Abstract
Purpose
Filgrastim is the generic name for recombinant methionyl human granulocyte colony-stimulating factor (r-metHuG-CSF). It is marketed under the brand name Neupogen® by Amgen. Since this product has lost patent protection, many biosimilar versions have been approved or are in the process of filing for market authorization throughout the world. Here we show that NMR spectroscopy can be used to assess the three-dimensional structure of the active ingredient in the formulated approved product Neupogen®.
Methods
Recombinant metHuG-CSF was prepared in E. coli and isotopically enriched with 13C and 15 N isotopes. NMR spectroscopy was used to study the effects of excipients on the conformation.
Results
The effects of pH variation on the amide chemical shifts suggest the presence of cation-pi interactions between His-79 and Trp-118, and His-156-Trp-58-His-52 that stabilizes the conformation at low pH. This may be associated with a small local conformational change. The NMR data showed that polysorbate does not interact significantly with filgrastim thus allowing the collection of spectra in the presence of 20 times the formulation concentration in the sample. However, at higher detergent concentrations a reduction of signal intensity is observed. Conclusions
The NMR fingerprint assay applied to filgrastim (Neupogen® and a CRS from the European Pharmacopeia (EP)) provided residue specific information of the structure of the drug substance. In addition to current methods, the ability to assess the conformation with a high degree of resolution can greatly facilitate comparability exercises.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neutrophils are the most abundant white blood cells that are an essential part of the innate immune system and provide an initial response to a bacterial infection (1). Neutropenia is a condition characterized by a low absolute neutrophil count (ANC). One of the many causes of a low ANC is the result of their destruction by chemotherapy. An effective treatment for neutropenia in cancer patients can be achieved using filgrastim, a recombinant methionyl granulocyte-colony stimulating factor (Met-G-CSF). The mature human G-CSF protein contains 174 amino acid residues that fold into a four-helix bundle (2,3). Part of the linker between helix-1 and helix-3 is folded into a small helix that is positioned perpendicular to the bundle axis. The primary sequence includes five cysteine residues where the last four form two disulfide bridges. Cysteine-17 is partially solvent-exposed (4).
The first filgrastim product marketed for treatment of neutropenia was Neupogen® from Amgen. Since this product lost market exclusivity, many sponsors have obtained, or are seeking, marketing authorization in various countries for their versions of filgrastim. In order to approve subsequent entry biologics (SEB, as they are being named in Canada) or biosimilars (European Union, United States and others), guidance documents have been developed by most regulatory agencies. The common guiding principle throughout all the various jurisdictions is that only a reduced set of clinical trials will be required, provided that similarity is demonstrated with a thorough comparability exercise with a recognized comparator (5). Comparability exercises are also required for approved products that undergo a significant change of their manufacturing process. These include an array of physico-chemical tests and biological assays to assess the similarity of various quality attributes between product and comparator. The three-dimensional structure of the drug substance is one of the most important quality attributes. Current methods used by manufacturers to characterize the three-dimensional structure of the drug substance are circular dichroism, fluorescence, Fourier transformed infrared spectroscopy, one dimensional proton NMR, and bioassays, to name only these. The assemblage of the various measurements from these techniques only provides a low-resolution assessment of this quality attribute. Our laboratory has proposed a simple two dimensional NMR method referred to as the NMR fingerprint assay (6) that can assess the three-dimensional structure of the drug substance of a therapeutic product at amino acid resolution. In this paper, we demonstrate the applicability of the method to the characterization of filgrastim-based products. The latter, whether they are from the innovator or subsequent entry products, are formulated using similar excipients at a low pH (4.0) to minimize the oxidation of the free cysteine residue. Therefore, the effects of various solution conditions have been studied to measure the perturbations induced on the spectroscopic fingerprint of the protein. In addition, the ability to distinguish wild type and mutants that have been engineered to enhance conformational stability (7) will be discussed.
Experimental
Protein Expression and Purification
The plasmid pET15b10-enterokinase-GCSF was constructed from a synthetic DNA fragment (BioS&T) corresponding to the mature polypeptide (174 amino acids) sequence of human granulocyte colony stimulating factor (accession number E01219) preceded by the sequence corresponding to an enterokinase cleavage site. The DNA sequence was codon optimized for expression in E. coli and Pichia pastoris. This synthetic DNA (enterokinase-GCSF) was inserted into a pET15b10 expression vector at the NdeI and BamHI sites. This vector is an in-house modified version of the pET15b vector (Novagen) with four extra histidine residues in the tag. Since filgrastim includes an additional methionine residue at the N-terminal, the plasmid pET15b10-enterokinase-Met-GCSF was obtained by site directed mutagenesis. Similarly, the A29G, A30G, A37G, and A29,30G mutants were obtained by site directed mutagenesis of the pET15b10-enterokinase-Met-GCSF construct.
