Abstract
Neuropathic pain is an unneglectable pain condition with limited treatment options owing to its enigmatic underlying mechanisms. Long noncoding RNA small nucleolar RNA host gene 5 (SNHG5) is involved in the progression of a spectrum of human cancers. However, its role in neuropathic pain remains undiscovered. In the present study, we established a mouse spinal nerve ligation (SNL) model, and a significant upregulation of SNHG5 was observed. Then we knocked down SNHG5 level in mouse L5 dorsal root ganglion (DRG) by delivering specific short hairpin RNA against SNHG5 with adenovirus vehicle. Mouse paw withdrawal threshold (PWT) and paw withdrawal latency (PWL) in response to mechanical stimuli was increased after SNHG5 knockdown, accompanied with decreased protein levels of glial fibrillary acidic protein (GFAP) and ionized calcium binding adapter molecule 1 (IBA-1). Besides, SNHG5 directly modulated the expression of miR-154-5p, which was downregulated in SNL mice. MiR-154-5p inhibition abolished the effect of SNHG5 knockdown on mouse behavioral tests and GFAP and IBA-1 levels. In addition, we validated that C-X-C motif chemokine 13 (CXCL13) was a novel downstream target of miR-154-5p, and CXCL13 level was positively related to that of SNHG5 in SNL mice. In conclusion, our study demonstrated that SNHG5 knockdown alleviated neuropathic pain and inhibited the activation of astrocytes and microglia by targeting the miR-154-5p/CXCL13 axis, which might be a novel therapeutic target for neuropathic treatment clinically.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neuropathic pain is defined as chronic pain condition results from a lesion or nervous system dysfunction [1]. Affecting approximately 8% of the general population, neuropathic pain severely impairs the life quality of patients and brings substantial healthcare cost to society [2, 3]. Neuropathic pain is a consequence of a spectrum of diseases, among which diabetes mellitus (DM), herpes zoster and human immunodeficiency virus (HIV) infection are most clinically common [4]. Current treatment of neuropathic pain is limited and challenging owing to its obscure etiology. Medication including antidepressants and anticonvulsants remain the first-line therapeutic option for neuropathic pain. However, only less than 50% of patients suffer from neuropathic pain experience pain relief, accompanied with side effects including sedation and dizziness [5, 6]. Consequently, a better understanding of the etiology to discover novel analgesics is of great importance to neuropathic pain management.
Long noncoding RNAs (lncRNAs) refer to non-protein-coding transcripts larger than 200 nucleotides. LncRNAs are involved in the development of a variety of human diseases including neuropathic pain [7]. For instance, lncRNA uc.48+ and NONRATT021972 knockdown alleviates DM-mediated neuropathic pain through targeting P2X3 receptor to suppress excitatory transmission [8, 9]. Kcna2 antisense RNA silencing reverses nerve injury and relieves neuropathic pain in rats [10]. LncRNA CCAT1 overexpression ameliorates neuropathic pain in rat bilateral sciatic nerve chronic constriction injuries model [11]. Recent studies have revealed that lncRNA small nucleolar RNA host gene 5 (SNHG5) was involved in the initiation and development of several human cancers including breast cancer, gastric cancer, glioma and colorectal cancer [12,13,14,15], yet its role in the onset and progression of neuropathic pain is still enigmatic. Bioinformatics prediction showed that lncRNA SNHG5 could directly bind to miR-154-5p (https://starbase.sysu.edu.cn/index.php). MiR-154-5p is down-regulated in chronic constriction injury rat model, and miR-154-5p overexpression alleviates neuropathic pain by targeting toll-like receptor 5 (TLR5) [16]. Besides, C-X-C motif chemokine 13 (CXCL13) is a putative downstream target of miR-154-5p by prediction on Targetscan database (https://www.targetscan.org/vert_72/). Therefore, SNHG5 might regulate neuropathic pain through competitive binding of miR-154-5p to increase CXCL13 expression.
In the present study, we examined the expression of SNHG5 and miR-154-5p and evaluated their effects on neuropathic pain in a mouse spinal nerve ligation (SNL) model. We also validated that CXCL13 is a downstream target of miR-154-5p which might be involved in neuropathic pain development. Our study may offer a potential therapeutic target for the treatment of neuropathic pain.
