Abstract
Purpose
The prognosis of patients with leptomeningeal metastasis (LM) remains poor. Circulating tumour DNA (ctDNA) has been proven to be abundantly present in cerebrospinal fluid (CSF); hence, its clinical implication as a biomarker needs to be further verified.
Methods
We conducted a retrospective study of 35 lung adenocarcinoma (LUAD) patients with LM, and matched CSF and plasma samples were collected from all patients. All paired samples underwent next-generation sequencing (NGS) of 139 lung cancer-associated genes. The clinical characteristics and genetic profiling of LM were analysed in association with survival prognosis.
Results
LM showed genetic heterogeneity, in which CSF had a higher detection rate of ctDNA (P = 0.003), a higher median mutation count (P < 0.0001), a higher frequency of driver mutations (P < 0.01), and more copy number variation (CNV) alterations (P < 0.001) than plasma. The mutation frequencies of the EGFR, TP53, CDKN2A, MYC and CDKN2B genes were easier to detect in CSF than in LUAD tissue (P < 0.05), possibly reflecting the underlying mechanism of LM metastasis. CSF ctDNA is helpful for analysing the mechanism of EGFR-TKI resistance. In cohort 1, which comprised patients who received 1/2 EGFR-TKIs before the diagnosis of LM, TP53 and CDKN2A were the most common EGFR-independent resistant mutations. In cohort 2, comprising those who progressed after osimertinib and developed LM, 7 patients (43.75%) had EGFR CNV detected in CSF but not plasma. Furthermore, patient characteristics and various genes were included for interactive survival analysis. Patients with EGFR-mutated LUAD (P = 0.042) had a higher median OS, and CSF ctDNA mutation with TERT (P = 0.013) indicated a lower median OS. Last, we reported an LM case in which CSF ctDNA dynamic changes were well correlated with clinical treatment.
Conclusions
CSF ctDNA could provide a more comprehensive genetic landscape of LM, indicating the potential metastasis-related and EGFR-TKI resistance mechanisms of LM patients. In addition, genotyping of CSF combined with clinical outcomes can predict the prognosis of LUAD patients with LM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leptomeningeal metastasis (LM) is a severe complication associated with metastatic solid tumours that is caused by neoplastic cells transferring to the pia mater, arachnoid, and subarachnoid space [1]. The incidence of LM is highest in melanoma, followed by advanced lung cancer, accounting for 3–5% [2]. Based on 2019 global cancer data, non-melanoma skin cancer, tracheal, bronchus, and lung (TBL) cancer, and colon and rectal cancer have the highest morbidity rates. Meanwhile, TBL cancer showed the highest mortality rate, with an age-standardized death rate of 25.18 [3].The incidence of LM in non-small cell lung cancer (NSCLC) patients is 3.4%, and that in EGFR-mutant patients is 9.4% [4]. The prognosis of patients with LM remains poor, with a median survival of less than 1 year despite treatments such as chemotherapy and whole-brain radiation therapy [5]. Since the first experiments targeting epidermal growth factor receptor (EGFR), targeted therapy has begun a new chapter based on an actionable molecular alteration in lung cancers [6]. Unfortunately, the vast majority of advanced NSCLC patients become resistant to current targeted treatments and eventually progress [7].
It is essential to characterize genomic diversity between primary and metastatic cancers. Nevertheless, tissue biopsies are invasive procedures, and some locations are not easy to access, such as the bones and brain [8]. However, tissue biopsy of LM is impractical due to diffuse distribution. On various occasions, circulating tumour DNA (ctDNA) is easier to collect serially than tissue biopsy. For patients who are harbouring difficult-to-biopsy neoplasms, such as pia mater or pia mater, ctDNA can provide critical molecular profiling and precision medicine treatment response in real time [9,10,11]. Alix-Panabières et al. elucidated that the detection and monitoring of ctDNA can provide effective information concerned with treatment decisions in cases of targeted therapy or immune checkpoint inhibition (ICI). ctDNA can be used to monitor the effect of targeted therapy and is a more suitable biomarker for liquid biopsy monitoring [8].
Due to the existence of the blood‒brain barrier (BBB), genotyping of the primary tumour and plasma does not represent the genetic alterations of cerebrospinal fluid (CSF) in LM. Currently, CSF is considered a crucial liquid biopsy medium for LM that provides an accessible and less invasive method to acquire information on genomic alterations [12]. CSF ctDNA exhibits unique genetic profiles and distinct resistance mechanisms during treatment in NSCLC patients with brain metastases (BMs), so it is crucial to test for acquired resistance at CNS progression [13, 14]. ctDNA is more abundantly present in the CSF than in plasma and has been shown to be a biomarker of treatment effect and tumour evolution in patients with LM [15,16,17]. To fully explore the effectiveness of CSF ctDNA representing genetic information in LM versus that of plasma ctDNA in our study, we compared the matched CSF and plasma from the same LM patients. However, few studies have analysed the correlation between ctDNA of CSF and/or plasma and survival outcome in patients with LM. In this study, we investigated the outcome of lung adenocarcinoma (LUAD) patients with LM in the real world and analysed the factors associated with their survival.
Materials and methods
Study design and patients
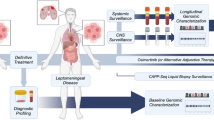
In this retrospective cohort study, we collected data from LUAD patients with LM who were enrolled in the Department of Neurology, The Second Hospital of Hebei Medical University (Hebei, China), from December 2015 to December 2022. The inclusion criteria were as follows: (1) > 18 and ≤ 75 years old; (2) diagnosis of LM according to the European Association of Neuro-Oncology-European Society for Medical Oncology (EANO-ESMO) clinical practice guidelines [18]; and (3) all LM patients underwent lumbar puncture. The exclusion criteria were as follows: (1) excessive missing clinical data; and (2) incomplete follow-up information. ctDNA was extracted from CSF and plasma samples and detected by a 139-gene next-generation sequencing (NGS) panel at the time of diagnosis of LM(Table S1). Finally, a total of 35 patients with LM were included for further analysis. Demographic, clinicopathological and therapies data were obtained for each patient. The study was approved by the Research Ethics Committee of The Second Hospital of Hebei Medical University. Included patients from our institute provided signed informed consent. This study was conducted in accordance with the principles of the Declaration of Helsinki.
To study clinical outcomes, overall survival (OS) was measured. OS was defined as the time from LM to death or last follow-up.
Preparation of CSF ctDNA, and plasma ctDNA
Freshly frozen CSF, and whole blood samples were collected for genomic profiling. Total DNA from freshly frozen CSF and plasma was extracted by QIAamp Circulating Nucleic Acid Kit (Qiagen, Germantown, MD, USA).
