1 kb regions upstream of the translational start codon (ATG) and were listed in Supplementary Table S1. PCR products were purified, sequenced and fused upstream of firefly LUC gene in the pGreenII0800-LUC vector, in which renilla (REN) LUC under the control of the 35S promoter was the endogenous control. The effector plasmid is pYJHA-BnaNAC55 and the control plasmid is pYJGFP. Dual LUC assay was performed as described previously. The binding ability of BnaNAC55 to various promoter regions was indicated by the ratio of LUC to REN. Triplicate transient assay measurements were included for each pair.
Statistical analysis
All experiments were repeated three times (three biological replicates). All data were statistically analyzed by using SPSS 16.0 software. The accession numbers of oilseed rape NAC55 genes in GenBank are KJ670121 and KJ670123, and promoter regions of N.benthamian genes reported in this study were deposited in the GenBank under the accession numbers KP747642–KP747651.
Results and discussion
Identification, cloning and analysis of NAC55 gene from oilseed rape
In a recent systematical study of NAC TF gene family in oilseed rape, we identified and cloned over 41 BnaNAC (B rassica na pus NAC, to differentiate them from B rassica ni gra) genes through mining the public expressed sequence tag (EST) database (Wang et al. 2015). We initiated to characterize each BnaNAC gene in the context of stress and ROS signaling, especially those BnaNAC genes classified as SNACs (stress-responsive NACs) to which BnaNAC55 belongs. RT-PCR cloning and sequence analysis indicate that two slightly different alleles are obtained (Supplementary Fig. S1). The translated proteins from both alleles contained 333 amino acids, which bore domains and motifs that were typical of NAC proteins (Fig. 1a). Further comparison of BnaNAC55 with Arabidopsis NAC genes demonstrated that it shows an identity of 82.5 and 82 % to ANAC055 at the nucleotide and amino acid residues levels, respectively. Moreover, the two alleles of BnaNAC55 show an identity of 98.2 and 98.8 % at the nucleotide and amino acid residues levels, respectively.
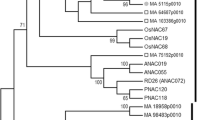
Domain organization, phylogenetic analysis and expression profiling of oilseed rape NAC55. a Schematic representation of BnaNAC55 protein. A highly conserved NAC domain is located at the N-terminal which is further divided into five conserved subdomains (a–e; shown in gray boxes). This region holds DNA-binding (DB) ability. The C-terminal region is more diverged and serves as a potential transcriptional regulatory (TR) domain. b Phylogenetic relationship of BnaNAC55 with those from representative species. The tree was reconstructed with amino acid sequences through the MP method implemented in MEGA6.06 with BnaNAC55 indicated by a filled diamond. The numbers on the nodes are percentages from a bootstrap analysis of 500 replicates. Gm, Glycine max; Hv, Hordeum vulgare; Os, Oryza sativa; Pp, Physcomitrella patents; Ptr, Populus trichocarpa; Ta, Triticum aestivum; Sl, Solanum lycopersicum; Sm, Selaginella moellendorffi; Zm, Zea may; Ph, Petunia hybrida. Tt, Triticum turgidum. c qRT-PCR analysis of BnaNAC55 in response to various treatments. Data is the mean of three biological replicates ± SE. Asterisks denote significant differences (compared to 1) by Student t test analysis (*p ≤ 0.05; **p ≤ 0.01)
We further examined conserved domains and motifs within the BnaNAC55 protein. From this alignment (Supplementary Fig. S1), it is clear that BnaNAC55 protein possesses a ca. 160 amino acid amino-terminal NAC domain, which is further divided into five highly conserved subdomains (A–E) (Puranik et al. 2012). These five subdomains are interspersed by short and variable sequences. In subdomain D, there is a short sequence containing conserved basic amino acids, which is the nuclear localization signal (NLS) (Supplementary Fig. S1).
To examine the evolutionary relationships of oilseed rape NAC55, we constructed a phylogenetic tree using a MP algorithm after performing an alignment of full-length amino acid sequences of NAC proteins from Arabidopsis, oilseed rape, rice, wheat, maize, soybean, poplar and other representative species. It can be seen that the BnaNAC55 forms a unique subclade with homologs from Arabidopsis (Fig. 1b).