Materials and Methods
Mouse SNL Model Establishment
Male 6–8-week old C57BL/6 mice were purchased from Guangdong Medical Laboratory Animal Center (China). All mice were housed in sterilized cages and allowed to drink and eat ad libitum. For mice in SNL group, surgery was performed to expose the L5 spinal nerve, followed by ligation with 4-0 silk thread. For mice in sham group, identical surgery was performed except for that the L5 spinal nerve was not ligated. All animal experiments were carried out according to the guidelines of the International Association for the Study of Pain and approved by the Laboratory Animal Ethics Committee of Guizhou Medical University (Approval No. 1900724).
Adenovirus Infection and Cell Transfection
The SNHG5 short hairpin RNA was synthesized and inserted into ADV3 adenovirus plasmid with CMV promoter (Genepharma, China) between EcoRI and BamHI, termed as Ad-SNHG5, and Ad-GFP (Genepharma, China) was used as a control. Ad-SNHG5, Ad-GFP, Ad-SNHG5 + NC antagomir, Ad-SNHG5 + miR-154-5p antagomir and Ad-GFP + NC antagomir were injected into the L5 dorsal root ganglion (DRG) of mice via canula seven days before surgery for three days (8 mice were used in each group; adenovirus titer was l × 1011 pfu/ml, 20 μl/day; the dosage of antagomir was 1 nmol/day) [10, 17, 18]. HEK293 cells were transfected with miR-154-5p mimics, miR-154-5p inhibitor, non-specific (NC) mimics or NC inhibitor using lipofectamine 2000 (Invitrogen, USA) according to the manufactures’ instruction.
In Situ Hybridization
SNHG5-specific and negative control locked nucleic acid-modified RNA probes, conjugated with digoxigenin at 3′ and 5′ terminals, were designed and purchased from Roche (Germany). Mouse L5 DRG was fixed with 4% paraformaldehyde and cryoprotected overnight. Then tissue sections were digested with proteinase K for 5 min and hybridized with modified RNA probes overnight at 50 ℃. Then tissue sections were sealed with blocking solution (Roche, Germany) for one hour at room temperature and incubated with alkaline phosphatase conjugated anti-digoxigenin antibody overnight at 4℃. Finally, tissue sections were stained with BM purple alkaline phosphatase substrate solution (Roche, Germany) for a week at room temperature and observed under a microscope [19].
Behavioral Tests
Von Frey filaments were used to measure the mouse paw withdrawal threshold (PWT) in response to mechanical stimuli (Muromachi Kikai, Japan) as previously described [18, 19]. And the paw withdrawal latency (PWL) was evaluated using a set of Plantar Test (Germany) according to the manufactures’ recommendation. The experimenter was blinded to mouse treatments. After behavioral tests, the mouse L5 DRG was removed for subsequent detections.
Quantitative Real-Time PCR
Mouse L5 DRG or HEK-293 cells were lysed using Trizol reagent (Thermofisher, USA) to extract total RNA. Total RNA was reverse-transcribed into cDNA using SuperScript IV reverse transcriptase (Thermofisher, USA). SYBR Green (Solarbio, China) was used to detect the mRNA level of SNHG4, which was normalized to GAPDH. The miR-154-5p level was detected using TaqMan MicroRNA assay kit (Thermofisher, USA), and small nuclear RNA U6 was used as an internal control [11]. The real-time PCR primers used in this study was listed in Table 1.
Western Blot
Mouse L5 DRG was lysed using RIPA lysis buffer (Beyotime, China). Total proteins were quantified and subjected to SDS-10% polyacrylamide gel electrophoresis. Then proteins were transferred onto a PVDF membrane (Millipore, USA) and blocked with 5% skim milk, followed by blotting with CXCL13 antibody (1:2000, Affinity Biosciences, USA) or GAPDH antibody (1:5000, Affinity Biosciences, USA) at 4℃ overnight. After rinsing with PBS buffer, the membrane was incubated with HRP-conjugated goat secondary antibody (1:5000, Affinity Biosciences, USA) for 60 min at 37℃. Then proteins were visualized using SuperSignal substrate (Thermofisher, USA) and protein bands were analyzed using Gel-Pro-Analyzer.