Expression and purification of 15 N-filgrastim and mutants was carried out essentially as described for labeled interferon α-2a (8) with minor modifications: cation-exchange chromatography was carried out at pH 4.0 on a Mono-S (GE-HealthCare) and a final polishing step was carried out using size-exclusion chromatography on HiLoad 26/60 Superdex 75 Prep Grade (GE-HealthCare).
Samples for NMR typically contained 0.017–0.4 mM of 15 N-Met-G-CSF in sodium deutero-acetate (pH 4.2) with 5% deuterium oxide for field frequency lock. Titrations of pH, salts, buffers and excipients (polysorbate-80 and sorbitol) were carried out in 15 N-labeled samples with protein concentration of 0.017, 0.2, or 0.4 mM as appropriate.
Sample Preparation for Neupogen and EP-CRS-Filgrastim
Neupogen® was purchased at a local pharmacy. A single vial contains 0.3 mg (32 μM) of filgrastim, 0.295 mg of acetate (10 mM), 0.02 mg of polysorbate-80 (0.03 mM), 25 mg of sorbitol (274 mM), and 0.0175 mg of sodium (1.52 mM) in 0.5 mL of water. Twenty vials were pooled and concentrated to a final volume of 0.4 mL using an Amicon ultra-4 centrifugal device (Millipore) with a 3 kDa molecular weight cutoff. Deuterium oxide (5% v/v) was added for field frequency lock. The EP Chemical Reference Standards for filgrastim (catalogue number Y0001173, batch 1) was obtained from the European Pharmacopeia. The EP-CRS-filgrastim was shipped frozen in sealed vials containing 0.2 mL of a 1.95 mg/mL solution. The content of 15 vials were diluted in 10 mL of 10 mM sodium deutero-acetate buffer at pH 4.0 and concentrated by ultra filtration. Three buffer exchanges were performed prior to final concentration to 0.3 mL. Assuming complete recovery of protein during buffer exchange, the final samples contained about 1.0 mM of filgrastim, 10 mM sodium deutero-acetate (pH 4.0) and 5% (v/v) of deuterium oxide for field frequency lock. Due to a contamination issue in our laboratory, the sample had to be cleaned with cation-exchange chromatography over a Mono-S resin (GE Healthcare), leading to the loss of two thirds of the original sample. The sample used in this study contained ~0.3 mM protein and showed extra peaks in the amide side-chain region.
NMR Spectroscopy
Two dimensional 1H-15N-heteronuclear single quantum coherence (HSQC) experiments with water flip-back pulse, gradient coherence selection and sensitivity enhancement were recorded on Bruker AVANCE III 600 MHz and AVANCE 700 MHz spectrometers (Milton, Canada) equipped with a cryogenic probehead. For all 2D-HSQC experiments, two dimensional data matrices of 1024 by 64 complex points were collected with spectral windows of 14 or 16 ppm and 32 ppm for proton and nitrogen, respectively. All spectra were recorded at 25°C with a relaxation delay of 1.0 s, an acquisition time of 100 millisecond per field induction decay (FID) and a total acquisition time of 15–60 min for 15 N-labeled samples or 90 h for natural abundance reference samples Neupogen® and CRS-EP. The latter required 2048 transients per FID. The residual water signal was deconvoluted with a polynomial function using the NMRPipe software (9) and each vector was multiplied by a shifted sine-square function prior to zero filling and Fourier transform. The indirect dimension was linear predicted to double the number of data points prior to multiplication of a cosine function followed by zero filling and Fourier transformation producing 2D maps with the following digital resolutions for maps collected at 600 MHz: 8.2 Hz/pt and 15.0 Hz/pt for proton and nitrogen, respectively, and for maps collected at 700 MHz: 9.6 Hz/pt and 17.5 Hz/pt for proton and nitrogen. Backbone chemical shift assignments were confirmed on a 13C,15 N-Met-GCSF sample using 2D 1H-15N HSQC, 3D HNCO, 3D HNCACB, and 3D CBCACONH from the report of Holak and coworkers (2,10). Resonance assignment of side-chain methyl groups was carried out with a set of four experiments: 3D HCCH-TOCSY, 3D HCCH-COSY, 3D CCC-TOCSY(CO)NH, and 3D HCC-TOCSY(CO)NH. Resonance assignment of aromatic side-chain was carried out with the HBCBCGCDHD and HBCBCGCDCEHE experiments. Data visualization was carried out using NMRView (11). All assignments were deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under the following accession numbers: 18291 (wild-type), 18293 (A29G), 18294 (A30G), and 18295 (A37G). Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311) (12).