NGS library preparation and sequencing data analysis
The Illumina libraries were carried out with KAPA Hyper Prep Kit (Kapa Biosystems, Woburn, MA, USA) according to the manufacturer’s protocol. Targeted panel sequencing was performed by SeqCap EZ System (Roche Nimblegen, Madison, WI, USA), and obtained by the Geneseeq Prime™ panel (Nanjing Geneseeq Technology Inc., Nanjing, JiangSu, China) which was covering 139 cancer-associated genes. On HiSeq X10 sequencing system (Illumina, San Diego, CA, USA), enriched libraries were sequenced with 150 bp pair-end reads.
Statistical analysis
Survival analysis was performed using Kaplan‒Meier method with log-rank P values and 95% confidence intervals (CIs) reported. All P values were 2-sided and were considered statistically significant at P < 0.05. Multivariate analysis was performed using Cox proportional hazard regression with inclusion of variables significant on univariate regression.
Associations between two variables were analyzed using Fisher’s exact, Chi-square tests or Wilcoxon test. Graph Pad Prism version 9.0, SPSS version 25.0 and R version 4.1.2 software used for statistical analyses and making graphs.
Results
Patient characteristics
We retrospectively profiled 35 LUAD patients with LM for analysis. All patients had matched CSF and plasma samples. The demographic and clinical characteristics of the included patients are shown in Tables 1 and S2. The majority of the patients denied a smoking history (77.14%), 18 patients (51.43%) were males, and the median age was 54, ranging from 30 to 75 years. EGFR mutations were found in 26 patients (74.29%) at the initial diagnosis of LUAD. Among them, 13 patients (37.14%) were diagnosed with LM and BMs simultaneously, and 23 patients (65.71%) had extracranial metastases. Most of the patients suffered from neurologic symptoms prior to LM diagnosis. The most common clinical manifestations were cerebral symptoms (97.14%), such as headache and dizziness. Posterior fossa symptoms, including various cranial neuropathies (51.43%) and spinal cord/root symptoms, such as motor and sensory dysfunction (13%), were also in evidence. Thirty-three patients (94.29%) received targeted therapy and other therapy combinations (angiogenesis inhibitors/ICIs/chemotherapy/radiotherapy). All patients underwent intrathecal methotrexate via lumbar puncture or Ommaya reservoir. Thirty-two patients (91.43%) displayed malignant cells in CSF. The diagnosis of LM was established by CSF cytology alone in 10 patients (28.57%) and by both MRI and CSF cytology in 21 patients (60%). LM developed in 33 patients (94.29%) during the course of lung cancer, and LUAD was detected in 2 patients (5.71%) at the time of LM diagnosis. CSF intracranial pressure was decreased (≤ 200 mmH2O) in 57.14% of patients with LM. CSF abnormal biochemical measures also occurred in some LM patients.
Genetic divergence of CSF and plasma samples
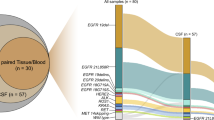
To delineate the genetic landscape, the somatic mutations from the matched CSF and plasma ctDNA of 35 patients were analysed by applying an NGS panel (Fig. 1). No somatic mutation was detected in the CSF and plasma of one patient. A total of 262 somatic variants of 103 mutated genes were detected in 35 CSF samples. Altogether, 85 somatic variants of 41 mutated genes were identified in 35 plasma samples, presenting a lower count than in CSF. For the detection of genomic alterations, only 37 (34.6%) were present in both plasma and CSF. However, 66 (61.7%) could be detected in CSF but could not be found in plasma, while 4 (3.7%) could be found in plasma but could not be detected in CSF (Fig. 2A). ctDNA was positive in 34 patients (97.14%) in CSF and in 25 patients (71.43%) in plasma, and the detection rate of ctDNA was significantly different in CSF and plasma (P = 0.003, Fig. 2B). The median mutation count of CSF was 5, which was significantly greater than that of plasma ctDNA (5 vs. 1, P < 0.0001, Fig. 2C). EGFR (29/35, 82.86%), TP53 (26/35, 74.29%) and CDKN2A (10/35, 28.57%) mutations frequently occurred in CSF, and coexisting TP53 and EGFR were found in 22 (22/35, 62.9%) CSF samples. In addition, the most frequently altered genes in plasma were EGFR (17/35, 48.57%) and TP53 (12/35, 34.29%). Based on statistical analysis, the mutation frequencies of EGFR, TP53 and CDKN2A were significantly higher in CSF than in plasma (P < 0.01, Fig. 2D). In addition, EGFR was the most frequent copy number variation (CNV) alterations in CSF, followed by CDKN2A (6/35, 17.1%) and MYC (6/35, 17.1%) mutations. However, only one CNV alteration was detected in the plasma sample. The percentage of CSF variants with CNV alterations was higher than that of plasma (83/262, 31.68% vs. 1/85, 1.18%, P < 0.001). Therefore, these results suggested that CSF had higher CNV alterations, and the importance of CNV in these alterations needs to be explored.
Genetic landscape of 35 LM patients. All patients matched CSF and plasma ctDNA. One patient was not shown, because no somatic mutation was detected in the CSF and plasma. These top bar display the number of genetic alterations of each patient, these right side-bar indicate the mutated genes, and these left side-bar present the percentage of genetic alterations. LM: leptomeningeal metastasis, ctDNA: circulating tumor DNA, CSF: cerebrospinal fluid
CSF and plasma ctDNA molecular characterization. A: venn diagram of mutated genes between CSF and plasma ctDNA analysis. B: the detection rate of ctDNA is statistically different between CSF and plasma. C: comparison of mutations count per sample between CSF and plasma. D: mutation frequency analysis of genes between CSF and plasma. E: frequency of CSF-private or plasma-private mutations versus shared mutations in paired CSF-plasma samples. CSF: cerebrospinal fluid; ctDNA: circulating tumor DNA. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
To further evaluate the genetic relevance and divergence between CSF and plasma, we analysed the shared and independently evolved mutations in each individual. In the 35 paired CSF-plasma pairs, a median 14.29% shared mutation rate was seen, which was relatively low. There were a few shared mutations found in 65.71% (23/35) of paired samples (Fig. 2E). Overall, LM showed genetic heterogeneity with slight differences in mutational prevalence between CSF and plasma. Plasma ctDNA was not reflective of LM status. All these data demonstrated a result of the low sensitivity of mutation detection via plasma biopsy. CSF might be a better ctDNA method to detect mutations of the tumour than liquid biopsy.