Immunofluorescence Assay
The expression levels of glial fibrillary acidic protein (GFAP) and ionized calcium binding adapter molecule 1 (IBA-1) were detected using immunofluorescence assay. Mouse L5 DRG was paraffin embedded and sliced into 5 μm sections, followed by dehydration at 60 °C incubator for 2 h. Then tissue sections were deparaffinized and boiled in antigen retrieval solution for 10 min. After blocking with goat serum (Solarbio, China) for 15 min at room temperature, tissue sections were washed and incubated with GFAP antibody (1:200, Abcam, UK) and Iba-1 antibody (1:200, Abcam, UK) at 4 °C overnight. Tissue sections were washed with PBS for three times and incubated with FITC-labeled goat anti-mouse IgG (1:200, Beyotime, China) for 90 min at room temperature Tissue sections were sealed with anti-fluorescence quenching reagent (Solarbio, China) and typical pictures were captured under a fluorescence microscope (OLYMPUS BX53, Japan, ×100 magnification).
Dual-Luciferase Reporter Assay
Dual-luciferase assay was performed to validate the interaction between SNHG5 and miR-154-5p. In brief, the mutant type of SNHG5 3′-untranslated region (UTR) was obtained by PCR using primers containing mutant seed sequence. Wildtype and mutant type of SNHG5 3′-UTR were cloned into pmirGLO (Promega, USA) plasmid, abbreviated as SNHG5-WT and SNHG5-MUT. 293T cells (Procell, China) were co-transfected with pmirGLO plasmids and NC/miR-154-5p agomir/inhibitor. Similar method was used to validate the interaction between miR-154-5p and CXCL13: the wildtype and mutant type of CXCL13 3′-UTR, termed as CXCL13-WT and CXCL13-MUT, were co-transfected with NC/miR-154-5p agomir into HEK-293 T cells. The correlation was assessed through calculating the ratio of firefly luciferase to renilla luciferase activities.
Statistical Analysis
Data were presented as means ± SD and were analyzed using GraphPad Prism 7. The effects of miR-154-5p mimics and inhibitor on miR-154-5p expression were analyzed using Students’ t-test. The expression levels of SNHG5, miR-154-5p, CXCL13, GFAP and IBA-1 were analyzed using one-way ANOVA combined with Bonferroni’s multiple comparisons test, and the rest of data were compared using two-way ANOVA combined with Bonferroni’s multiple comparisons test. All experiments were repeated at least three times and p values less than 0.05 were regarded as statistically significant.
Results
SNHG5 was Up-Regulated in a Mouse SNL Model
Mouse SNL model was established as previously described. Mouse PWT and PWL in response to mechanical stimuli were significantly reduced in SNL model from day 3 after surgery and remained lower (Fig. 1a, b). The SNHG5 level in mouse spinal cord was remarkably up-regulated since the third day after SNL model establishment, and SNHG5 level was over four times higher in SNL group than that in sham group 14 days after surgery (Fig. 1c). In situ hybridization assay displayed similar results that SNHG5 expression was dramatically elevated in L5 DRG in SNL group compared with that in sham group (Fig. 1d). These results showed that SNHG5 level was up-regulated in a mouse SNL model.
SNHG5 was up-regulated in a mouse SNL model. a, b Mouse PWT and PWL were evaluated using a Von Frey filaments a set of Plantar Test according to the manufactures’ instruction. c Relative SNHG5 level in mouse L5 DRG at day 3, day 7 and day 14 was measured using quantitative real-time PCR. d The SNHG5 expression was further assessed using in situ hybridization (Bar = 50 μm). SNL group: surgery was performed to expose the L5 spinal nerve, followed by ligation with 4-0 silk thread. Sham group, identical surgery was performed except for that the L5 spinal nerve was not ligated. PWT paw withdrawal threshold, PWL paw withdrawal latency, *p < 0.05, **p < 0.01
SNHG5 Knockdown Alleviated Neuropathic Pain in a Mouse SNL Model
Two adenoviruses carrying SNHG5 shRNA were injected into mouse L5 DRG, and SNHG5 level was significantly knocked down in both sham group and SNL group (Fig. 2a). Ad-SNHG5 2# resulted in higher efficiency in SNHG5 knockdown and was chosen for subsequent experiments. SNHG5 knockdown conspicuously increased PWT and PWL in SNL group at day 14 after animal model establishment, while no obvious effect of SNHG5 knockdown was observed on sham group (Fig. 2b, c). These results suggested that SNHG5 knockdown alleviated neuropathic pain in a mouse SNL model.