Expression of BnaNAC55 responds to multiple stress treatments
To elucidate the functions of BnaNAC55 in the context of abiotic, biotic and hormone stimuli, its expression pattern was studied in response to a variety of stress conditions using quantitative real-time PCR (qRT-PCR). The stress conditions applied to oilseed rape seedlings included abscisic acid (ABA), salicylic acid (SA), Paraquat [methyl viologen (MV)], high salinity (200 mM NaCl), dehydration (12.5 % PEG8000), jasmonic acid (JA), 1-aminocyclopropane-1-carboxylate (ACC), cold (4 °C), heat (37 °C), a necrotrophic fungal pathogen Sclerotinia sclerotiorum and its secreted virulence factor oxalic acid (OA). Among these stress treatments, ABA is a well-known stress hormone and MV is a herbicide triggering ROS burst. JA and SA are two known plant defense hormones, with JA mediating responses to necrotrophs and SA regulating responses to biotrophic pathogens and systemic acquired resistance. ACC was used to mock the effect of ethylene, which is an important hormone in many processes including fruit ripening and leaf senescence. We subjected oilseed rape seedlings to moderate stress treatments and measured the response at two time-points to better monitor the transcript changes of the investigated BnaNAC55 gene. Data of three independent biological replicates showed that expression of BnaNAC55 was significantly up-regulated by ABA and JA at the 1 h time-point, and by ACC, heat, and S. sclerotiorum treatments at the 24 h time-point (Fig. 1c). In contrast, cold treatment significantly down-regulated BnaNAC55 expression at both 1 and 24 h time-points. The other stress or hormone treatments did not significantly change the expression of BnaNAC55 at the transcript level. Taken together, these data indicate that BnaNAC55 gene participates in transduction of multiple stresses, including abiotic, biotic stresses and hormone stimuli.
BnaNAC55 has transcriptional activation activity
To characterize the function of a NAC protein in oilseed rape, it is necessary to know its ability to activate or repress transcription. To this end, we analyzed the ability of BnaNAC55 TF to activate reporter genes LacZ, TRP1, LEU1 and ADE2 in budding yeast (S. cerevisiae). Firstly, the coding region of BnaNAC55 gene was fused to the GAL4 DNA-binding domain to examine its ability to activate transcription from the GAL4 upstream activation sequence (UAS) and thereby promote yeast growth. The yeast cells transformed with the pGBKT7-BnaNAC55 plasmid or control plasmid pGBKT7 grew well on SD-WL medium, while on SD-WLH medium supplemented with 5 mM 3-AT or on SD-LWHA, only yeast containing pGBKT7-BnaNAC55 plasmid could grow (Fig. 2a). X-Gal staining also indicated that another reporter gene LacZ was expressed. Therefore, BnaNAC55 protein has transcriptional activation activity, while the empty vector (EV) showed no transcriptional activation activity, as expected. A previous study with Arabidopsis ANAC055 using a similar approach demonstrated that full-length ANAC055 has no transcriptional activation activity while its C-terminal regulatory domain has transactivation activity in yeast (Bu et al. 2008). We previously also observed this type of difference between oilseed rape and Arabidopsis in a few NAC orthologs (Wang et al. 2015).
Analysis of transactivational activity, subcellular localization and interaction of BnaNAC55. a The yeast cells of strain AH109 harboring the indicated plasmids were grown on either the nonselective (SD-WL) or selective (SD-WLH + 5 mM 3-AT and SD-LWHA) media, followed by β-galactosidase assay (X-Gal staining). Decreasing cell densities in the dilution series are illustrated by narrowing triangles. EV represents empty pGBKT7 vector. b Effects of BnaNAC055 and GFP control on firefly LUC gene expression controlled by triple tandem repeats of NACRS element as revealed by dual LUC assay. Asterisks denote significant differences by Student t test analysis (**p ≤ 0.01). c Subcellular localization of BnaNAC55 protein in N. benthamiana cells using green fluorescence protein (GFP). The upper and lower panels represent BnaNAC55 and GFP alone, respectively. In each panel, the extreme left is GFP fluorescence, the middle bright field and the right an overlay of the two images as indicated on the top of the picture. Bar 50 μm. d Bimolecular fluorescence complementation (BiFC) analysis of interactions between BnaNAC55 and BnaNAC55 in N. benthamiana leaf cells. The coding region of BnaNAC55 was fused to both the N- and C-terminal halves of YFP, respectively. The fluorescence of YFP was observed by confocal laser microscopy. The left panel is YFP fluorescence, the middle bright field and the right represents an overlay of the two images. Bar 50 μm
The cis-DNA elements for several NAC proteins have been identified and it is known that NACs of SNAC subfamily can bind to the NAC recognition site (NACRS) containing the core CATGTG motif (Puranik et al. 2012). We therefore investigated if BnaNAC55 and ANAC055 can bind to this element in dual LUC assay system. Triple tandem repeats of NACRS sequences were synthesized and inserted, together with the CaMV35S minimal promoter, upstream of firefly LUC gene (Supplementary Fig. S2A). The effector plasmids were pYJHA-NACs. The results showed that BnaNAC55 showed significant transactivation activity at two time-points examined (Fig. 2b), while ANAC055 showed repressor activity (Supplementary Fig. S2B). This suggests differences in regulatory mechanism exist between oilseed rape and Arabidopsis orthologous genes, highlighting the necessity of studying oilseed rape NAC genes directly, rather than making inferences about their functions.
BnaNAC55 localizes in nuclei and form homodimers
To investigate the subcellular localization of BnaNAC55 protein, its coding region was fused upstream of GFP and expressed in leaves of N. benthamiana. We found that in leaf cells harboring the fusion proteins of BnaNAC55-GFP, GFP signals were present in the nucleus only (Fig. 2c), which is in agreement with its role as a TF. As a control, we also examined the subcellular localization of the GFP protein in leaf cells, and green signals were present obviously both in cytosol and nuclei (Fig. 2c).