The potential metastatic mechanism of LM
To assess the potential metastatic mechanism of LM, we compared CSF/plasma samples with LUAD tissue, which was from a China Pancancer study and is a cancer genomic study of a large-scale Asian population [19]. The majority of patients in this study were from China, including 1572 LUAD cases, mostly early cancer specimens (Table S3). There was no significant difference between age and sex compared with our patients. Of note, the mutation frequencies of the EGFR, TP53, CDKN2A, MYC and CDKN2B genes in CSF were significantly higher than those in LUAD tissue, and these genes were easier to detect in CSF (P < 0.05, Fig. 3A). However, the mutation frequencies of EGFR, TP53 and KRAS in LUAD tissue were higher than those in plasma, but there were no significant differences. There was a significant increase in MDM2 mutations in plasma compared with LUAD tissues (P < 0.05, Fig. 3B). Subsequent analysis of CNV showed higher frequencies of EGFR, CDKN2A, MYC, CDKN2B, TP53, RICTOR, NTRK1, RB1 and MET in CSF than in LUAD tissue (P < 0.05, Fig. 3C). The high occurrence rate of CNV in CSF ctDNA suggested universal genome instability in LM. CSF-specific mutations revealed differences between LM and LUAD tissues. EGFR, TP53, CDKN2A, MYC and CDKN2B mutations might be potential mechanisms of metastasis.
Comparison of mutation frequency of mutated genes between diffrent groups. A: Diferences in mutation frequency between CSF and LUAD tissue. B: Diferences in mutation frequency between plasma and LUAD tissue. C: Diferences in mutation frequency of CNV alterations between CSF and LUAD tissue. CSF: cerebrospinal fluid; LUAD: lung adenocarcinoma; CNV: copy number variation. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
Mechanisms of resistance to EGFR-TKIs
We categorized patients with EGFR mutations that were detected in CSF, plasma or LUAD tissue into 2 cohorts based on osimertinib administration. In cohort 1 (baseline), a total of 16 patients did not receive osimertinib before the diagnosis of LM with baseline CSF and plasma genotyping. In cohort 2 (resistance), a total of 16 patients progressed after osimertinib and developed LM with CSF and plasma genotyping (Table S4). In cohort 1, icotinib was the most common EGFR tyrosine kinase inhibitor (EGFR-TKI). In cohort 2, gefitinib was the most common EGFR-TKI before the application of osimertinib. In cohort 2, 1 patient received osimertinib as first-line therapy, 13 patients as second-line therapy, and 2 patients as third-line treatment. In cohort 1, 87.5% (14/16) of the patients had EGFR exon 19 deletion or 21 L858R mutation in CSF, whereas only 50% (8/16) of the patients had these mutations in the matching plasma. In cohort 2, 81.25% (13/16) of the patients had EGFR exon 19 deletion or 21 L858R mutation in CSF, whereas only 50% (8/16) of the patients had these mutations in the matching plasma.
The genes varied among the different cohorts (Fig. S1). EGFR T790M is the most common mechanism of resistance following treatment with 1st- or 2nd-generation EGFR-TKIs. However, in cohort 1 (Fig. S1A), an EGFR-dependent resistance mechanism of the T790M mutation was found in 2 patients receiving gefitinib or icotinib in plasma but not CSF. TP53 and CDKN2A were the most common EGFR-independent resistant mutations. In addition, alterations in the CDK4, CDKN2A, CDKN2B, and MYC genes were identified in CSF. In cohort 2 (Fig. S1B), a total of 7 patients (43.75%) had EGFR CNV detected in CSF but not plasma after osimertinib administration. Interestingly, alterations of EGFR exon 20 T790M, exon 18 G719A and exon 21 insertion were found to occur simultaneously in one patient. However, we found EGFR-independent resistance mechanisms of CDKN2A, CDK4, MET, KRAS, and PIK3CA mutations and NTRK1 CNV in 5 patients. We did not perform similar histologic and phenotypic transformation assessments on the tissue samples to determine whether there was SCLC transformation due to condition limitations.
Prognostic factors of LM
To further explore the survival prognostic factors of different patients, patient characteristics and various genes were included for interactive survival analysis. The median OS was 14.4 months, and the median time interval between diagnosis of primary tumours and LM was 21.5 months. Patients with BMs, EGFR-mutated LUAD and decreased intracranial pressure were associated with longer median OS (BMs, 35.7 months vs. 8.4 months, P = 0.044; EGFR-mutated LUAD, 23.5 months vs. 4.5 months, P = 0.003; decreased intracranial pressure, 25.4 months vs. 6.3 months, P = 0.031, Fig. 4A–C). Regardless of whether the enhanced brain MRI was positive, patients with negative CSF cytology still survived (median OS not reached, median follow-up 18.2 months) and survived significantly longer than those with positive CSF cytology (median OS 11.1 months, P = 0.035, Fig. S2A). This suggested that floating tumour cells in CSF may lead to a poorer survival prognosis. Based on the genotyping of CSF, the Kaplan‒Meier method revealed that telomerase reverse transcriptase (TERT), NFE2L2, PKHD1 and POLD1 mutations were associated with shortened median OS (TERT, 1.8 months vs. 16.6 months, P = 0.001; NFE2L2, 2.6 months vs. 14.4 months, P = 0.003; PKHD1, 4.5 months vs. 16.6 months, P = 0.044; POLD1, 3.6 months vs. 14.4 months, P = 0.032, Fig. 4D, S2B-D).
Genetic alterations and clinical features associated with OS. Kaplan‒Meier analysis of OS in patients, A: With and without BMs. B: EGFR-mutated LUAD and non-EGFR-mutated LUAD. C: Decreased intracranial pressure and increased intracranial pressure. D: With and without TERT mutation in CSF. E: Multivariate Cox proportional hazards regression analysis, EGFR-mutated LUAD and CSF TERT mutant remained independent survival factors. OS: overall survival; EGFR: epidermal growth factor receptor; BMs: brain metastases; CSF: cerebrospinal fluid; TERT: telomerase reverse transcriptase
The co-occurrence of gene mutations may influence prognosis. Therefore, we found that patients with concurrent EGFR and TP53 mutations or concurrent EGFR and TP53 wild-type had a longer median OS than those with TP53 mutations (EGFR and TP53 mutation vs. TP53 mutation, 16.6 months vs. 3.6 months, P = 0.003; EGFR and TP53 wild-type vs. TP53 mutation, NA vs. 3.6 months, P = 0.049, Fig. S2E). The genetic profile of plasma was also analysed, and TOP2A and KMT2C mutations indicated lower median OS (TOP2A, 3.6 months vs. 14.4 months, P = 0.034; KMT2C, 3.6 months vs. 14.4 months, P = 0.034, Fig. S2G/H). Then, we compared the OS of different EGFR mutation states in CSF and plasma, respectively, but no significant difference was found (Fig. S2F/I).