SNHG5 knockdown alleviated neuropathic pain in a mouse SNL model. Adenoviruses were injected into the L5 DRG of mice for three days (n = 6; adenovirus titer was l × 1011 pfu/ml, 20 μl/day). a The SNHG5 level in mouse L5 DRG was measured using quantitative real-time PCR after adenoviruses injection. b, c Mouse PWT and PWL were assessed using a commercial Von Frey filaments a set of Plantar Test. PWT paw withdrawal threshold, PWL paw withdrawal latency, **p < 0.01
SNHG5 Knockdown Inhibited Astrocyte and Microglia Activation in Spinal Cord in a Mouse SNL Model
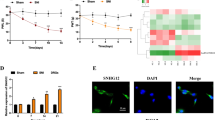
We further examined astrocyte and microglia activation in L5 DRG in SNL mouse after SNHG5 knockdown by immunofluorescence assay. SNHG5 knockdown significantly decreased the protein levels of GFAP and IBA-1 in SNL group. However, no obvious effect of Ad-SNHG5 infection on GFAP and IBA-1 expression was observed in sham group (Fig. 3). This finding indicated that astrocyte and microglia activation in SNL mice was inhibited after SNHG5 knockdown.
SNHG5 is an Upstream Regulator of miR-154-5p
Bioinformatics prediction (https://starbase.sysu.edu.cn/index.php) showed that SNHG5 is an upstream regulator of miR-154-5p. The binding site of SNHG5 on miR-154-5p was shown in Fig. 4a. MiR-154-5p mimics we used significantly elevated the miR-154-5p level, while miR-154-5p inhibitor transfection remarkably reduced the miR-154-5p level in HEK-293 cell (Fig. 4b). Then we validated the interaction between SNHG5 and miR-154-5p using luciferase reporter assay. MiR-154-5p mimics dramatically inhibited the luciferase activity in SNHG5-WT group yet had no obvious effect on SNHG5-MUT group. On the contrary, miR-154-5p inhibitor transfection remarkably increased the luciferase activity in SNHG5-WT group but had no significant impact on SNHG5-MUT group (Fig. 4c). We further examined miR-154-5p level in mouse L5 DRG. The level of miR-154-5p was significantly decreased since day 3 and reduced by more than a half on day 14 after surgery in SNL group compared with sham group (Fig. 4d). Besides, Ad-SNHG5 infection elevated the miR-154-5p level in mouse L5 DRG in SNL mice compared with Ad-GFP control (Fig. 4e). These findings suggested that SNHG5 is an upstream regulator of miR-154-5p.
SNHG5 is an upstream regulator of miR-154-5p. a The biding site of SNHG5 on miR-154-5p was shown. b Relative miR-154-5p level in HEK-293 cell after miR-154-5p mimics or miR-154-5p inhibitor transfection was measured using quantitative real-time PCR. c The interaction between miR-154-5p and SNHG5 was validated using luciferase reporter assay. d, e Relative miR-154-5p level in mouse L5 DRG after animal model established and adenoviruses infection was measured using quantitative real-time PCR, **p < 0.01
CXCL13 is a Downstream Target of miR-154-5p
Bioinformatics prediction on Targetscan database (https://www.targetscan.org/vert_72/) showed that CXCL13 is a putative downstream target of miR-154-5p. The binding site of miR-154-5p on CXCL13 was shown in Fig. 5a. Luciferase reporter assay showed that miR-154-5p mimics significantly suppressed the luciferase activity in CXCL13-WT group compared with miR-NC, yet had no significant effect on that in CXCL13-MUT group (Fig. 5b). Then we examined the protein level of CXCL13 in SNL mouse by western blot. The CXCL13 level was significantly elevated since day 3 and reached more than eight times higher at day 14 than that in sham group (Fig. 5c). MiR-154-5p agomir injection remarkably increased the miR-154-5p level (Fig. 5d), yet conspicuously reduced the CXCL13 protein level in SNL mice (Fig. 5e). These results revealed that CXCL13 is a downstream target of miR-154-5p.
