Abstract
Reactive nitrogen species, such as nitric oxide (NO), exert their biological activity in large part through post-translational modification of cysteine residues, forming S-nitrosothiols. This chemical reaction proceeds via a process that we and our colleagues have termed protein S-nitrosylation. Under conditions of normal NO production, S-nitrosylation regulates the activity of many normal proteins. However, in degenerative conditions characterized by nitrosative stress, increased levels of NO lead to aberrant S-nitrosylation that contributes to the pathology of the disease. Thus, S-nitrosylation has been implicated in a wide range of cellular mechanisms, including mitochondrial function, proteostasis, transcriptional regulation, synaptic activity, and cell survival. In recent years, the research area of protein S-nitrosylation has become prominent due to improvements in the detection systems as well as the demonstration that protein S-nitrosylation plays a critical role in the pathogenesis of neurodegenerative and other neurological disorders. To further promote our understanding of how protein S-nitrosylation affects cellular systems, guidelines for the design and conduct of research on S-nitrosylated (or SNO-)proteins would be highly desirable, especially for those newly entering the field. In this review article, we provide a strategic overview of designing experimental approaches to study protein S-nitrosylation. We specifically focus on methods that can provide critical data to demonstrate that an S-nitrosylated protein plays a (patho-)physiologically-relevant role in a biological process. Hence, the implementation of the approaches described herein will contribute to further advancement of the study of S-nitrosylated proteins, not only in neuroscience but also in other research fields.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background on Protein S-Nitrosylation
In mammalian cells, three nitric oxide synthases (NOSs), namely NOS1 (or neuronal NOS), NOS2 (or inducible NOS) and NOS3 (endothelial NOS), are responsible for NO production from arginine [1]. In large part, the biological effects of NO are mediated through a redox reaction, S-nitrosation, leading to the post-translational modification of S-nitrosylation, which affects protein function in many ways analogous to phosphorylation. In this reaction, an NO moiety interacts with the thiol (–SH) group, or perhaps more properly thiolate anion (–S−), of a cysteine residue [2, 3]. NO at either physiological or pathophysiological levels reacts with specific thiol groups. Several chemical mechanisms by which only a subset of cysteine residues form S-nitrosothiols (or SNO-proteins) in vivo have been proposed [2–4]. Among these, the presence of the ‘SNO motif’ near the target cysteine residues is perhaps the most accepted concept [2, 5–7]. The SNO motif typically contains acidic/basic amino acids flanking the critical cysteine residue that facilitate deprotonation of the thiol group to produce thiolate anion (RS−), thus promoting the reaction of a nitrosonium cation (NO+) intermediate with the target thiol (Fig. 1). Consequently, S-nitrosylation influences protein activity, protein–protein interactions, and cellular localization of the target protein. For instance, basal levels of NO mediate a number of normal, physiological processes, such as synaptic plasticity and neuronal survival, via S-nitrosylation-dependent regulation of NMDA-type glutamate receptors, HDAC2, and caspases [8–11]. In contrast, risk factors for neurodegenerative diseases, including aging, exposure to environmental toxins, and neuroinflammation, can lead to elevated and prolonged generation of NO in the brain; this will trigger aberrant S-nitrosylation of neuronal proteins, which normally are not S-nitrosylated by low/basal levels of NO. These aberrantly nitrosylated proteins include GAPDH, protein disulfide isomerase (PDI), myocyte enhancer factor 2 (MEF2), dynamin related protein 1 (Drp1), and X-linked inhibitor of apoptosis (XIAP). Such aberrant nitrosylation can contribute to mitochondrial dysfunction, protein misfolding, synaptic loss, and increased neuronal cell death [12–16].
Possible mechanisms by which cysteine thiol residues form S-nitrosothiols in vivo. Pathway 1: As an active intermediate, nitrosonium cation [NO+], with brackets indicated the reaction intermediate rather than free NO+, may be generated from a metal ion-dependent oxidation of NO. [NO+] reacts with a thiolate anion (R–S−) to form an S-nitrosothiol (R–SNO). Pathway 2: Radical recombination of ·NO with thiyl radical (RS·) yields an S-nitrosothiol. Pathway 3: Transnitrosylation from an S-nitrosothiol (R2–SNO), such as another S-nitrosylated protein, GSNO or SNOC. Here, the NO group is transferred from one thiol to another
The demonstration by our group and others of the (patho-)physiological relevance of protein S-nitrosylation has attracted an increasing number of scientists to this research field. However, the complex chemistry of S-nitrosothiol formation, the fact that air (in which most experiments are performed) is such as oxidizing environment compared to the brain and other tissues in vivo, and the very demanding procedures that are required to detect protein S-nitrosylation in biological samples often leads to experimental artifacts. As a result, many publications have reported non-physiological or artifactually S-nitrosylated proteins that are detected only in the presence of very high concentrations of exogenous NO. Moreover, many studies suffer from a lack of evidence for the causative role of SNO-proteins in a particular cell phenomenon; that is, they only show a correlation between SNO-protein formation and the cellular events of interest. Hence, there is clearly a need for consensus guidelines that can help design experimental procedures for studying protein S-nitrosylation. Here we provide scientific and technical instructions to help lay out an experimental flow diagram in order to facilitate proper identification and characterization of novel SNO-proteins that form under either physiological or pathological conditions in vivo (Fig. 2).