Results and Discussion
Sample Preparation
The procedure for the preparation of labeled filgrastim takes advantage of the poly-histidine tag to accomplish on-column refolding under oxidative conditions and affinity purification in a single step. Samples for NMR studies are thus obtained by cleavage of the tag and further purifications using cation exchange and size exclusion chromatography. Almost all backbone resonances were assigned using a doubly labeled sample (13C,15 N-Met-GCSF). Examination of the 2D-1H,15N-HSQC spectrum (Fig. 1) indicated the presence of a number of unassigned resonances. The majority of these belong to proton-nitrogen pairs on arginine, lysine and histidine side-chains. The low pH of the sample causes the chemical exchange of their protons with the solvent to be significantly reduced. In addition, extra resonances have been observed and result from the formation of an oxidated form of filgrastim. See supplemental information (Supplemental information and accompanying Supplemental Figs. 1 and 2). Considering that the resonance assignment allowed us to track all resonance belonging to the properly folded protein, the presence of this, or these, minor oxidized species can be treated as an impurity that will not interfere with the study on the effects of excipients, pH, and temperature on the NMR spectra. Further oxidation was not observed at pH 4.0. Doubly labeled samples (13C,15 N), used for the resonance assignment procedure, showed no significant changes over the period of several months to a year.
Effects of Sample Conditions on NMR Fingerprint; to Deformulate or not
The resonances of proton nitrogen amide pairs are very sensitive to solution conditions. This is the result of the involvement of amide protons in the formation of hydrogen bonds and their ability to chemically exchange with protons from the bulk water. Therefore, amide resonances can be ‘too sensitive’ and thus they may not be very reliable probes of conformational changes. However, when no changes in the resonance pattern is observed, it can be readily concluded that there are no changes of the protein conformation.
Various factors can modify the local magnetic environment of any given residue on the protein, hence affecting the resonance frequency. These include: change of the conformation (local or global), temperature, and changes of the solution conditions such as ionic strength (ion concentration and types of ions), pH, co-solutes (polyols used as cryoprotectant, such as sorbitol, mannitol, and sugars, and non-denaturing detergents such as polysorbates). Identifying changes on the spectra related to these factors was useful to determine whether filgrastim products required the isolation of the drug substance or if they could be studied in the presence of excipients. In addition, the ability to correlate differences between two spectra to any of these changes is very important in the context of a comparability study. For example, small changes in the spectra, that could be attributed to changes of the conformation, may simply be the results of small differences of pH or ionic strengths between a product and its comparator. Starting from the list of excipients in the formulation of Neupogen®, we performed a series of titrations to study the effects of acetate buffer concentration, pH, temperature, sorbitol concentration, polysorbate-80 concentration and ionic strength with various salts such as sodium and potassium as cations, and chloride, phosphate, and sulfate as anions.
Temperature Variation
The resolution of NMR spectra of proteins can sometimes be improved by raising the temperature. This increases the tumbling of protein molecules leading to narrower peaks, with the possible caveat of a decrease in sample stability. In addition, it is well documented that amide chemical shifts vary with change of temperature (13). Since amides are involved in hydrogen bonding with either a carbonyl acceptor from another residue or the bulk water, higher sample temperatures induce larger thermal fluctuations resulting in longer average hydrogen bond distances. As amide chemical shifts are highly dependent on these distances, resonances move almost linearly with temperature, provided that the latter is 15 degrees lower than the unfolding temperature (13). Therefore, we performed a study on the effects of temperature on the spectra to investigate whether a higher temperature could provide higher resolution spectra by either narrowing the resonances and/or moving the amide resonances, such as to better resolve overlapping regions of the spectrum. Considering that the melting temperature (Tm) of filgrastim is 60°C (14), we collected a series of two-dimensional spectra in the range of 25–37°C with 2.5°C increments. An overlay of all spectra (see Supplemental Fig. 3) does not indicate a significant narrowing of the resonances, nor an improvement of peak resolution.