Next, BiFC assay was employed to examine whether BnaNAC55 protein could form homodimers in plant cells. As a result, leaf cells expressing BnaNAC55, which was fused to both the N-terminal half and C-terminal half the YFP reporter gene, displayed strong yellow signals in nuclei (Fig. 2d), indicating that BnaNAC55 protein can interact with itself and form homodimers.
Expression of BnaNAC55 induces ROS accumulation and cell death
In the aforementioned GFP subcellular assay using N. benthamiana, we noticed that expression of BnaNAC55 in leaves led to (HR)-like symptom within 2 d after infiltration. HR is a common feature of plant immune responses and a type of PCD. Activation of cell death is one of the aspects of plant defense responses where ROS play a crucial role (Coll et al. 2011). Consequently, we initiated further experiments to investigate the potential connection between BnaNAC55 and cell death signaling. In this regard, we performed 3, 3′-diaminobenzidine (DAB) staining of the agroinfiltrated leaves and identified obvious staining, indicative of ROS accumulation (data not shown). To further explore the roles of BnaNAC55 gene in inducing ROS accumulation and cell death, we constructed the coding region, N-terminal NAC domain (BnaNAC55-∆C) or the C-terminal regulatory domain (BnaNAC55-∆N) of BnaNAC55 into another binary vector pYJHA individually under the control of CaMV 35S promoter (Fig. 3b). Then we performed individual agroinfiltration into N. benthamiana leaves and evaluated the phenotype daily for a total of 7 d. Interestingly, expression of BnaNAC55 indeed caused pathogen-independent cell death compared with GFP vector control, beginning 2 days post-infiltration (dpi) and proceeded as expected (Fig. 3a). However, expression of either the N-terminal NAC domain or C-terminal regulatory domain did not induce HR-like cell death. To examine the role of ROS, especially H2O2 during cell death, we performed DAB staining and the results showed there was staining in sites expressing BnaNAC55 beginning at 2 dpi and continued till 7 dpi, but not in sites expressing BnaNAC55-∆N, BnaNAC55-∆C or GFP control (Fig. 3a). Semi-quantitative RT-PCR and Western blot assays indicate that both GFP and BnaNAC55 as well its truncated fragments were expressed in the respective leaf tissues after agroinfiltration (Supplementary Fig. S3). Moreover, we examined the electrolyte leakage of leaf discs taken from leaves expressing BnaNAC55 or its truncated versions and the GFP alone. The results showed that a significant increase in ion leakage was visible with leaf sites expressing BnaNAC55 at 2 dpi in contrast to that of leaves expressing either the N-terminal NAC domain or C-terminal regulatory domain and, the GFP gene alone (Fig. 3c), which further demonstrates that HR-like cell death associated with hydrogen peroxide production is triggered by expression of BnaNAC55.
Overexpression of BnaNAC55 induced pathogen-independent cell death in N. benthamiana leaves. a Symptoms of N. benthamiana leaf areas expressing BnaNAC55, its two truncated fragments or GFP control gene from 1 to 7 days post-infiltration (dpi). The left, middle and right panels represent the front, back sides and DAB staining, respectively. Leaves were infiltrated with agrobacterium carrying individual plasmids. b Schematic representation of domain structures of BnaNAC55 protein. The DNA-binding NAC domain (open boxes) is located at the N-terminal region and the transcriptional regulatory domain is located at the C-terminal region. The numbers of total amino acid (AA) residues of full-length protein or truncated fragments are shown at the right side of each structure. Numbers at the bottom row indicate residue positions. c Measurement of electrolyte leakage in leaf discs expressing BnaNAC55, its truncated versions and GFP at 2, 4, 6 and 7 dpi. d Quantification of MDA contents in leaf discs expressing BnaNAC55, its truncated versions and GFP at 1, 2, 3, 5 and 7 dpi. Values represent the means of three independent assays for each time-point ± SE. Asterisks denote significant differences by Student t test analysis (**p ≤ 0.01)
Considering that intracellular ROS accumulation could cause lipid peroxidation, which damages cell membranes and therefore the cells, we therefore monitored the content of MDA formed by the decomposition of polyunsaturated fatty acids. The data showed that in leaf discs expressing the full-length BnaNAC55, the MDA concentration was significantly higher than that in the GFP control at 2, 3, 5 and 7 dpi (Fig. 3d), although the difference was insignificant at 1 dpi. This further indicates that the aforementioned cell death is associated with ROS-induced cell membrane breakage.