Proposed experimental scheme to study protein S-nitrosylation. Initially, presence of a S-nitrosylated protein (SNO-protein) of interest can be detected by the biotin-switch assay (best if performed in human tissue for relevance), mass spectrometry (MS), or other methods. In order to observe SNO-protein formation under physiological conditions, a dose-response with NO donors (e.g., SNOC or GSNO) or, even better, endogenous NO should be employed. For the mapping of S-nitrosylation site(s), site-directed mutagenesis is necessary of the putative critical cysteine residue, followed by MS, NMR, and crystal structure analysis. Lastly, in order to demonstrate that a specific SNO-protein significantly contributes to a biological process, functional assays should be explored preferably with a non-nitrosylatable cysteine mutant. Determining the endogenous enzymatic system (S-nitrosylase and denitrosylase) for a given SNO-protein is also a critical experimental avenue
Detection of SNO-Proteins
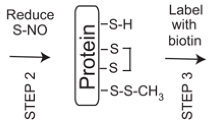
Many experimental methods have been developed for the detection of SNO-proteins. Among these techniques, the biotin-switch assay originally devised by Sami Jaffrey and Solomon Snyder is perhaps the most commonly used assay to verify formation of a SNO-protein in cells or tissues [17] (Fig. 3). This assay relies on the ability of ascorbate to selectively reduce S-nitrosothiol to a free sulfhydryl group [8], which is subsequently conjugated with a biotin adduct. The biotinylated proteins can then be enriched with avidin biochemistry and analyzed by conventional western blotting with antibodies against the target proteins or by mass spectrometry (MS) or other methods after trypsin digestion. As a proof-of-concept experiment, one can employ the biotin-switch assay and assess the presence of SNO-proteins in cells (or tissue lysates) exposed to an NO donor. However, caution should be exercised when using NO donating molecules, because the majority of NO donors commercially available are non-physiological donors with extremely long half-lives. Here we propose the use of naturally-occurring NO donors, such as S-nitrosoglutathione (GSNO) and S-nitrosocysteine (SNOC) [18], to induce S-nitrosylation of cellular proteins. For adoption of these NO donors into experimental systems, we refer readers to published articles [19–21]. Another important consideration is the concentration of NO donor used in the study. Physiological concentrations of NO are believed to be less than 1 μM [22, 23]. In order to avoid non-physiological, artifactual generation of SNO-proteins, cells should exposed to <200 μM exogenous SNOC or GSNO and a careful dose-response curve for producing S-nitrosylated protein should be performed. Importantly, the biotin-switch assay should be employed to verify that ‘endogenously produced NO’ can give rise to the formation of SNO-proteins in cellular or animal models of the disease of interest (if possible, in patient-derived tissue samples), strengthening the notion that the SNO-proteins play a patho-physiological role. When detecting endogenous SNO-proteins, proper controls should be included in the biotin-switch assay to validate the quality of the result, as this assay can generate a SNO-independent biotinylation (i.e., false positive) result [24]. For example, omission of ascorbate or ‘pre-photolysis of the S–NO bond’ should attenuate the biotinylation of S-nitrosylated cysteine residues, validating that the target protein was indeed S-nitrosylated. For troubleshooting the biotin-switch assay, readers are referred to the excellent articles by Stamler and colleagues [24, 25]. Other important assays for SNO-proteins include, for example, the 2,3-diaminonapththalene (DAN) assay on purified or recombinant protein, and chemiluminescent assays [2, 12, 19].