Variation of pH
Recombinant protein therapeutic products are usually formulated at a pH near physiological pH in order to avoid a significant pH change upon injection in patients. Filgrastim products are formulated at a lower pH (4.0–5.0), using weak buffering agents such as acetate or glutamate buffers, in order to minimize oxidation of the free sulfydryl of cysteine (Cys-17). In addition, it has been shown that the protein conformation is more stable at this lower pH (15,16). Moreover, Narhi et al. have observed a quenching of fluorescence at low pH, which they attributed to a change in protein conformation that increased solvent exposition of tryptophan side-chains (17). In view of these important effects of pH for this product, we investigated the effects of pH by NMR. A series of NMR spectra were recorded at various pH values (2.8 to 6.2) to monitor which amide resonances were most affected by the ionization state of histidine and acidic residues (aspartate and glutamate). The overlay of the eight spectra (see Supplemental Fig. 4a) shows a number of well-resolved resonances that experience large shifts upon varying pH. These are Ser-76, His-79, Ser-80, Glu-93, Glu-98, Asp-104, Trp-118, and Ala-127. While it was expected to observe significant changes for residues with titratable side-chains such as aspartate, glutamate and histidine as their carboxylic or imidazole group becomes protonated with the addition of acid, the large changes displayed by His-79 and Ser-80, among others, suggested the possibility of a conformational change. Examination of the three-dimensional structures determined by X-ray crystallography (crystallization at pH 4.8, pdbid: 2D9Q) (18) and NMR spectroscopy in solution at pH 3.5 (pdbid: 1GNC) (2) revealed that three histidine (His-52, His-79 and His156) residues are in close proximity with tryptophan residues (Trp-58 and Trp-118). In fact, a minor conformational change of the local structure, if any, may be needed to allow Trp-58 to be interacting with His-156 (Xray) and/or His-52 (NMR), and Trp-118 to interact with His-79. Since the latter residues are located in very crowded region of the spectra, it was not possible to assess their chemical shift perturbation directly from the overlay. Therefore, extracting resonances of these residues generated a simplified overlay spectrum (Fig. 2a). This shows that His-79 and Trp-118 are most significantly perturbed with pH, while the chemical shifts of the His-156-Trp-58-His-52 triad are perturbed to a lesser extent. The magnitude of the chemical shift perturbation for the above-mentioned histidine and tryptophan residues, and their proximity in the structure, suggests the formation of cation-π interactions (19). These, observed in M2 ion channels, act as a gating mechanism in channel activation (20). In this system, a histidine side chain becomes protonated at low pH and interacts with a nearby tryptophan leading to a conformational change that opens the gate of the channel. Based on the magnitude of the chemical shift perturbations observed in filgrastim with pH, two cation-π interactions are possible. The first interaction would involve His-79 and Trp-118, while the second interaction would involve two or three residues: His-156 and/or His-52 and Trp-58. The presence of these interactions at low pH could readily explain the gain in conformational stability observed by Narhi et al. (17). A similar interaction between a protonated histidine and a tryptophan that produced a gain in stability has been observed in Barnase, whereby the change of free energy of unfolding was lowered by 1.5 kcal in mutants lacking the interacting tryptophan residue (21). The presence of a number of large chemical shift perturbations for residues that do not have an ionizable side chain suggests that the formation of a cation-π interaction may be associated with local conformational changes. The latter can simply be a reorientation of the side-chain of interacting residues to properly position them for optimal interaction, or a change of the relative orientation of secondary structure elements such as helices and loops.
(a) Simplified overlay spectrum of the amide proton-nitrogen pairs affected by change in pH.. The full spectral overlay is shown in supplemental Fig. 4. (b) Overlay of the methyl region from the 2D 1H-13C-HSQC, recorded at three pH values (4.3, 5.0, and 6.4). Contrary to the nitrogen experiment, fewer residues show chemical shift perturbation with pH, these are Ala-91, Ala-127 Ile-117 (both γ and δ methyl signals) and Leu-168. (c). Overlay of the aromatic regions of the 2D 1H-13C-HSQC. For clarity, the signal intensity of the tryptophan regions was raised by a factor of 1.5 to allow better visualization. Resonance assignment of particular residues could not be achieved.