To further confirm the accumulation of H2O2, we measured the content of it using an improved protocol. We observed significantly higher content of H2O2 in leaf discs expressing BnaNAC55 than that of GFP, beginning at 3 dpi and lasting till 7 dpi tested (Fig. 4a). We then examined the concentration of chlorophyll and anthocyanins in leaf discs expressing different genes or fragments. Anthocyanins are present in a wide range of plant species and are responsible for the purple coloration of plant parts. Anthocyanins are also important antioxidants and can help protect plants from damage by ROS (Nagata et al. 2003). The results showed that in leaf discs expressing BnaNAC55, the concentration of chlorophyll decreased significantly compared to GFP control or the two truncated versions at the three time-points (Fig. 4b). In addition, this decrease in total chlorophyll in leaf tissues expressing BnaNAC55 proceeded as the symptoms developed more seriously, although a slight decrease in the control leaf tissues was also observed, possibly as result of agrobacterium infiltration. We also quantified the anthocyanin concentration in different leaf tissues and the results showed that expressing BnaNAC55 significantly increased anthocyanin accumulation (Fig. 4c).
Quantitative analysis of physiological indexes in N. benthamiana leaves expressing BnaNAC55 and DNA ladder assay. a Quantification of hydrogen peroxide contents in leaf discs expressing BnaNAC55 and GFP at various time-points. b Quantification of chlorophyll contents in leaf discs expressing BnaNAC55, its truncated versions and GFP at 3, 5 and 7 dpi. c Quantification of anthocyanin accumulation in leaf tissues expressing BnaNAC55 or GFP genes. Values represent the means of three independent assays for each time-point ± SE. Identical and different letters represent non-and significant differences (p ≤ 0.05). d DNA ladder assay of cell death induced by expression of BnaNAC55 gene. Genomic DNA was extracted from leaf discs expressing full-length, N- or C-terminal BnaNAC55 gene or GFP gene (control) at specified time points. Lanes 1–5 represent 10 μg of genomic DNA extracted from leaf discs at 3, 4, 5, 6 and 7 dpi, respectively. After electrophoresis, DNA was visualized with EB staining. M is the commercial 1 kb DNA marker
Because chromatin condensation and DNA fragmentation occur in naturally senescent tissues or during cell death, we further examined the change in nuclear DNA in leaf discs expressing BnaNAC55, its truncated fragments or GFP. The results showed that nuclear DNA extracted from leaf discs expressing full-length BnaNAC55 showed fragmentation with an obvious smear of shorter DNA fragments in the agarose gel, whereas DNA from tissues expressing the GFP gene alone, or the N-terminal or C-terminal half of BnaNAC55 did not (Fig. 4d).
Next, we examined nuclear DNA fragmentation as a hallmark of cell death in leaf cells using the TUNEL assay, which detects single- and double-strand DNA breaks by addition of fluorescent dUTP to free 3′ DNA termini (Kang et al. 2012). For this assay, β-glucuronidase (GUS) gene was used as the control, because the excitation wavelength (488 nm) of GFP and fluorescein (FITC) overlaps and so GFP interferes with TUNEL assay (data not shown). We also confirmed that expression of GUS did not cause any symptom of HR-like cell death (data not shown). At either 1 or 2 dpi, we did not observe obvious signal of FITC in leaf tissues expressing BnaNAC55 (Supplementary Fig. S4). Beginning at 3 dpi, leaf discs expressing full-length BnaNAC55 exhibited strong fluorescent signals of fluorescein in the epidermal nuclei based on confocal microscopy, suggesting single- or double-stranded DNA breaks in the nuclei (Fig. 5a, c). The signal was not detectable in control leaf cells expressing GUS gene (Fig. 5b, d). Overall, both the DNA ladder and TUNEL assays support that there was internucleosomal degradation of genomic DNA in BnaNAC55-expressing leaf tissues undergoing cell death.
BnaNAC55 expression also induces cell death in oilseed rape protoplasts
To confirm that BnaNAC55 induced HR-like cell death in oilseed rape, we used a transient assay system in oilseed rape protoplasts. Firstly, we constructed two transient expression plasmids by introducing expression cassettes of GFP (control) and BnaNAC55 under the control of a constitutive CaMV35S promoter into the pUC19 backbone. Secondly, we introduced the two expression vectors into protoplasts prepared from wild-type oilseed rape leaves, and then examined cell death using Evans blue dye, a marker of PM integrity (Kaneda et al. 2009). As expected, we identified that in protoplasts transformed with BnaNAC55 plasmids, a portion of protoplasts displayed PM shrinkage and loss of PM integrity (Fig. 6a). As a result, the dead protoplasts accumulated the dye, whereas in protoplasts expressing the GFP gene alone, all protoplasts excluded the dye (Fig. 6a). In addition, the transformed protoplasts were separately stained in FDA solution and intact protoplasts could be stained, while dead ones were not (Fig. 6b). Overall, the death rate in protoplasts of oilseed rape transformed with BnaNAC55 plasmids was 16 %, while that in GFP control was almost undetectable (Fig. 6c).