Steps in the biotin-switch assay for the detection of S-nitrosylated proteins. Step 1: After preparation of cell or tissue lysates, free thiols are blocked by incubation with methymethanethiosulfonate (MMTS) under denaturing conditions. The unreacted MMTS is then removed by acetone precipitation. Step 2: S-nitrosothiols are selectively reduced by ascorbate, and then the reformed free thiols are labeled with N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP). Step 3: The biotinylated proteins are collected by a biotin-avidin affinity purification procedure, e.g., on beads. Step 4: Proteins eluted from the beads are separated by SDS-PAGE and subjected to immunoblot analysis. Alternatively, ‘proteins on beads’ or ‘proteins eluted from beads’ can be digested by trypsin for MS analysis
Note that commercially available antibodies against S-nitrosylated cysteine often suffer from non-specific binding. Thus, if one decides to use SNO-antibodies, experiments should include carefully-designed negative controls to verify that the candidate protein is really S-nitrosylated. Moreover, the recruitment of a confirmatory experiment with another assay, such as the biotin-switch method, is strongly encouraged. In our hands and those of many of our colleagues, commercially available SNO-antibodies are in general not reliable and may yield artifactual results.
Mapping SNO Site(s) on Proteins
As alluded to earlier, endogenously produced NO can modify specific cysteine residues. In order to determine the target cysteine residue(s), researchers often adopt a site-directed mutagenesis technique, specifically substituting cysteine resides with another conservatively-changed amino acid, such as alanine or serine. When a critical cysteine residue is mutated, this should significantly diminish the SNO signal on the biotin-switch assay, thus indicating the SNO site. However, some caveats are known to be associated with this approach. Mutation of a cysteine residue may perturb the overall structure of the protein, potentially altering the sensitivity of other cysteine residues to S-nitrosylation. In addition, because the biotin-switch assay is not a quantitative method, if a protein contains two or more S-nitrosylation sites, the removal of a single SNO site may not significantly decrease the SNO signal. To overcome these potential problems, mass spectrometry (MS), NMR structure, and crystal structure analysis of wild-type SNO-protein (non-mutated with non-nitrosylatable mutant as a control) may be alternative approaches, albeit these assays often require the presence of special instruments and core facilities. Typically, purified proteins are used for these alternative analyses, as it is extremely challenging to observe endogenous S-nitrosylation of a protein in a ‘top-down’ fashion. Additionally, concerning MS analysis, a more mild, electron transfer dissociation technique (ETD), in lieu of the more harsh, conventional collision-induced dissociation technique (CID), should be explored because of the lability of SNO modifications [14]. NO is often a good ‘leaving group’ and thus it may also be replaced by reactive oxygen species (ROS), leading to sulfenated (RSOH), sulfinated (ROS2H), or sulfonated (RSO3H) adducts; these may be physiologically or pathologically relevant substitutions depending on the conditions under which they were observed [16, 26].
Effects of S-Nitrosylation on Protein Function
After determining the SNO-site, the next step is to study the effect of S-nitrosylation on protein activity. If the target protein possesses known catalytic properties, these experiments might entail assays of enzymatic activity in vitro and in situ. Alternatively, proteins functioning as ion channels or carriers, transcription factors, chaperones, and binding partners can all be affected by S-nitrosylation. S-Nitrosylation can inhibit or increase protein activity, depending on the target. For example, NO may modify a catalytically-active cysteine to form an S-nitrosothiol, abrogating its enzymatic activity [9, 11, 16, 26]. Alternatively, S-nitrosylation can positively or negatively regulate protein activity in an allosteric manner [12, 15]. In addition, one should evaluate if the SNO-protein influences downstream biological signaling. One approach involves expressing a non-nitrosylatable cysteine mutant in specific cells in vitro or in the whole animal. In particular, in cases of allosteric regulation of protein function by S-nitrosylation, many studies have shown that mutation of the target cysteine residue typically spares basal activity. The prediction would be that cells expressing non-nitrosylatable mutant would remain unresponsive to NO compared to wild-type transfected cells, strongly suggesting a causal link between SNO-protein formation and the biological processes in question (see References [12, 15] as examples). Recent advancements in gene editing technologies, including ZFNs (zinc-finger nucleases), TALENs, and CRISPR/Cas9 [27], provide tools to introduce site-specific genome modifications with high precision. Future studies should investigate the role of endogenous nitrosylation using these techniques to introduce the non-nitrosylatable mutant.
In contrast to allosteric modulation, when analyzing a SNO-protein whose S-nitrosylation occurs on an active-site cysteine, mutation may not provide a mechanistic link between SNO-protein formation and downstream effect because mutation of this cysteine itself leads to the generation of an activity-dead enzyme in the absence of NO. As an alternative, researchers are encouraged to employ additional approaches. For instance, RNAi-mediated knockdown of the target protein may mimic the effects of S-nitrosylation. Reduced expression of target protein by genetic alteration may render cells or animals insensitive to a particular effect of NO due to lack of the target cysteine.