In order to determine the nature and amplitude of this local conformational change, it is appropriate to inspect the chemical shifts of methyl groups from alanine, valine, leucine, and isoleucine side-chains as a function of pH. The proton and carbon-13 chemical shift pairs of these groups are better probes for conformational variations than proton-nitrogen pairs. The latter are influenced by the local structure; in addition, these chemical shifts are also modulated by chemical exchange, change of hydrogen bond length, temperature, etc. In other words, observation of a methyl resonance shifts can be more confidently correlated to a change of the local structure. They can be seen as the tips of the side-chain ‘fingers’ interacting with adjacent hydrophobic residues. A series of 2D 1H,13C-HSQC, recorded at three pH values (4.3, 5.0 and 6.4), are shown as an overlay in Fig. 2b, where the well-resolved methyl signals have been assigned. During data collection at pH 6.4, the sample showed signs of precipitation that resulted in a small loss of protein. This prevented us to record data at higher pH values. Upon lowering the pH, an overshoot brought the pH down to 1.5. We decided to record a spectrum at that pH, but the spectrum showed a significant loss of signal intensity and is of poor quality. In contrast to the 1H,15N-HSQC where many perturbations are observed, only five residues are experiencing chemical shift perturbations with pH variation. In comparison, the amide chemical shift of Thr-115 is moderately perturbed with pH, while the methyl shifts are very little if any. These are Ala-91, Ile-95, Ala-127 Ile-117 (both γ and δ methyl signals) and Leu-157. Their position on the structure is indicated on Fig. 3. Among these, Ile-117 and Leu-157 are in close vicinity to His-79-Trp-118 and His-156 and/or His-52 and Trp-58, respectively, with Ile-117 δ methyl showing a large perturbation. This suggests that the delta methyl group of Ile-117 is pointing at the His-79-Trp-118 pair and is perturbed by the formation of the cation-π interaction. It is noteworthy that Ala-127 located on the CD loop above the H79-W118 pair is showing the second largest perturbation, while Ala-114 that is under Trp-118 by one helix turn is unperturbed. Moreover, Ala-91 (largest perturbation) and Ile-95 (small perturbation) are not in the vicinity of neither cation-π interactions. This supports a very small change of the orientation of helix C that does not induce an observable chemical shift perturbation on residues on that helix, such as Ala-114 or Ala-111, but gets slightly ‘amplified’ when transmitted to the CD loop, in a manner similar to a whip. A small reorientation of the handle (Helix C) is amplified by the lash (CD loop). Thus a small reorientation of the CD loop could reposition Phe-140, the neighbor of Ala-91 and Ile-95. Holak and coworkers have shown, using heteronuclear NOE measurements (2), that the CD loop is the most flexible section of the structure, thus a small change in the orientation of the C helix may impact the dynamics of that loop, or its average conformation, and the position of Phe-140. Analysis of the aromatic resonances from the overlays of the 2D 1H,13C-HSQC (Fig. 2c) showed very little perturbations of chemical shifts associated to phenylalanine residues, but only those resonances attributed to tryptophan and histidine side-chains are perturbed with pH. The spectrum collected at pH 1.5 (see Supplemental Fig. 4b) showed that the Hε1Cε1 correlations overlay with these resonances at pH 4.3 thus confirming that all histidines side chains are fully protonated at pH 4.0. Therefore, the analysis of the chemical shift perturbation suggests that the formation of cation-π interactions is accompanied by a small reorientation of the histidine and tryptophan side-chain, which may be a simple rotation of a bond to maximize the interaction. This does not support a conformation with buried tryptophan residues that become exposed upon lowering pH, in other words, nothing significant to explain the increase of tryptophan fluorescence quenching upon lowering pH. This is more likely the result of the proximity of a protonated histidine residue for both Trp-118 and Trp-58. Compared to the neutral histidine, the protonated form has been shown to be a more efficient quencher of tryptophan fluorescence (22).
Residues experiencing chemical shift perturbations (amide or proton-carbon correlations) upon pH variations are indicated in the structure (a single model from the NMR ensemble of 10 structure, pdbID: 1GNC). The regions of the structure where the two tryptophan residues can interact with histidine residues are shown in boxes.
Variation of Ionic Strength
After performing the pH titration, the sample was returned to pH 4.0. The spectra showed small differences near Ser-80, suggesting that variation of the pH may have altered the sample. Therefore we investigated whether this was an irreversible change of the protein or the effects of a different ionic strength from the generation of salt during the titration. This question could easily be addressed by studying the effects of ionic strength. Throughout the study, the pH was monitored to ensure that no significant change (less than 0.03 pH unit) occurred with the addition of various salts. As salt concentration was increased, the chemical shifts of His-79, Ser-80 and Trp-118 varied, but to a lesser extent when compared to the pH effects (see Supplemental Fig. 5). It is noteworthy that as little as a 10 mM increase in ionic strength induces chemical shift changes on Ser-80. This indicates that the perturbations of these chemical shifts after the pH titration are indeed the result of generating sodium chloride from the neutralization of the acid by the base and vice versa. The effects of increasing the ionic strength on the amide shift of His-79 and Ser-80, when compared to the chemical shift perturbations induced by pH variations, are too small to induce a significant conformational change. As observed in barnase, the data support that high salt concentrations do not significantly mask the cation-π interaction (21).