CXCL13 is a downstream target of miR-154-5p. a The binding site of miR-154-5p on CXCL13 was shown. b The interaction between miR-154-5p and CXCL13 was verified using luciferase activity. c The CXCL13 protein level was detected at day 3, day 7 and day 14 after SNL model establishment. d Relative miR-154-5p level in mouse L5 DRG after miR-154-5p agomir or NC agomir injection was measured using quantitative real-time PCR. e The CXCL13 protein level in mouse L5 DRG after miR-154-5p agomir or NC agomir injection was detected using western blot, **p < 0.01
SNHG5 Knockdown Alleviated Neuropathic Pain by Inhibiting Astrocyte and Microglia Activation Through miR-154-5p
We tested the efficiency of miR-154-5p antagomir using quantitative real-time PCR and found that miR-154-5p antagomir showed significant inhibitory effect on miR-154-5p expression (Fig. 6a). Ad-SNHG5 infection significantly inhibited CXCL13 expression while miR-154-5p antagomir abolished the SNHG5 knockdown induced CXCL13 down-regulation in SNL mice (Fig. 6b). Ad-SNHG5 infection increased the PWT and PWL in SNL mice at day 14 after surgery, whereas simultaneous miR-154-5p inhibition partially abrogated such impact (Fig. 6c). Besides, SNHG5 knockdown decreased the protein levels of GFAP and IBA-1 in SNL mice, while miR-154-5p inhibition partially reversed such effect (Fig. 6d). Together, these findings suggest that SNHG5 knockdown ameliorated neuropathic pain by suppressing astrocyte and microglia activation through miR-154-5p.
SNHG5 knockdown alleviated neuropathic pain and inhibited astrocyte activation through miR-154-5p. a The inhibitory effect of miR-154-5p antagomir was assessed using quantitative real-time PCR. b The protein level of CXCL13 in mouse L5 DRG after Ad-SNHG5 or miR-154-5p antagomir injection was examined by western blot assay. c Mouse PWT and PWL were assessed using a commercial Von Frey filaments a set of Plantar Test. d The protein levels of GFAP and IBA-1 after Ad-SNHG5 or miR-154-5p antagomir injection were assessed using immunofluorescence assay. PWT paw withdrawal threshold, PWL paw withdrawal latency, **p < 0.01
Discussion
The dysregulated expression of lncRNAs in injured nerve, DRG and spinal cord has generated increasing interests in neuropathic pain research. To date, several lncRNAs including Kcna2 AS RNA, MRAK009713, NONRATT021972, BC168687 and uc.48+ have been reported to participate in the regulation of neuropathic pain [9, 10, 20, 21]. Although exploring the roles of lncRNAs in the pathogenesis of this unneglectable disorder may offer novel therapeutic options, our investigation is still at its preliminary stage. More lncRNAs remain to be discovered and more intensive researches are required for their clinical application [22]. Jiang et al. investigated the role of SNHG5 in rats with spinal cord injury (SCI) and found that SNHG5 was up-regulated in the spinal cord of SCI rats, and SNHG5 over-expression aggravated SCI and facilitated the activation of astrocytes and microglia by targeting Krüppel-like factor 4 (KLF4) [23]. Astrocyte and microglia activation is positively related to neuropathic pain [24]. These previous studies inspired us to investigate the role of SNHG5 in neuropathic pain. In the present study, we first examined the expression level of SNHG5 in the mouse L5 DRG. PWT and PWL detections revealed that we successfully established a mouse neuropathic pain model, and a dramatic up-regulation of SNHG5 was observed. Then we delivered SNHG5 short hairpin RNAs via adenovirus vehicle into mouse L5 DRG and observed a conspicuously increased PWT and PWL, indicating that SNHG5 knockdown could efficiently alleviate neuropathic pain. Besides, SNHG5 knockdown also reduced levels of GFAP and IBA-1, typical markers for astrocyte and microglia activation, in SNL mice but not in sham mice. These results indicated that SNHG5 knockdown could efficiently ameliorate neuropathic pain and had no effect on normal mice. Therefore, SNHG5 knockdown is a potential therapeutic strategy for neuropathic pain treatment. However, more preclinical investigations are required to validate this strategy.