To assess the potential biomedical importance of genetic mutations, we examined their correlations with clinical variables. In further multivariate Cox proportional hazards regression analysis, patients with EGFR-mutated LUAD (P = 0.042, 95% CI 0.09–0.96 months, Fig. 4E) had a higher median OS, and CSF TERT mutation (P = 0.013, 95% CI 1.49–29.35 months) indicated a lower median OS. This result suggested that EGFR-mutated LUAD and CSF TERT mutation remained independent survival factors after adjusting for other patient characteristics.
CSF ctDNA guided the treatment of LM
Finally, in a case presentation, dynamic changes in CSF ctDNA during the treatment of LM were revealed (Fig. S3A, B). In a case presentation, the female patient had a headache for 3 months and underwent a LUAD resection 3 years ago. The patient was diagnosed with LM and BMs because of meningeal enhancement and insular lobe metastatic tumours on brain MRI and malignant cells detected by CSF cytology (CSF1). The patient received gefitinib and antiangiogenic therapy for LM based on EGFR p.L858R detected in CSF.
Nine months later, her Karnofsky performance scale (KPS) and Eastern Cooperative Oncology Group performance score (ECOG PS) progressed dramatically, and CSF ctDNA showed a higher frequency of EGFR p.L858R. Considering the progression, the targeted therapy was changed to osimertinib 80 mg once a day. Twenty-two months later, she returned with worsening headaches, dizziness, and back pain, which was associated with worse KPS and ECOG PS. CSF cytology revealed a large number of tumour cells (CSF3). The genetic profiles of CSF ctDNA demonstrated EGFR-independent resistance mutations (Fig. S3B). Unfortunately, the mutation was never detected in plasma. Moreover, a double dose of osimertinib (160 mg) was administered due to changes in the driver genes of CSF ctDNA. It was suggested that CSF ctDNA plays a potential role in the diagnosis and monitoring of LM.
Discussion
Liquid biopsy is an alternative method to detect mutations in patients if tumour samples are not available. For LM patients, CSF and plasma can be easily obtained. In this research, we characterized the genomic alteration of ctDNA in the CSF and compared it to matched plasma samples. In our study, we revealed that CSF genetic profiles had a unique role in identifying patients with LUAD with LM and that plasma genetic profiles failed to do so. At the time of initial diagnosis of LM, more abundant genotypes were detected in CSF than in plasma in addition to driver mutations. Moreover, we demonstrated that CSF ctDNA has a significantly higher detection rate than plasma. This result is consistent with a previous study that showed that CSF ctDNA exhibits a more comprehensive genetic landscape of CNS metastases than plasma [14, 16, 17, 20]. In previous studies, scholars have compared the detection rate of EGFR and found that it is higher in CSF than in plasma. LM is more common in patients with EGFR mutations than in those with wild-type EGFR [4]. Moreover, in EGFR-mutated patients, the detection rate of EGFR was 67.6–100% in CSF and 36.4–73.1% in plasma [16]. In our research, EGFR was the most frequently mutated gene in CSF, accounting for 82.86%, which was higher than that in matched plasma. This is also consistent with Li’s findings [21], suggesting that patients with EGFR mutations may be more susceptible to BMs. In short, the available data revealed heterogeneous genetic profiles between CSF and plasma, with good concordance in driver mutations.
In TRACERx research, subclonal whole genome doubling (WGD) was detected in 29% of lung tumours. These data demonstrate that WGD and copy number heterogeneity were associated with shorter disease-free survival and distant metastases, respectively. WGD and copy number instability are important factors of relapse in NSCLC patients, which guide the evolution of clinical cancer [22]. Numerous CNVs were identified in CSF ctDNA, and they were significantly more than those in plasma in our study. Several studies have indicated that CNVs are enriched in the CSF of CNS metastases [14, 23]. Therefore, we can speculate that CNVs in CSF may cause distant metastasis of tumours and is a major type of mutation that causes LM, exhibiting a difference relative to plasma.
Studies have demonstrated the genetic heterogeneity between the original tumour and the metastatic lesions in the same patient [24]. Genetically distinct subclones of the primary tumour result from somatic evolution of the tumour genome and thus have distinct biologic properties and therapeutic individualization. When tumour cells metastasize, they escape from the primary site, spread and proliferate in secondary locations, and can also evade immune surveillance, eventually forming metastatic lesions. This may lead to the introduction of considerable genomic heterogeneity between the final metastatic cell and the primary cancer [25]. It has been unclear to what extent the genotyping of LMs differs from the genotyping of primary cancers. Previous research demonstrated that overexpression of MYC, MMP13 or YAP, which are enriched for focal amplification in BMs, can each contribute to brain metastasis formation. TP53, CDKN2A, and TERT are abundant in a variety of metastatic cancers. While TP53 mutations are strongly associated with genetic instability, CDK2NA and TERT play a key role in regulating cell proliferation, and both pathways are frequently perturbed in metastatic tumours. In conclusion, these mutated genes may disturb pancancer hallmarks of tumorigenesis, hence improving the aggressiveness of the tumour [26, 27]. To further investigate the importance and evolutionary process of LM genetic alterations, we compared CSF/plasma samples with primary tumour samples. The EGFR, TP53, MYC, CDKN2A and CDKN2B genes in CSF were significantly higher than those in LUAD tissue. Therefore, genomic characterization of the CSF of patients with LM represents a feasible strategy to find a potential method for the detection of metastasis.
This conclusion was reached in the study of Nanjo et al. that the T790M mutation was less frequent in leptomeningeal than in extracranial specimens by biopsy of patients with lung cancer tissue and leptomeningeal metastases [28]. This study confirmed this idea. After the patients became resistant to 1st- or 2nd-generation EGFR-TKIs, the T790M mutation was identified in plasma but not in CSF, which may be due to the differential expression of the T790M mutation in CNS and extra-CNS lesions.