Titration of Polysorbate-80
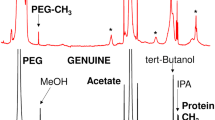
Detergents are used in product formulations to minimize protein unfolding at air-liquid and solid–liquid interfaces, and to inhibit protein aggregation by coating hydrophobic patches at the protein surface (23). Filgrastim products are formulated with the non-denaturing detergents polysorbate-80 (PS-80) or polysorbate-20. These can bring challenges to the application of spectroscopic methods, in particular NMR spectroscopy, to assess the conformation of the drug substance. The relatively low sensitivity of NMR often requires the use of more than one dose in order to obtain a sufficient concentration of the protein (6). The sample can either be prepared from the isolation of the drug substance, or from the direct concentration an appropriate number of vials to obtain the desired amount. For example, to obtain a sample of Neupogen® with a target protein concentration of 1 mM requires 20 vials containing each 300 μg of drug substance. Concentration of this many vials by ultra-filtration, for an NMR sample with final volume of 300 μL, results in a protein concentration near 1 mM. However, since the formulated product contains 0.03 mM of PS-80 with a critical micelle concentration (CMC) of 0.012 mM, the ultra-filtration process also increases the concentration of the detergent twenty-fold. Since the concentration is a more simple approach to prepare samples and does not require de-formulation, we tested whether the concentration of PS-80 impacts the NMR analysis or the conformation.
Therefore, we performed a series of PS-80 additions to obtain a final concentration of 0.03 mM (1X the formulation concentration) of PS-80, 0.4 mM (~13X), and 2.4 mM (80X), into a sample of 0.4 mM of 15 N-labelled filgrastim. The NMR spectra did not show significant chemical shift perturbations or line broadening in the 1X and 13X spectra (Fig. 4a). At 80X, some chemical shift perturbations are observed as well as line-broadening effects of all resonances are observed (Fig 4b). In addition, very broad resonances can be seen in the centre of the 2D map. Broadening of the resonances indicates that the tumbling rate of protein molecules is reduced at high concentrations of PS-80, probably due to an increase in the viscosity of the sample, and/or the presence of weak non-specific interactions between filgrastim and detergent micelles. Moreover, the observation of extra resonances, at very high concentrations of PS-80, suggests the presence of protein-detergent interactions. Under these conditions, it was not possible to conclude whether monomers or micelles are the interacting species. In an attempt to shed some light, we performed a titration of PS-80 (0X, 0.5X the CMC and 1X, 5X, 10X, 20X the PS-80 formulation) on a sample at the formulation concentration, i.e., 17 μM of 15 N-filgrastim. Here again, no chemical shifts perturbations were observed up to 20X the formulation concentration of PS-80. This results from the impossibility to saturate low affinity binding sites with large excess of PS-80 monomers. However, using 1D plots of the 1H-15N HSQC (Fig. 4c), a decrease of signal intensity suggesting that PS-80 micelles does influence the tumbling of the protein and induce line broadening. A lineshape analysis of two well-resolved residues (Gly-28 and Gln-11) from the overlay of four 2D spectra shown in Fig. 3a and b and the 2D spectrum of Neupogen® also show that the linewidth at half-height does not change with increasing concentration of PS-80. The lack of chemical shift perturbation in equimolar detergent-protein quantities and the effects on signal intensities as the detergent concentration is high, suggest the presence of weak interactions. These can be better characterized with a more in-depth NMR study that would be outside the scope of this paper. The above observations help to develop guidelines for the feasibility of NMR analyses of formulated filgrastim products. The effects of the detergent on signal intensities limit the number of filgrastim doses that can be concentrated to prepare an NMR sample. We have successfully made samples of 1 mM Neupogen® corresponding to 20X the formulation concentration of PS-80 that produced very high quality NMR spectra (Fig. 5). While we did not test higher concentrations, we feel that a maximum of 2 mM filgrastim, corresponding to 40 doses may be the practical upper limit. These sample conditions, i.e., relatively high protein concentrations and long acquisition times (2–4 days) do not present any problems. Filgrastim samples are stables under NMR conditions for many months and modern NMR spectrometers are very stable to allow such measurements.
Effects of Polysorbate-80 (a) 0.4 mM 15 N-met-GCSF and 10X formulation concentration of PS-80; (b) 0.4 mM 15 N-met-GCSF and 80X formulation concentration of PS-80. (c) One-dimensional 1H-15N HSQC of 0.03 mM 15 N-filgrastim. (d) Lineshapes of two cross-peaks from Gly-28 and Gln-11 at four concentrations of PS-80 (green: 1X, blue: 13X, red: 20X, and black: 80X the formulation concentration) are superimposed. The 20X line is from the Neupogen® sample. The average linewidth at half-height (Δν1/2) is illustrated with the black dotted lines.