Expression of BnaNAC55 gene in oilseed rape protoplasts induced cell death. a Detection of cell death through Evans blue staining. Protoplasts were transfected with the pUC19-BnaNAC55 and pUC19-GFP (control) plasmids, respectively. After 18 h, viability of protoplasts was assayed with Evans blue staining. Red arrow indicates a dead protoplast. Chlorophyll means auto-fluorescence of chloroplasts with red circle representing the site of a dark dead protoplast. b Detection of cell viability through FDA staining. Red arrow indicates a dead protoplast and red circle represents the site of a dead protoplast. c Percentages of dead protoplasts transfected with pUC19-GFP (control) or pUC19-BnaNAC55 plasmids. 18 h after transfection, dead protoplasts were scored using a light microscope. The values with error bars were mean values of three independent assays ± SE
NbNAC55 but not ANAC055 showed a similar function to BnaNAC55 in inducing ROS accumulation and cell death
To further compare the functions of BnaNAC55 and ANAC055, we also analyzed if ANAC055 has a similar role in inducing ROS accumulation and cell death, and the results showed that expression of ANAC055 in leaves of N. benthamiana did not have significant change compared to the GFP control (Supplementary Fig. S5A). A similar measurement of relative conductivities at three time-points also supports this (Supplementary Fig. S5B). Examination of the ANAC055 and GFP protein expression in leaf discs showed expected results (Supplementary Fig. S5C). These evidences support our previous conclusion that role of BnaNAC55 in modulating ROS accumulation and cell death is different from its Arabidopsis counterpart.
Similarly, we mined the sequenced draft genome of N.benthamiana and cloned the cDNA sequence of a putative ortholog of BnaNAC55, which is named as NbNAC55. Sequence analysis demonstrated that amino acid sequence of NbNAC55 is highly similar to BnaNAC55 at the N-terminal NAC domains, while the C-termini are quite divergent (Supplementary Fig. S1). Overexpression of NbNAC55 showed enhanced ROS accumulation and cell death, which is supported by DAB staining and increased relative conductivities, compared to the GFP control (Supplementary Fig. S6A and B). To investigate if silencing of NbNAC55 has an opposite effect, we employed virus-induced gene silencing technique (Supplementary methods) and gene-specific fragments were used to construct the silencing plasmids. A qRT-PCR assay confirmed that transcript level of NbNAC55 was decreased by 65 and 70 % at 15 and 20 dpi, respectively (Supplementary Fig. S6C). Scoring of ROS- and cell death-related phenotype showed that there is no significant difference between the newly emerged true leaves infiltrated with pTRV2-NbNAC55 or pTRV2-GUS at both 15 and 20 dpi (Supplementary Fig. S6C), which may be explained by the functional redundancy between NAC genes in N. benthamiana.
BnaNAC55 modulates ROS and defense-related gene transcription
Our data presented above showed that ROS accumulation and cell death are correlated with expression of BnaNAC55 gene. To determine how BnaNAC55 modulates these two processes, we first employed qRT-PCR to examine the expression of an array of downstream marker genes. ROS are produced via diverse biosynthetic pathways, the most prominent of which is the NOX (Rboh)-mediated pathway. Previous study has demonstrated that NbRbohA and NbRbohB, the orthologs of Arabidopsis RbohF and RbohD, respectively, mediate H2O2 accumulation (Yoshioka et al. 2003). qRT-PCR assays of three biological replicates showed that the transcript levels of NbRbohA and NbRbohB were approximately twofold higher in leaf discs expressing BnaNAC55 than that in the GFP control (Fig. 7a). Next we examined the transcript changes of a variety of genes encoding antioxidant metabolic enzymes, such as APX, CAT, GPX, GST, glutathione reductase (GR) and (SOD) (Apel and Hirt 2004) as well as alternative oxidase (AOX) lowering mitochondrial ROS production in plant cells (Maxwell et al. 1999). The results demonstrated that transcript levels of NbAOX1a, NbCAT3, NbGPX2, NbGR2, NbGST and NbSOD were most prominently increased with an average of two to over thirty fold higher in leaf discs expressing BnaNAC55 compared to the GFP control (Fig. 7a). The expression level of NbAPX1 gene was slightly increased as a result of BnaNAC55 expression. In addition, this increase in gene induction was observed at either 2 or 3 dpi or both time-points. We therefore propose that the up-regulation of the activities of ROS-scavenging enzymes could help protect plants against oxidative damage due to ROS accumulation. However, the transcription of NbAXP4, NbtAPX and NbCAT1 was slightly down-regulated or did not show any significant change, suggesting that these three genes may not act downstream of BnaNAC55 TF. Thirdly, the transcript levels of plant-specific genes encoding enzymes functioning as executors of cell death, such as VPEs (Hatsugai et al. 2004; Hara-Nishimura et al. 2005) were also investigated. Besides, two metacaspase genes MC4 and MC8 (also known as MCP2d and MCP2e, respectively), encoding cysteine proteases structurally related to animal caspases, were also included in our analysis. In Arabidopsis, MC4 is the predominant and constitutively expressed member of type II metacaspases and plays a positive regulatory role in biotic and abiotic stress-induced PCD (Vercammen et al. 2004; Watanabe and Lam 2011), while MC8 modulates PCD induced by ultraviolet light and H2O2 (He et al. 2008). Our results demonstrated that transcript levels of none of the five genes showed significant induction at either 2 or 3 dpi (Fig. 7a). Instead, the transcript abundance of NbMC4 gene was slightly decreased in leaf discs undergoing cell death. These data suggest that these protease-encoding genes are not directly related to BnaNAC55-mediated cell death.