In the study by Zheng et al., EGFR C797S mutation, MET dysregulation, and TP53 plus RB1 co-occurrence were possible resistance mechanisms of LM in the progression of osimertinib of CSF in NSCLC [29]. Unfortunately, the C797S mutation was not found in our study of cohort 2 because the lack of sufficient samples hindered further discussion. The mechanisms of resistance to osimertinib progression in LM patients may be found in the CSF, such as EGFR CNV. Furthermore, another study also revealed that EGFR amplification is the resistance mechanism associated with EGFR-TKIs [30]. From the AURA3 trial, EGFR mutation was one of the most common acquired resistance mechanisms detected, followed by MET amplification [31]. In cohort 2, we found that EGFR CNV occurred in 7 patients and MET mutations in 3 patients after the diagnosis of LM, which partly accounted for the progressive disease of LM. In our study, we found cell cycle pathway alterations (CDKN2A, CDKN2B, CDK4, and CDK6) after osimertinib administration. In previous studies, altered cell cycle genes were found to be possibly involved in the mechanism of resistance to osimertinib as the 1st- or 2nd-line therapy [32]. PIK3CA amplification or mutations promote tumour infiltration and activate the PI3K/AKT/mTOR pathway, which suggests that PI3K/AKT/mTOR pathway activation might be related to resistance to 3rd-generation EGFR-TKIs [32, 33]. Similar to this report, in our 2 cohorts, the PIK3CA mutation was present in the osimertinib-resistant cohort but not in the osimertinib-naive cohort.
Tissue biopsy of the primary tumour to determine the presence of SCLC transformation is also one of the resistance mechanisms to osimertinib [34]. However, the lack of matched primary cancer tissue genetic profiles limits further clarification of the results. The discovery of these important mechanisms of acquired resistance to EGFR-TKIs could facilitate precise treatments for such patients after disease progression.
In our study, a multivariate analysis indicated that the presence of EGFR-mutated LUAD was an independent favourable predictor of survival, whereas TERT mutation in CSF was an independent predictor of poor survival after excluding other confounding factors. Patients with advanced LUAD who harboured EGFR mutations had significantly longer OS than those without EGFR mutations after treatment with EGFR-TKIs [35]. Suda et al. reviewed that the better prognosis of patients with EGFR mutations may be related to the use of EGFR-TKIs [36]. This has been confirmed by other studies showing that EGFR-TKIs after LM diagnosis were independent favourable predictors of survival [37]. There is no doubt that all 26 patients with EGFR-mutated LUAD were treated with EGFR-TKIs before the diagnosis of LM. The TERT gene represents a ribonucleoprotease that is essential for the replication of chromosome termini and telomere elongation in eukaryotes. The study suggests that targeting TERT promoter (pTERT) mutations may serve as a viable approach for cancer therapy [38, 39]. Previous studies have suggested that TERT mutations are associated with a poor prognosis in tumours, such as thyroid malignancies, melanoma and gliomas [40,41,42,43]. Yang et al. found that TERT mutations were detected in 11% of patients with NSCLC, and TERT mutation carrier status was an independent risk factor for poor prognosis [44]. Likewise, TERT mutations were found in the CSF of 11.43% of patients with LM with LUAD, and these patients had a worse prognosis in our study. This had not been reported to be associated with survival in LM. Thus, we believe that TERT mutation may have clinical value as a potential biomarker for disease monitoring [45, 46].
In the previous literature, TP53 mutation and EGFR/TP53 comutation have been considered poor prognostic factors in LUAD patients [47]. In our research, patients with TP53 mutations in CSF showed shorter OS than those in the other groups (P < 0.05). Dual EGFR/TP53 mutation was associated with inferior OS compared with dual EGFR/TP53 wild-type, although these results were not statistically significant. In this study, CDKN2A was common in CSF samples, accounting for 28.6%, regardless of prognosis. This is consistent with the findings of Yang et al. [48].
In the CSF circulation, disseminated cancer cells can float freely within the CSF or attach to the meninges and can be captured by CSF cytological examination or appear as linear or nodular enhancement on MRI [49]. Remsik et al. found that floating cells were more invasive in vivo than adherent or mixed cells in a mouse model, which was further manifested by the rapid development of neurological symptoms and reduced survival. Remarkably, they found that patients diagnosed with positive CSF cytology only demonstrated substantially diminished survival after LM diagnosis through clinical case collection [50]. Additionally, in the current study, patients with negative CSF cytology still survived and survived significantly longer than those with positive CSF cytology, regardless of whether the enhanced brain MRI was positive.
Finally, we demonstrated with a specific case that dynamic changes in CSF ctDNA at different stages could better predict intracranial tumour responses and track clonal evolution in LM patients.
There are several limitations in our study. First, this was a retrospective study with a small sample size. Second, matched primary lung cancer tissues of the patients were unavailable, and the NGS data were obtained from CSF- or plasma-derived ctDNA without the analysis of matched tumour tissue DNA. Third, there is a lack of observation of CSF tumour markers, and we will continue this study in future observations. However, this is still a rare study that involved exploration of exactly matched CSF and plasma genetic information and analysis of survival prognosis in LUAD patients with LM.
Conclusions
In conclusion, our findings demonstrate that CSF is a more sensitive and reliable liquid biopsy medium than plasma for LM. CSF ctDNA could provide a more comprehensive genetic landscape of LM, which reveals the potential metastasis-related mechanisms of malignant tumours and the resistance mechanisms to EGFR-TKIs and guides clinical strategies. In addition, EGFR-mutated LUAD was associated with better OS, and CSF TERT mutation was associated with poorer OS. These indicators may have clinical value as potential novel biomarkers for disease monitoring.
Data availability
The human sequence data from patients are not publicly available due to restrictions on participant privacy. The data that supports the findings of this study are available on reasonable request from the corresponding authors. The China Pan-cancer study (https://www.cbioportal.org/study/summary?id=pan_origimed_2020) was downloaded from cBioPortal.