The “competing endogenous RNAs (ceRNA)” hypothesis that noncoding RNAs communicate with each other through microRNA response elements, is validated to be a crucial regulatory network in numerous physiological processes [25]. MiR-154-5p modulates the progression of a spectrum of human cancers by targeting specific target genes [12, 26,27,28]. Wei et al. first demonstrated that lncRNA X inactive-specific transcript (XIST) aggravated neuropathic pain by sponging miR-154-5p to increase TLR5 expression, implying its critical role in neuropathic pain progression [16]. In this study, we found that miR-154-5p was down-regulated in L5 DRG in mouse SNL model. We further validated that SNHG5 directly regulates the expression of miR-154-5p in mouse SNL model, which was highly in accordance with previous study in breast cancer [12]. Besides, SNHG5 knockdown mediated neuropathic pain alleviation was abolished by miR-154-5p inhibition, accompanied with promoted astrocyte and microglia activation. These findings suggested that SNHG5 knockdown alleviates neuropathic pain by regulating miR-154-5p. In addition, CXCL13 is up-regulated in SNL animal model and induces neuropathic pain by activating spinal astrocyte [29]. In the present study, CXCL13 was validated to be a downstream target of miR-154-5p. The expression of CXCL13 was also positively regulated by SNHG5 according to our results, which suggest that SNHG5 modulates neuropathic pain by regulating the miR-154-5p/CXCL13 axis. We may investigate the exact interaction between SNHG5 and CXCL13 in the progression of neuropathic pain in our future work.
In conclusion, our results demonstrated that lncRNA SNHG5 knockdown alleviated neuropathic pain and inhibited the astrocyte and microglia activation through regulating the miR-154-5p/CXCL13 axis. Our findings might offer a potential therapeutic target for the clinical treatment of neuropathic pain.
Ethics Approval
All animal experiments were carried out according to the guidelines of the International Association for the Study of Pain and approved by the Laboratory Animal Ethics Committee of Guizhou Medical University (Approval No. 1900724).
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Treede RD, Jensen TS, Campbell JN, Cruccu G, Dostrovsky JO, Griffin JW, Hansson P, Hughes R, Nurmikko T, Serra J (2008) Neuropathic pain: redefinition and a grading system for clinical and research purposes. Neurology 70(18):1630–1635. https://doi.org/10.1212/01.wnl.0000282763.29778.59
McDermott AM, Toelle TR, Rowbotham DJ, Schaefer CP, Dukes EM (2006) The burden of neuropathic pain: results from a cross-sectional survey. Eur J Pain 10(2):127–135. https://doi.org/10.1016/j.ejpain.2005.01.014
van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N (2014) Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain 155(4):654–662. https://doi.org/10.1016/j.pain.2013.11.013
Nightingale S (2012) The neuropathic pain market. Nat Rev Drug Discov 11(2):101–102. https://doi.org/10.1038/nrd3624
O'Connor AB, Dworkin RH (2009) Treatment of neuropathic pain: an overview of recent guidelines. Am J Med 122(10 Suppl):S22–32. https://doi.org/10.1016/j.amjmed.2009.04.007
Lin CC, Tsai MC, Lee CT, Sun MH, Huang TL (2018) Antidepressant treatment increased serum miR-183 and miR-212 levels in patients with major depressive disorder. Psychiatry Res 270:232–237. https://doi.org/10.1016/j.psychres.2018.09.025
Chen X, Yan GY (2013) Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 29(20):2617–2624. https://doi.org/10.1093/bioinformatics/btt426
Wang S, Xu H, Zou L, Xie J, Wu H, Wu B, Yi Z, Lv Q, Zhang X, Ying M et al (2016) LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal 12(1):139–148. https://doi.org/10.1007/s1130-2015-9488-x
Peng H, Zou L, Xie J, Wu H, Wu B, Zhu G, Lv Q, Zhang X, Liu S, Li G et al (2017) lncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Mol Neurobiol 54(1):511–523. https://doi.org/10.1007/s12035-015-9632-1
Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V et al (2013) A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci 16(8):1024–1031. https://doi.org/10.1038/nn.3438