Effects of Mutations: NMR vs Bioassays
Various chemical modifications, such as oxidation, or deamidation, to name only these, can affect the quality of recombinant protein drug products. These modifications can be detrimental to biological function, or have no effects at all. Bristow and coworkers (24) have produced a number of helix-destabilizing mutants (single point A29G, A30G and A37G, and double, and triple point) near the binding interface (see Supplemental Fig. 6). Their results showed that a bioassay can better distinguish between mutants, but one (A37G), while the physico-chemical method (CD) chosen could not distinguish any of the mutants. We then prepared the three single and a double mutants, (the triple mutants were not stable under NMR conditions) and studied them by NMR. All four mutants produced distinctive 2D spectra (Supplemental Fig. 7). The near complete resonance assignments of the backbone atoms were carried out for the single mutants to identify the effects of mutations on the 2D spectra. Mutation of any of the three alanine residues to a glycine residue resulted in the disappearance of the corresponding amide signal from the alanine with the appearance of a new signal for the glycine amide. In addition, the mutations produced a number of chemical shift perturbations of the amide resonances of surrounding residues, as a result of the change in local magnetic environment at the mutation site. Therefore, all four mutants have unique NMR fingerprints that readily allow their distinction between themselves and the wild type.
Case study: Neupogen® and Chemical Reference Standard
In order to demonstrate the usefulness of NMR fingerprints in a comparability study, we recorded 2D-NMR spectra of three filgrastim samples: Neupogen®, the EP-CRS and 15 N-filgrastim prepared in our laboratory. Note that we were able to record high-resolution data in approximately 83 h on the EP-CRS sample containing only 0.3 mM of filgrastim. The spectral overlay of all three samples (see Supplemental Fig. 8) shows small chemical shift differences that can be attributed to the small difference of pH, and the presence of excipients in the Neupogen® sample. Based on the above pH, salts, and detergents effects on the spectra, we can conclude that it is highly unlikely that the three proteins show any conformational differences, but that they are highly similar.
Conclusion
Application of high-resolution techniques, such as NMR spectroscopy, to the assessment of the higher order structure of recombinant protein therapeutics can raise many concerns. It is often feared that such techniques may bring about differences that may not be easily explained, and that, in the end are not clinically significant. This study has shown that solution conditions and sample temperature influence the chemical shifts of amide resonances. In particular, the effects of pH on the NMR spectra are pretty striking and may suggest large perturbations of the conformation, as suggested by earlier studies with fluorescence and CD. However, our data is more indicative of a stabilization of the conformation via a cation-π interaction that may be associated with a small local structural change. In other words, this study has allowed us to provide a better handle of the magnitude of the changes observed and to suggest that they are not significant. Yet they produced significant changes in fluorescence.
The small case study illustrated the feasibility of using NMR fingerprints in the context of a comparability exercise. A more thorough study on the applicability and validation of NMR fingerprints to filgrastim products from the marketplace can be conducted in a round robin study.
Abbreviations
- ANC:
-
Absolute neutrophil count
- CMC:
-
Critical micelle concentration
- CRS:
-
Chemical Reference Standard
- EP:
-
European Pharmacopeia
- HSQC:
-
Heteronuclear single quantum coherence
- IMAC:
-
Immobilized metal affinity chromatography
- Met-G-CSF:
-
Recombinant methionyl granulocyte-colony stimulating factor
- PS-80:
-
Polysorbate-80
- SEB:
-
Subsequent entry biologics
References
Parham P. The Immune System 2nd Ed. New York: Garland Science; 2005. p. 227–76.
Zink T, Ross A, Lüers K, Cieslar C, Rudolph R, Holak TA. Structure and dynamics of the human granulocyte colony-stimulating factor determined by NMR spectroscopy. Loop mobility in a four-helix-bundle protein. Biochemistry. 1994;33:8453–63.
Hill CP, Osslund TD, Eisenberg D. The structure of granulocyte-colony-stimulating factor and its relationship to other growth factors. Proc Natl Acad Sci U S A. 1993;90:5167–71.
Arakawa T, Prestrelski SJ, Narhi LO, Boone TC, Kenney WC. Cysteine 17 of recombinant human granulocyte-colony stimulating factor is partially solvent-exposed. J Protein Chem. 1993;12:525–31.