Abbreviations
- BBB:
-
Blood–brain barrier
- BMs:
-
Brain metastases
- CNV:
-
Copy number variation
- CNS:
-
Central nervous system
- CSF:
-
Cerebrospinal fluid
- ctDNA:
-
Circulating tumour DNA
- EGFR:
-
Epidermal growth factor receptor
- EGFR-TKI:
-
EGFR-tyrosine kinase inhibitor
- LUAD:
-
Lung adenocarcinoma
- LM:
-
Leptomeningeal metastases
- MRI:
-
Magnetic resonance imaging
- NGS:
-
Next-generation sequencing
- NSCLC:
-
Non-small cell lung cancer
- OS:
-
Overall survival
References
Graber JJ, Kesari S (2018) Leptomeningeal metastases. Curr Treat Options Oncol 19:3. https://doi.org/10.1007/s11864-018-0518-0
Remon J, Le Rhun E, Besse B (2017) Leptomeningeal carcinomatosis in non-small cell lung cancer patients: a continuing challenge in the personalized treatment era. Cancer Treat Rev 53:128–137. https://doi.org/10.1016/j.ctrv.2016.12.006
Lin L, Li Z, Yan L, Liu Y, Yang H, Li H (2021) Global, regional, and national cancer incidence and death for 29 cancer groups in 2019 and trends analysis of the global cancer burden 1990–2019. J Hematol Oncol. https://doi.org/10.1186/s13045-021-01213-z
Li YS, Jiang BY, Yang JJ, Tu HY, Zhou Q, Guo WB, Yan HH, Wu YL (2016) Leptomeningeal Metastases in Patients with NSCLC with EGFR Mutations. J Thoracic Oncol : Off Pub Inter Assoc Study Lung Cancer 11:1962–1969. https://doi.org/10.1016/j.jtho.2016.06.029
Cheng H, Perez-Soler R (2018) Leptomeningeal metastases in non-small-cell lung cancer. Lancet Oncol 19:e43–e55. https://doi.org/10.1016/S1470-2045(17)30689-7
Herbst RS, Maddox AM, Rothenberg ML, Small EJ, Rubin EH, Baselga J, Rojo F, Hong WK, Swaisland H, Averbuch SD, Ochs J, LoRusso PM (2002) Selective oral epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 is generally well-tolerated and has activity in non-small-cell lung cancer and other solid tumors: results of a phase I trial. J Clin Oncol : Off J Am Societ Clin Oncol 20:3815–3825. https://doi.org/10.1200/jco.2002.03.038
Wang M, Herbst RS, Boshoff C (2021) Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med 27:1345–1356. https://doi.org/10.1038/s41591-021-01450-2
Alix-Panabieres C, Pantel K (2021) Liquid biopsy: from discovery to clinical application. Cancer Discov 11:858–873. https://doi.org/10.1158/2159-8290.CD-20-1311
Nikanjam M, Kato S, Kurzrock R (2022) Liquid biopsy: current technology and clinical applications. J Hematol Oncol 15:131. https://doi.org/10.1186/s13045-022-01351-y
Zhao Y, He J, Zou Y, Guo X, Cui J, Guo L, Bu H (2019) Evaluating the cerebrospinal fluid ctDNA detection by next-generation sequencing in the diagnosis of meningeal Carcinomatosis. BMC Neurol 19:331. https://doi.org/10.1186/s12883-019-1554-5
Zhao Y, He J, Cui J, Meng Z, Zou Y, Guo X, Chen X, Wang X, Yan L, Han W, Li C, Guo L, Bu H (2020) Detection of genes mutations in cerebrospinal fluid circulating tumor DNA from neoplastic meningitis patients using next generation sequencing. BMC Cancer 20:690. https://doi.org/10.1186/s12885-020-07172-x
Offin M, Chabon JJ, Razavi P, Isbell JM, Rudin CM, Diehn M, Li BT (2017) Capturing genomic evolution of lung cancers through liquid biopsy for circulating tumor DNA. Journal of oncology 2017:4517834. https://doi.org/10.1155/2017/4517834
Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S, Omuro A, Lin X, Fleisher M, Grommes C, Panageas KS, Meng F, Selcuklu SD, Ogilvie S, Distefano N, Shagabayeva L, Rosenblum M, DeAngelis LM, Viale A, Mellinghoff IK, Berger MF (2016) Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncology : Off J Am Societ Clin Oncol 34:2404–2415. https://doi.org/10.1200/jco.2016.66.6487
Li M, Chen J, Zhang B, Yu J, Wang N, Li D, Shao Y, Zhu D, Liang C, Ma Y, Ou Q, Hou X, Chen L (2022) Dynamic monitoring of cerebrospinal fluid circulating tumor DNA to identify unique genetic profiles of brain metastatic tumors and better predict intracranial tumor responses in non-small cell lung cancer patients with brain metastases: a prospective cohort study (GASTO 1028). BMC Med 20:398. https://doi.org/10.1186/s12916-022-02595-8
De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, Oliveira M, Arias A, Raventos C, Tang J, Guerini-Rocco E, Martínez-Sáez E, Lois S, Marín O, de la Cruz X, Piscuoglio S, Towers R, Vivancos A, Peg V, Ramon Y, Cajal S, Carles J, Rodon J, González-Cao M, Tabernero J, Felip E, Sahuquillo J, Berger MF, Cortes J, ReisFilho JS, Seoane J (2015) Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nature Commun 6:8839. https://doi.org/10.1038/ncomms9839
Gao T, Chen F, Li M (2023) Sequencing of cerebrospinal fluid in non-small-cell lung cancer patients with leptomeningeal metastasis: a systematic review. Cancer Med 12:2248–2261. https://doi.org/10.1002/cam4.5163
Nie N, Zhou H, Zhang K, Liu L, Luo N, Wang R, Li X, Zhu M, Hu C, Wang Y, Liu Z, Li L, He Y (2022) Genotyping of cerebrospinal fluid in lung cancer patients with leptomeningeal metastasis. Thorac Cancer 13:2574–2583. https://doi.org/10.1111/1759-7714.14592
Le Rhun E, Weller M, Brandsma D, Van den Bent M, de Azambuja E, Henriksson R, Boulanger T, Peters S, Watts C, Wick W, Wesseling P, Rudà R, Preusser M (2017) EANO–ESMO Clinical practice guidelines for diagnosis, treatment and follow-up of patients with leptomeningeal metastasis from solid tumours. Annals Oncology. https://doi.org/10.1093/annonc/mdx221