qRT-PCR analysis of changes at the transcript levels of ROS- and defense-related marker genes. a, b Real-time quantitative RT-PCR analyses were performed to determine transcript levels of marker genes in control and BnaNAC55 leaves at 2 and 3 dpi. Each value represents the mean ± SE of three biological replicates. The PP2A, L23 and F-box mRNA levels were used as the endogenous control. Asterisks denote significant differences (compared to 1) by Student t test analysis (*p ≤ 0.05; **p ≤ 0.01)
Considering the facts that accumulation of ROS is involved in plant defense response and HR-like cell death is a common feature of defense response (Heller and Tudzynski 2011), we further examined the expression of a set of genes implicated in plant defense response and marker genes of HR-like cell death. These include pathogenesis-related (PR)2, PR5, Harpin inducing (HIN)1, and cysteine protease (CP)23 genes, which are all highly induced during HR-like cell death (Pontier et al. 1999). A cytochrome P450 gene, CYP76C2, whose expression was shown to be induced during hypersensitive and developmental cell death (Godiard et al. 1998), was also included in the assay. Moreover, PHYTOALEXIN DEFICIENT4 (PAD4), encoding a lipase-like enzyme that is important for SA signaling and functions in resistance (R) gene-mediated basal plant disease resistance (Vogelmann et al. 2012), was assayed together. The results showed that, among these genes, only CYP76C2, NbHIN1, NbPR2 and NbPR5 genes were transcriptionally induced in BnaNAC55-expressing leaf discs at one or two time-points (Fig. 7b). Expression of NbCP23 was slightly repressed or did not show significant change at the 2 and 3 dpi, respectively. Furthermore, three marker genes that are rapidly and transiently up-regulated upon pathogen-associated molecular pattern (PAMP) treatment in N. benthamiana, namely NbACRE31, NbACRE132 and NbCYP71D20 (Heese et al. 2007) were examined. qRT-PCR data showed that transcript abundance of NbACRE31 and NbCYP71D20 were induced by BnaNAC55 expression at 2 and/or 3 dpi (Fig. 7b). However, no significant change was observed with NbACRE132. Lastly, expression of a subset of marker genes of leaf senescence and ROS-induced cell death was also monitored. These include homologs of Arabidopsis senescence-associated gene (SAG) 12, SAG101, NAP/ANAC029 (Guo and Gan 2006), WRKY53 (Miao et al. 2004) and ZAT12 (Davletova et al. 2005). We found that expression of NbSAG101 and NbNAP increased significantly as a result of BnaNAC55 expression at one or two time-points tested, and that expression of NbSAG12 was also increased at 3 dpi. The most prominent change occurs to NbZAT12, whose transcript level was induced by over 50-folds at both 2 and 3 dpi. In Arabidopsis, the zinc-finger TF ZAT12 plays a central role in reactive oxygen and abiotic stress signaling (Rizhsky et al. 2004; Davletova et al. 2005). However, the transcript abundance of NbWRKY53 did not show any significant change (Fig. 7b).
The next question is whether BnaNAC55 directly regulates the transcription of the above marker genes. Since BnaNAC55 is a transcriptional activator, we are interested in characterizing those genes that are up-regulated as a result of BnaNAC55 expression. Therefore, we first used PCR to clone the ca. 1 kb upstream regions (relative to the translational start codon ATG, containing promoter and 5′UTR) of each up-regulated genes using high-fidelity polymerase. We succeeded in cloning the upstream regions of NbRbohB, NbAOX1a, NbGPX2, NbGST, NbSOD, NbCYP76C2, NbHIN1, NbPR2, NbNAP and NbZAT12 genes, but failed to clone those of NbRbohA and NbCAT3 gene. Next, we performed a transcriptional fusion of these promoter regions to the upstream regions to the firefly LUC gene (Fig. 8a). The different combinations of reporter vectors and an effector vector pYJHA-BnaNAC55 were individually co-infiltrated into the leaf epidermis of N. benthamiana. Another effector vector, pCsGFPBT, was used as the negative control. The renilla (REN) LUC driven by the constitutive CaMV 35S promoter was included to monitor transformation efficiency (Hellens et al. 2005). To better monitor the transcriptional regulation, two time-points (2 and 3 dpi) after agroinfiltration were included in this assay. The results demonstrated that co-infiltration of the pYJHA-BnaNAC55 and pProNbAOX1a-LUC vectors activated LUC reporter gene expression by 6.4 and 2.8 folds at the 2 and 3 dpi, respectively, compared with the GFP control (Fig. 8b). Similarly, the ability of upstream region of NbHIN1 to activate LUC expression was increased by BnaNAC55 to 18.9 and 15.2 folds at the 2 and 3 dpi, respectively (Fig. 8c). Besides, a significant increase in LUC activity was also observed with the promoter regions of NbPR2, NbCYP76C2 and NbZAT12 genes (Fig. 8d–f). These observations demonstrate that the BnaNAC55 TF could activate transcription of LUC reporter gene driven by the promoters of NbAOX1a, NbHIN1, NbPR2, NbCYP76C2 and NbZAT12 in planta. In contrast, co-infiltration of the reporter vector pYJHA-BnaNAC55 with either pProNbRbohB-LUC, pProNbGST-LUC, ProNbPR5-LUC, pProNbSOD-LUC, pProNbNAP-LUC or pProNbGPX2-LUC vectors did not significantly influence the LUC reporter gene expression (Fig. 8g, h, data not shown), which suggests that BnaNAC55 is not directly related to those gene expression and other unknown factors may participate in the activation of those gene transcription.