Conclusions
The goal of this ‘COMMENTS’ article is to provide an overview of experimental procedures to investigate protein S-nitrosylation. Strategic considerations for experimental design are presented in an attempt to standardize the procedures for investigating SNO-proteins and avoiding artifacts. In the coming years, a number of improved or novel methods are expected to emerge, allowing further refinement of the approach to redox-mediated chemical biology in general and S-nitrosylation in particular.
Abbreviations
- GSNO:
-
S-Nitrosoglutathione
- MS:
-
Mass spectrometry
- NO:
-
Nitric oxide
- NOS:
-
NO synthase
- SNO:
-
S-Nitrosylation or S-nitrosothiol
- SNOC:
-
S-Nitrosocysteine
References
Martinez-Ruiz A, Cadenas S, Lamas S (2011) Nitric oxide signaling: classical, less classical and nonclassical mechanisms. Free Radic Biol Med 51:17–29
Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6:150–166
Nakamura T, Tu S, Akhtar MW, Sunico CR, Okamoto S, Lipton SA (2013) Aberrant protein S-nitrosylation in neurodegenerative diseases. Neuron 78:596–614
Smith BC, Marletta MA (2012) Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol 16:498–506
Seth D, Stamler JS (2011) The SNO-proteome: causation and classifications. Curr Opin Chem Biol 15:129–136
Stamler JS, Lamas S, Fang FC (2001) Nitrosylation. The prototypic redox-based signaling mechanism. Cell 106:675–683
Stamler JS, Toone EJ, Lipton SA, Sucher NJ (1997) (S)NO signals: translocation, regulation, and a consensus motif. Neuron 18:691–696
Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS (1993) A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature 364:626–632
Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS (1999) Fas-induced caspase denitrosylation. Science 284:651–654
Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A (2008) S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature 455:411–415
Tenneti L, D’Emilia DM, Lipton SA (1997) Suppression of neuronal apoptosis by S-nitrosylation of caspases. Neurosci Lett 236:139–142
Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA (2009) S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science 324:102–105
Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7:665–674
Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, Clemente AT, Okamoto S, Salvesen GS, Riek R, Yates JR III, Lipton SA (2010) Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell 39:184–195
Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR 3rd, Nakanishi N, Andreyev AY, Okamoto S, Jaenisch R, Ambasudhan R, Lipton SA (2013) Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell 155:1351–1364
Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA (2006) S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441:513–517
Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3:193–197
Seneviratne U, Godoy LC, Wishnok JS, Wogan GN, Tannenbaum SR (2013) Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs. J Am Chem Soc 135:7693–7704
Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins ME, Sugarbaker DJ, Chee C, Singel DJ et al (1993) Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc Natl Acad Sci USA 90:10957–10961
Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, Lipton SA (1992) Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron 8:1087–1099
Zhang Y, Hogg N (2005) S-Nitrosothiols: cellular formation and transport. Free Radic Biol Med 38:831–838
Hirota Y, Ishida H, Genka C, Obama R, Matsuyama S, Nakazawa H (2001) Physiological concentration of nitric oxide induces positive inotropic effects through cGMP pathway in isolated rat ventricular myocytes. Jpn J Physiol 51:455–461
Pinsky DJ, Patton S, Mesaros S, Brovkovych V, Kubaszewski E, Grunfeld S, Malinski T (1997) Mechanical transduction of nitric oxide synthesis in the beating heart. Circ Res 81:372–379
Forrester MT, Foster MW, Benhar M, Stamler JS (2009) Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med 46:119–126
Forrester MT, Foster MW, Stamler JS (2007) Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem 282:13977–13983
Shi ZQ, Sunico CR, McKercher SR, Cui J, Feng GS, Nakamura T, Lipton SA (2013) S-Nitrosylated SHP-2 contributes to NMDA receptor-mediated excitotoxicity in acute ischemic stroke. Proc Natl Acad Sci USA 110:3137–3142
Gaj T, Gersbach CA, Barbas CF 3rd (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Acknowledgments
This work was supported in part by NIH Grants P30 NS076411, R01 NS086890, R01 ES017462, P01 HD029587, and R21 NS080799 (SAL), the Brain and Behavior Research Foundation (SAL), and the Michael J. Fox Foundation (SAL and TN).
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: In honor of Dr. Philip Beart.
Rights and permissions
About this article
Cite this article
Nakamura, T., Lipton, S.A. Nitrosative Stress in the Nervous System: Guidelines for Designing Experimental Strategies to Study Protein S-Nitrosylation. Neurochem Res 41, 510–514 (2016). https://doi.org/10.1007/s11064-015-1640-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-015-1640-z