Dou L, Lin H, Wang K, Zhu G, Zou X, Chang E, Zhu Y (2017) Long non-coding RNA CCAT1 modulates neuropathic pain progression through sponging miR-155. Oncotarget 8(52):89949–89957. https://doi.org/10.18632/oncotarget.21192
Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y, Cao XC (2019) SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids 17:138–149. https://doi.org/10.1016/j.omtn.2019.05.013
Li M, Zhang YY, Shang J, Xu YD (2019) LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci 23(10):4185–4191. https://doi.org/10.26355/eurrev_201905_17921
Li X, Liu L, Luo Y, Cui S, Chen W, Zeng A, Shi Y, Luo L (2019) Long non-coding RNA SNHG5 promotes glioma progression via miR-205/E2F3 axis. Biosci Rep. https://doi.org/10.1042/BSR20190668
Zhang M, Li Y, Wang H, Yu W, Lin S, Guo J (2019) LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther 20(4):524–536
Wei M, Li L, Zhang Y, Zhang ZJ, Liu HL, Bao HG (2018) LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. J Cell Biochem. https://doi.org/10.1002/jcb.27088
Jiang H, Guo S, Xiao D, Bian X, Wang J, Wang Y, Zhou H, Cai J, Zheng Z (2017) Arginine deiminase expressed in vivo, driven by human telomerase reverse transcriptase promoter, displays high hepatoma targeting and oncolytic efficiency. Oncotarget 8(23):37694–37704. https://doi.org/10.18632/oncotarget.17032
Sakai A, Suzuki H (2013) Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem Biophys Res Commun 435(2):176–181. https://doi.org/10.1016/j.bbrc.2013.04.089
Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H (2013) miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain 136(Pt 9):2738–2750. https://doi.org/10.1093/brain/awt191
Li G, Jiang H, Zheng C, Zhu G, Xu Y, Sheng X, Wu B, Guo J, Zhu S, Zhan Y et al (2017) Long noncoding RNA MRAK009713 is a novel regulator of neuropathic pain in rats. Pain 158(10):2042–2052. https://doi.org/10.1097/j.pain.0000000000001013
Liu CL, Deng ZY, Du ER, Xu CS (2018) Long noncoding RNA BC168687 small interfering RNA reduces high glucose and high free fatty acidinduced expression of P2X7 receptors in satellite glial cells. Mol Med Rep 17(4):5851–5859. https://doi.org/10.3892/mmr.2018.8601
Wu S, Bono J, Tao YX (2019) Long noncoding RNA (lncRNA): a target in neuropathic pain. Expert Opin Ther Targets 23(1):15–20
Jiang ZS, Zhang JR (2018) LncRNA SNHG5 enhances astrocytes and microglia viability via upregulating KLF4 in spinal cord injury. Int J Biol Macromol 120(Pt A):66–72. https://doi.org/10.1016/j.ijbiomac.2018.08.002
Colburn RW, Rickman AJ, DeLeo JA (1999) The effect of site and type of nerve injury on spinal glial activation and neuropathic pain behavior. Exp Neurol 157(2):289–304. https://doi.org/10.1006/exnr.1999.7065
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358. https://doi.org/10.1016/j.cell.2011.07.014
Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, Xu X, Xu T, Hu X, Sun L et al (2018) The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer 17(1):018–0894
Wang X, Sun S, Tong X, Ma Q, Di H, Fu T, Sun Z, Cai Y, Fan W, Wu Q et al (2017) MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res 1676:69–76. https://doi.org/10.1016/j.brainres.2017.08.014
Huang J, Wu J, Li Y, Li X, Yang T, Yang Q, Jiang Y (2014) Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res Int 364316(10):20
Jiang BC, Cao DL, Zhang X, Zhang ZJ, He LN, Li CH, Zhang WW, Wu XB, Berta T, Ji RR et al (2016) CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J Clin Invest 126(2):745–761. https://doi.org/10.1172/JCI81950
Funding
This work was supported by the Innovation and Entrepreneurship Training Program for College Students in Guizhou Province (Grant No. 20195200924).
Author information
Authors and Affiliations
Contributions
XHZ and XGZ conceived and designed the experiments, MC and YY analyzed and interpreted the results of the experiments, WQZ, XNL and JLW performed the experiments.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, M., Yang, Y., Zhang, W. et al. Long Noncoding RNA SNHG5 Knockdown Alleviates Neuropathic Pain by Targeting the miR-154-5p/CXCL13 Axis. Neurochem Res 45, 1566–1575 (2020). https://doi.org/10.1007/s11064-020-03021-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-020-03021-2