Wu L, Yao H, Chen H, Wang A, Guo K, Gou W, Yu Y, Li X, Yao M, Yuan S, Pang F, Hu J, Chen L, Liu W, Yao J, Zhang S, Dong X, Wang W, Hu J, Ling Q, Ding S, Wei Y, Li Q, Cao W, Wang S, Di Y, Feng F, Zhao G, Zhang J, Huang L, Xu J, Yan W, Tong Z, Jiang D, Ji T, Li Q, Xu L, He H, Shang L, Liu J, Wang K, Wu D, Shen J, Liu Y, Zhang T, Liang C, Wang Y, Shang Y, Guo J, Liang G, Xu S, Liu J, Wang K, Wang M (2022) Landscape of somatic alterations in large-scale solid tumors from an Asian population. Nature Communications. https://doi.org/10.1038/s41467-022-31780-9
Chen X, Bai K, Zhang Y, Xu Y, Huo Y, Wang S, Zou Y, Qi X, Guo R, Ou Q, Liu D, Yin S, Chen S, Bu H (2023) Genomic alterations of cerebrospinal fluid cell-free DNA in leptomeningeal metastases of gastric cancer. J Transl Med 21:296. https://doi.org/10.1186/s12967-023-04077-8
Li Y-S, Zheng M-M, Jiang B-Y, Tu H-Y, Yang J-J, Zhang X-C, Wu Y-L (2020) Association of cerebrospinal fluid tumor DNA genotyping with survival among patients with lung adenocarcinoma and central nervous system metastases. JAMA Network Open. https://doi.org/10.1001/jamanetworkopen.2020.9077
Frankell AM, Dietzen M, Al Bakir M, Lim EL, Karasaki T, Ward S, Veeriah S, Colliver E, Huebner A, Bunkum A, Hill MS, Grigoriadis K, Moore DA, Black JRM, Liu WK, Thol K, Pich O, Watkins TBK, Naceur-Lombardelli C, Cook DE, Salgado R, Wilson GA, Bailey C, Angelova M, Bentham R, Martinez-Ruiz C, Abbosh C, Nicholson AG, Le Quesne J, Biswas D, Rosenthal R, Puttick C, Hessey S, Lee C, Prymas P, Toncheva A, Smith J, Xing W, Nicod J, Price G, Kerr KM, Naidu B, Middleton G, Blyth KG, Fennell DA, Forster MD, Lee SM, Falzon M, Hewish M, Shackcloth MJ, Lim E, Benafif S, Russell P, Boleti E, Krebs MG, Lester JF, Papadatos-Pastos D, Ahmad T, Thakrar RM, Lawrence D, Navani N, Janes SM, Dive C, Blackhall FH, Summers Y, Cave J, Marafioti T, Herrero J, Quezada SA, Peggs KS, Schwarz RF, Van Loo P, Miedema DM, Birkbak NJ, Hiley CT, Hackshaw A, Zaccaria S, Consortium TR, Jamal-Hanjani M, McGranahan N, Swanton C, (2023) The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature 616: 525-533. https://doi.org/10.1038/s41586-023-05783-5
Li YS, Jiang BY, Yang JJ, Zhang XC, Zhang Z, Ye JY, Zhong WZ, Tu HY, Chen HJ, Wang Z, Xu CR, Wang BC, Du HJ, Chuai S, Han-Zhang H, Su J, Zhou Q, Yang XN, Guo WB, Yan HH, Liu YH, Yan LX, Huang B, Zheng MM, Wu YL (2018) Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann Oncol 29:945–952. https://doi.org/10.1093/annonc/mdy009
Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366:883–892. https://doi.org/10.1056/NEJMoa1113205
Hunter KW, Amin R, Deasy S, Ha NH, Wakefield L (2018) Genetic insights into the morass of metastatic heterogeneity. Nat Rev Cancer 18:211–223. https://doi.org/10.1038/nrc.2017.126
Martínez-Jiménez F, Movasati A, Brunner SR, Nguyen L, Priestley P, Cuppen E, Van Hoeck A (2023) Pan-cancer whole-genome comparison of primary and metastatic solid tumours. Nature. https://doi.org/10.1038/s41586-023-06054-z
Varn FS, Johnson KC, Martinek J, Huse JT, Nasrallah MP, Wesseling P, Cooper LAD, Malta TM, Wade TE, Sabedot TS, Brat D, Gould PV, Wöehrer A, Aldape K, Ismail A, Sivajothi SK, Barthel FP, Kim H, Kocakavuk E, Ahmed N, White K, Datta I, Moon HE, Pollock S, Goldfarb C, Lee GH, Garofano L, Anderson KJ, Nehar-Belaid D, Barnholtz-Sloan JS, Bakas S, Byrne AT, D’Angelo F, Gan HK, Khasraw M, Migliozzi S, Ormond DR, Paek SH, Van Meir EG, Walenkamp AME, Watts C, Weiss T, Weller M, Palucka K, Stead LF, Poisson LM, Noushmehr H, Iavarone A, Verhaak RGW (2022) Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell 185:2184-2199.e2116. https://doi.org/10.1016/j.cell.2022.04.038
Nanjo S, Arai S, Wang W, Takeuchi S, Yamada T, Hata A, Katakami N, Okada Y, Yano S (2017) MET copy number gain is associated with Gefitinib resistance in leptomeningeal carcinomatosis of EGFR-mutant lung cancer. Mol Cancer Ther 16:506–515. https://doi.org/10.1158/1535-7163.Mct-16-0522
Zheng MM, Li YS, Tu HY, Jiang BY, Yang JJ, Zhou Q, Xu CR, Yang XR, Wu YL (2021) Genotyping of cerebrospinal fluid associated With osimertinib response and resistance for leptomeningeal Metastases in EGFR-Mutated NSCLC. J Thoracic Oncol : Off Pub Inter Assoc Study Lung Cancer 16:250–258. https://doi.org/10.1016/j.jtho.2020.10.008
Knebel FH, Bettoni F, Shimada AK, Cruz M, Alessi JV, Negrão MV, Reis LFL, Katz A, Camargo AA (2017) Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer 108:238–241. https://doi.org/10.1016/j.lungcan.2017.04.004
Chmielecki J, Mok T, Wu YL, Han JY, Ahn MJ, Ramalingam SS, John T, Okamoto I, Yang JC, Shepherd FA, Bulusu KC, Laus G, Collins B, Barrett JC, Hartmaier RJ, Papadimitrakopoulou V (2023) Analysis of acquired resistance mechanisms to osimertinib in patients with EGFR-mutated advanced non-small cell lung cancer from the AURA3 trial. Nat Commun 14:1071. https://doi.org/10.1038/s41467-023-35962-x
He J, Huang Z, Han L, Gong Y, Xie C (2021) Mechanisms and management of 3rd-generation EGFR-TKI resistance in advanced non-small cell lung cancer (Review). Int J Oncol. https://doi.org/10.3892/ijo.2021.5270
Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, Feeney N, Sholl LM, Dahlberg SE, Redig AJ, Kwiatkowski DJ, Rabin MS, Paweletz CP, Thress KS, Jänne PA (2018) Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 4:1527–1534. https://doi.org/10.1001/jamaoncol.2018.2969
Minari R, Bordi P, Del Re M, Facchinetti F, Mazzoni F, Barbieri F, Camerini A, Comin CE, Gnetti L, Azzoni C, Nizzoli R, Bortesi B, Rofi E, Petreni P, Campanini N, Rossi G, Danesi R, Tiseo M (2018) Primary resistance to osimertinib due to SCLC transformation: Issue of T790M determination on liquid re-biopsy. Lung Cancer 115:21–27. https://doi.org/10.1016/j.lungcan.2017.11.011