EMEA Comparability of biotechnological/biological products subject to changes in their manufacturing process. CPMP/ICH/5721/03, 13 (2006). Canada, H. GUIDANCE FOR SPONSORS: Information and Submission Requirements for Subsequent Entry Biologics (SEBs). 2010/03/05. Health Canada - Publications, 17 (2010). FDA, U. Guidance for Industry: Quality Considerations in Demonstrating Biosimilarity to a Reference Protein Product. Office of Communications US FDA UCM291134, 1-20 (2012). FDA, U. Guidance for Industry: Scientific Considerations in Demonstrating Biosimilarity to a Reference Product. Office of Communications US FDA UCM291128, 1-25 (2012).
Aubin Y, Gingras G, Sauvé S. Assessment of the three-dimensional structure of recombinant protein therapeutics by nmr fingerprinting: demonstration on rhGM-CSF. Anal Chem. 2008;80:2623–7.
Bishop B, Koay DC, Sartorelli AC, Reagan L. Reengineering granulocyte colony-stimulating factor for enhanced stability. J Biol Chem. 2001;276(36):33465–70.
Panjwani N, Hodgson DJ, Sauvé S, Aubin Y. Assessment of the effects of pH, formulation and deformulation on the conformation of interferon alpha-2 by NMR. J Pharm Sci. 2010;99:3334–42.
Delaglio F, Grzesiek S, Vuiter GW, Zhu G, Pfeifer J, Bax A. NMRpipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93.
Zink T, Ross A, Ambrosius D, Rudolph R, Holak TA. Secondary structure of human granulocyte colony-stimulating factor derived from NMR spectroscopy. FEBS Lett. 1992;314:435–9.
Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–14.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12.
Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J Biomol NMR. 1997;9:359–69.
Kolvenbach CG, Narhi LO, Philo JS, Li T, Zhang M, Arakawa T. Granulocyte-colony stimulating factor maintains a thermally stable, compact, partially folded structure at pH 2. J Pept Res. 1997;50:310–8.
Ricci MS, Sarkar CA, Fallon EM, Lauffenburger DA, Brems DN. pH Dependence of structural stability of interleukin-2 and granulocyte colony-stimulating factor. Protein Sci. 2003;12:1030–8.
Herman AC, Boone TC, Lu HS. Characterization, formulation and stability of Neupogen (Filgrastim), a recombinant human granulocyte-colony stimulating factor. In: Pearlman R, Wang J, editors. Formulation, characterization and stability of protein drugs. New York: Plenum Press; 1996. p. 303–28.
Narhi LO, Kenney WC, Arakawa T. Conformational changes of recombinant human granulocyte- colony stimulating factor induced by pH and guanidine hydrochloride. J Protein Chem. 1991;10:359–67.
Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, et al. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. Proc Natl Acad Sci U S A. 2006;103:3135–40.
Mecozzi S, West Jr AP, Dougherty DA. Cation-π interactions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc Natl Acad Sci U S A. 1996;93:10566–71.
Okada A, Miura T, Takeuchi H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 Ion channel from influenza a virus. Biochemistry. 2001;40:6053–60.
Loewenthal R, Sancho J, Fersht AR. Histidine-aromatic interactions in barnase. Elevation of histidine pKa and contribution to protein stability. J Mol Biol. 1992;224:759–70.
Van Gilst M, Hudson BS. Histidine-tryptophan interactions in T4 lysozyme: anomalous pH dependence of fluorescence. Biophys Chem. 1996;63:17–25. references cited therein.
Chou DK, Krishnamurthy R, Randolph TW, Carpenter JF, Manning MC. Effects of Tween 20 and Tween 80 on the stability of Albutropin during agitation. J Pharm Sci. 2005;94(6):1368–81.
Bristow AF, Bird C, Bolgiano B, Thorpe R. Regulatory requirements for therapeutic proteins: the relationship between the conformation and biological activity of filgrastim. Pharmeur Bio Sci Notes. 2012;2012:103–17.
ACKNOWLEDGMENTS AND DISCLOSURES
The authors would like to thank Dr. Adrian Bristow (NIBSC) for stimulating discussions and for sharing his results on the mutants prior to publication, and Drs. Michel Girard and Michael Johnston for critical reading of the manuscript. The mass spectrometry support by Drs Daryl Smith and Terry Cyr is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 12913 kb)
Rights and permissions
About this article
Cite this article
Aubin, Y., Hodgson, D.J., Thach, W.B. et al. Monitoring Effects of Excipients, Formulation Parameters and Mutations on the High Order Structure of Filgrastim by NMR. Pharm Res 32, 3365–3375 (2015). https://doi.org/10.1007/s11095-015-1713-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-015-1713-3