BnaNAC55 activates the transcription of multiple ROS- and defense-related marker genes in a dual luciferase (LUC) assay. a Schematic representation of the double-reporter and effector plasmids used in the dual LUC assay. The double-reporter plasmid contains the respective promoter regions fused to LUC and Renilla (REN) luciferase driven by CaMV35S. The effector plasmid contains the BnaNAC55 driven by the CaMV35S. b–h BnaNAC55 activates the promoter regions of respective genes in N.benthamiana leaves. The ability of BnaNAC55 TF to activate the reporter LUC gene was indicated by the ratio of LUC to REN. Error bars indicate the SE of three biological replicates. Asterisks denote significant differences by Student t test analysis (*p ≤ 0.05; **p ≤ 0.01)
Conclusion
NAC TFs are plant-specific and play important roles in plant development, abiotic stress and defense responses (Nakashima et al. 2012; Puranik et al. 2012). Over the past decade or longer, studies have indicated that ROS, such as hydrogen peroxide, are not only toxic by-products of aerobic metabolism with cellular levels strictly controlled, but they also function as signaling agents regulating many biological processes including stress adaptation and PCD in plants (Apel and Hirt 2004; De Pinto et al. 2012). However whether and how NAC TFs trigger ROS burst and cell death remain elusive.
In the present study, we described the functional characterization of NAC55 TF gene isolated from oilseed rape, one of the most important oil crops in China and world-wide. BnNAC55 responded to multiple stress and hormone treatments as well as caused ROS accumulation and cell death in N. benthamiana leaves. The expression profiling data suggested that NAC55 TF may participate in the cross-talk of different signaling pathways under abiotic and biotic stress conditions (Fig. 1). Moreover, we found that there was a difference in transcriptional activity and function of orthologous NAC genes between oilseed rape and Arabidopsis as ANAC055 was found to function as a transcriptional repressor while BnaNAC55 is a transcriptional activator (Fig. 2), which highlights the limitations of applying conclusions from Arabidopsis to oilseed rape. More importantly, we identified BnaNAC55 as a novel regulator of ROS level and cell death in leaves of N. benthamiana (Fig. 3). In addition, quantification of physiological indexes indicated that BnaNAC55 expression led to accumulation of H2O2, degradation of chlorophyll, accumulation of anthocyanins (Fig. 4). Besides, an increase in MDA content supports that BnaNAC55 expression indeed induced lipid peroxidation as a result of ROS accumulation. Further, the role of BnaNAC55 in inducing PCD was confirmed by nuclear DNA fragmentation (Fig. 5). The function of BnaNAC55 in inducing cell death was also confirmed with oilseed rape protoplasts (Fig. 6), since oilseed rape leaves are recalcitrant to agroinfiltration. A bioinformatic analysis and literature search indicated that BnaNAC55 is a novel gene modulating ROS accumulation and HR-like cell death in plants, as difference in the context of ROS and cell death exists between BnaNAC55 and ANAC055. We further monitored the expression of a variety of marker genes encoding ROS-producing and ROS-scavenging enzymes as well as proteins mediating plant defense or senescence and, detected significant changes with quite a few genes (Fig. 7). Lastly, the role of BnaNAC55 on activating the promoter activities of five genes encoding a ROS-scavenging enzyme or mediating defense response was confirmed through a dual LUC system in planta. Taken together, our work has clearly demonstrated that BnaNAC55 is an important gene that modulates ROS accumulation and cell death in plants.