Takano T, Fukui T, Ohe Y, Tsuta K, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Furuta K, Tamura T (2008) EGFR mutations predict survival benefit from gefitinib in patients with advanced lung adenocarcinoma: a historical comparison of patients treated before and after gefitinib approval in Japan. J Clin Oncol : Off J Am Soc Clin Oncol 26:5589–5595. https://doi.org/10.1200/jco.2008.16.7254
Suda K, Mitsudomi T (2015) Role of EGFR mutations in lung cancers: prognosis and tumor chemosensitivity. Arch Toxicol 89:1227–1240. https://doi.org/10.1007/s00204-015-1524-7
Yan W, Jing W, An N, Tian Y, Guo D, Kong L, Zhu H, Yu J (2019) The clinical characteristic and prognostic factors of leptomeningeal metastasis in patients with non-small-cell lung cancer-a retrospective study from one single cancer institute. Cancer Med 8:2769–2776. https://doi.org/10.1002/cam4.2156
Powter B, Jeffreys SA, Sareen H, Cooper A, Brungs D, Po J, Roberts T, Koh ES, Scott KF, Sajinovic M, Vessey JY, de Souza P, Becker TM (2021) Human TERT promoter mutations as a prognostic biomarker in glioma. J Cancer Res Clin Oncol 147:1007–1017. https://doi.org/10.1007/s00432-021-03536-3
Li X, Qian X, Wang B, Xia Y, Zheng Y, Du L, Xu D, Xing D, DePinho RA, Lu Z (2020) Programmable base editing of mutated TERT promoter inhibits brain tumour growth. Nat Cell Biol 22:282–288. https://doi.org/10.1038/s41556-020-0471-6
Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, Carrera C, Schimming T, Möller I, Schwamborn M, Sucker A, Hillen U, Badenas C, Malvehy J, Zimmer L, Scherag A, Puig S, Schadendorf D (2014) TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Nat Cancer Institute. https://doi.org/10.1093/jnci/dju246
Yang TT, Yu S, Ke CK, Cheng ST (2023) The Genomic Landscape of melanoma and Its therapeutic Implications. Genes. https://doi.org/10.3390/genes14051021
Kim TH, Kim YE, Ahn S, Kim JY, Ki CS, Oh YL, Kim K, Yun JW, Park WY, Choe JH, Kim JH, Kim JS, Kim SW, Chung JH (2016) TERT promoter mutations and long-term survival in patients with thyroid cancer. Endocr Relat Cancer 23:813–823. https://doi.org/10.1530/erc-16-0219
Chan AK, Yao Y, Zhang Z, Chung NY, Liu JS, Li KK, Shi Z, Chan DT, Poon WS, Zhou L, Ng HK (2015) TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 28: 177–186. https://doi.org/10.1038/modpathol.2014.94
Yang L, Wang M, Li N, Yan L-D, Zhou W, Yu Z-Q, Peng X-C, Cai J, Yang Y-H (2023) TERT mutations in non-small cell lung cancer clinicopathologic features and prognostic implications. Clin Med Insight Oncol. https://doi.org/10.1177/11795549221140781
Li H, Li J, Zhang C, Zhang C, Wang H (2020) TERT mutations correlate with higher TMB value and unique tumor microenvironment and may be a potential biomarker for anti-CTLA4 treatment. Cancer Med 9:7151–7160. https://doi.org/10.1002/cam4.3376
Groeneweg JW, Roze JF, Peters EDJ, Sereno F, Brink AGJ, Paijens ST, Nijman HW, van Meurs HS, van Lonkhuijzen L, Piek JMJ, Lok CAR, Monroe GR, van Haaften GW, Zweemer RP (2021) FOXL2 and TERT promoter mutation detection in circulating tumor DNA of adult granulosa cell tumors as biomarker for disease monitoring. Gynecol Oncol 162:413–420. https://doi.org/10.1016/j.ygyno.2021.05.027
Wang F, Zhao N, Gao G, Deng HB, Wang ZH, Deng LL, Yang Y, Lu C (2020) Prognostic value of TP53 co-mutation status combined with EGFR mutation in patients with lung adenocarcinoma. J Cancer Res Clin Oncol 146:2851–2859. https://doi.org/10.1007/s00432-020-03340-5
Yang H, Wen L, Pan Y, Shan C, Hong W, Wang H, Zhou C, Cai L, Zhou C (2022) Gene alternation of cerebrospinal fluid in patients with leptomeningeal metastases of lung adenocarcinoma using next-generation sequencing. BMC Cancer 22:580. https://doi.org/10.1186/s12885-022-09597-y
Lin X, Fleisher M, Rosenblum M, Lin O, Boire A, Briggs S, Bensman Y, Hurtado B, Shagabayeva L, DeAngelis LM, Panageas KS, Omuro A, Pentsova EI (2017) Cerebrospinal fluid circulating tumor cells: a novel tool to diagnose leptomeningeal metastases from epithelial tumors. Neuro Oncol 19:1248–1254. https://doi.org/10.1093/neuonc/nox066
Remsik J, Chi Y, Tong X, Sener U, Derderian C, Park A, Saadeh F, Bale T, Boire A (2022) Leptomeningeal metastatic cells adopt two phenotypic states. Cancer Rep (Hoboken) 5:e1236. https://doi.org/10.1002/cnr2.1236
Acknowledgements
We would like to thank all the patients and family members who gave their consents on presenting the data in this study, as well as the investigators and research staff involved. This work was supported by Natural Science Foundation of Hebei Province of China (No. H2020206126) and Central Government Guide Local Science and Technology Development Fund Project (Science and Technology Innovation Base Project) (No. 236Z7753G).
Author information
Authors and Affiliations
Contributions
KXB and HB conceived and designed experiments; Data acquiring and analyzing were performed by JJJ, YY, YJL and SHF; KXB, XC and XJQ contributed to writing—original draft preparation; YZ, YLZ, YYL, LLY and JL contributed to visualization; HB contributed to project administration and acquired funding. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bai, K., Chen, X., Qi, X. et al. Cerebrospinal fluid circulating tumour DNA genotyping and survival analysis in lung adenocarcinoma with leptomeningeal metastases. J Neurooncol 165, 149–160 (2023). https://doi.org/10.1007/s11060-023-04471-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-023-04471-8