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Bonneau L, Ge Y, Drury GE, Gallois P (2008) What happened to plant caspases? J Exp Bot 59:491–499
Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18:756–767
Chen B, Niu F, Liu WZ, Yang B, Zhang J, Ma J, Cheng H, Han F, Jiang YQ (2016) Identification, cloning and characterization of R2R3-MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive-like cell death. DNA Res 23:101–114
Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18:1247–1256
Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139:847–856
De Pinto MC, Locato V, De Gara L (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35:234–244
Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43:745–757
Faria JA, Reis PA, Reis MT, Rosado GL, Pinheiro GL, Mendes GC, Fontes EP (2011) The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress-and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol 11:129
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168:17–20
Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28:1091–1101
Godiard L, Sauviac L, Dalbin N, Liaubet L, Callard D, Czernic P, Marco Y (1998) CYP76C2, an Arabidopsis thaliana cytochrome P450 gene expressed during hypersensitive and developmental cell death. FEBS Lett 438:245–249
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M (2005) Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol 8:404–408
Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305:855–858
He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P (2008) Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem 283:774–783
Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104:12217–12222
Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13
Heller J, Tudzynski P (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol 49:369–390
Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che FS (2009) The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. Embo J 28:926–936
Kang YW, Jeon Y, Pai HS (2012) Characterization of cell death induced by NbBPS1 silencing in Nicotiana benthamiana. Mol Cells 34:185–191
Lam E (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5:305–315
Liang W-W, Yang B, Yu B-J, Zhou Z-Z, Li C, Sun Y, Zhang Y, Jia M, Wu F-F, Zhang H-F, Wang B-Y, Deyholos M, Jiang Y-Q (2013) Identification and analysis of MKK and MPK gene families in Canola (Brassica napus L.) BMC Genom 14:392
Liu D, Shi L, Han C, Yu J, Li D, Zhang Y (2012) Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7:e46451
Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63:289–305
Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96:8271–8276
Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragao FJ, Fontes EP (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress-and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110:19627–19632
Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55:853–867
Nagata T, Todoriki S, Masumizu T, Suda I, Furuta S, Du Z, Kikuchi S (2003) Levels of active oxygen species are controlled by ascorbic acid and anthocyanin in Arabidopsis. J Agric Food Chem 51:2992–2999
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103
Neff M, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118:27–35
Niu F, Wang B, Wu F, Yan J, Li L, Wang C, Wang Y, Yang B, Jiang YQ (2014) Canola (Brassica napus L.) NAC103 transcription factor gene is a novel player inducing reactive oxygen species accumulation and cell death in plants. Biochem Biophys Res Commun 454:30–35
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Pontier D, Gan S, Amasino RM, Roby D, Lam E (1999) Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol Biol 39:1243–1255
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279:11736–11743
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Sun Y, Wang C, Yang B, Wu F, Hao X, Liang W, Niu F, Yan J, Zhang H, Wang B, Deyholos M, Jiang Y-Q (2014) Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J Exp Bot 65:2171–2188
Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35:259–270
Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8:397–403
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Van Breusegem F, Dat JF (2006) Reactive oxygen species in plant cell death. Plant Physiol 141:384–390
Van Hautegem T, Waters AJ, Goodrich J, Nowack MK (2015) Only in dying, life: programmed cell death during plant development. Trends Plant Sci 20:102–113
Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inze D, Van Breusegem F (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 279:45329–45336
Vogelmann K, Drechsel G, Bergler J, Subert C, Philippar K, Soll J, Engelmann JC, Engelsdorf T, Voll LM, Hoth S (2012) Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159:1477–1487
Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56:505–516
Wang X, Basnayake BM, Zhang H, Li G, Li W, Virk N, Mengiste T, Song F (2009) The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant Microbe Interact 22:1227–1238
Wang B, Guo X, Wang C, Ma J, Niu F, Zhang H, Yang B, Liang W, Han F, Jiang YQ (2015) Identification and characterization of plant-specific NAC gene family in canola (Brassica napus L.) reveal novel members involved in cell death. Plant Mol Biol 87:395–411
Watanabe N, Lam E (2011) Arabidopsis metacaspase 2 d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J 66:969–982
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, Xie Q (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290
Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX (2014) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10:e1004243
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JD, Doke N (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15:706–718
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China [31471153 to Y.-Q. J. and 31301648 to B.Y.]. We thank profs. Jorg Kudla (Universitat Munster, Germany) for providing the BiFC vectors, Jian-Ye Chen (South China Agricultural University) for the LUC vector, Yule Liu (Tsinghua University) for providing VIGS vectors, and Tianyong Zhao (NWAFU) for equipment use. We also thank Wu-Zhen Liu for technical help in ROS quantification.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2016_502_MOESM2_ESM.pdf
Supplementary Fig. S1. Multiple sequence alignment and comparison of NAC55 protein from oilseed rape and representative species. (PDF 24 KB)
11103_2016_502_MOESM4_ESM.doc
Supplementary Fig. S3. Detection of BnaNAC55 expression in agroinfiltrated leaf tissues of N. benthamiana through semi-quantitative RT-PCR and Western blot. (DOC 2215 KB)
11103_2016_502_MOESM7_ESM.pdf
Supplementary Fig. S6. Examination of phenotype of NbNAC55 expression and silencing in N. benthamiana leaves. (PDF 160 KB)
Rights and permissions
About this article
Cite this article
Niu, F., Wang, C., Yan, J. et al. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol Biol 92, 89–104 (2016). https://doi.org/10.1007/s11103-016-0502-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0502-7