Abstract
Introduction
Machine learning methods have been introduced as a computer aided diagnostic tool, with applications to glioma characterisation on MRI. Such an algorithmic approach may provide a useful adjunct for a rapid and accurate diagnosis of a glioma. The aim of this study is to devise a machine learning algorithm that may be used by radiologists in routine practice to aid diagnosis of both: WHO grade and IDH mutation status in de novo gliomas.
Methods
To evaluate the status quo, we interrogated the accuracy of neuroradiology reports in relation to WHO grade: grade II 96.49% (95% confidence intervals [CI] 0.88, 0.99); III 36.51% (95% CI 0.24, 0.50); IV 72.9% (95% CI 0.67, 0.78). We derived five MRI parameters from the same diagnostic brain scans, in under two minutes per case, and then supplied these data to a random forest algorithm.
Results
Machine learning resulted in a high level of accuracy in prediction of tumour grade: grade II/III; area under the receiver operating characteristic curve (AUC) = 98%, sensitivity = 0.82, specificity = 0.94; grade II/IV; AUC = 100%, sensitivity = 1.0, specificity = 1.0; grade III/IV; AUC = 97%, sensitivity = 0.83, specificity = 0.97. Furthermore, machine learning also facilitated the discrimination of IDH status: AUC of 88%, sensitivity = 0.81, specificity = 0.77.
Conclusions
These data demonstrate the ability of machine learning to accurately classify diffuse gliomas by both WHO grade and IDH status from routine MRI alone—without significant image processing, which may facilitate usage as a diagnostic adjunct in clinical practice.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Gliomas represent 80% of malignant brain tumours diagnosed [1], and represent a disproportionate cause of cancer related morbidity and mortality [2]. However, a dichotomy exists between a radiologist’s reporting of a newly diagnosed glioma based on morphology alone, and a pathologist’s diagnosis incorporating both morphology and genetics [3], especially as the isocytrate dehydrogenase (IDH) status of a glioma has significant therapeutic and diagnostic implications [4, 5].
Technological advances have led to the acquisition of higher resolution magnetic resonance images (MRI), enabling the visualisation of smaller structures and better characterisation of abnormalities, such as diffuse gliomas. However, this comes at the cost of producing an increasing number of images per patient. In this environment of an ever increasing workload, machine learning may provide a useful adjunct to a radiologist’s diagnostic toolkit [6, 7]. As radiology is a data driven specialty, it is ideally placed to incorporate such technologies into routine practice. Machine learning may extract information from images, heretofore, not apparent on visual inspection [6]. Such an algorithmic approach has provided useful insights in the radiological abnormalities observed in gliomas [8,9,10].
A radiologist’s interpretation of the grade of a glioma is of importance, particularly in instances where biopsy is not possible, or a non-representative biopsy is obtained and one wishes to formulate a germane management plan based on WHO grade [11]. However, prior studies have demonstrated the potential variability in a neuroradiologist’s grading of a glioma [12]. Such efforts may be confounded by: the heterogeneity of the tissue in a glioma [13], or contrast enhancement in a low grade glioma, with absent enhancement in a higher grade tumour [14]. Therefore, machine learning offers an advantage in such instances, and prior studies have demonstrated that quantitative MRI measures such as: DWI [15, 16], spectroscopy [17], or markers of cerebral blood volume [18], as input of such techniques discriminated WHO grade II and III gliomas may be discriminated from grade IV gliomas. The level of accuracy, recorded using the area under the receiver operator curve (AUC), in discriminating between glioma grade has varied from 0.84 [16] to 0.94 [18]. However, in each instance significant post-processing of the images was required to incorporate the imaging data into a machine learning algorithm.
IDH mutations occur in 70–90% of WHO grade II and III gliomas [19], following determination of WHO grade from MRI it would be desirable to also predict the IDH status of a newly diagnosed glioma. IDH is of prognostic value, as patients with an IDH mutation have a better response to treatment [20]. Furthermore, a recent study suggested that degree of resection—partial in high grade and total in low grade—relates to survival in IDH wildtype and mutant gliomas respectively [21]. Radiologically, IDH mutant gliomas have a preponderance for the frontal lobe [22], and may also have a more well defined border [23]. These and other observations have prompted efforts to characterise the IDH status of gliomas radiologically, through the use of machine learning [24,25,26,27]. Analogous to determination of WHO grade, such studies have employed quantitative MRI, such as spectroscopy [24, 27], or significant off-line processing of the images with textural analysis [25, 26], neither of which may be apposite to a radiologist’s routine clinical practice.
This current study aims to evaluate the discriminative power of machine learning, to evaluate whether such an approach may provide a useful diagnostic adjunct in the radiological diagnosis of a glioma’s grade and IDH status. Ab initio, this study was based on the premise that all data acquired for the algorithms used should be obtained from minimal image processing of diagnostic MRI scans, and that the image analysis could be performed within a reasonable time constraint, reflecting routine clinical practice.
Methods
Patients
Patients with a histopathologically proven diffuse glioma (grade II–IV) were identified from a single neuropathological database in our centre. We selected patients from 2015 to 2017, without any a priori knowledge of the radiological abnormalities in each case. All patients included had a de novo diagnosis of a glioma, i.e. patients with a recurrent tumour, or those who had an extension of a prior surgery were not included. All patients gave written informed consent for their data to be recorded in our neuro-oncology database. The study was approved by Beaumont Hospital medical ethics committee and carried out in accordance with approved guidelines.
Neuropathology and molecular genetic studies
We obtained a histopathological diagnosis of each glioma from the surgical specimen. In each instance the glioma was graded (II–IV), according to the current WHO diagnostic criteria [3]. We retrospectively applied the 2016 WHO criteria to earlier cases (i.e. from 2015), so that all cases included were diagnosed uniformly.
We stained all samples initially with an immunohistochemical stain for IDH1-R132H mutation [28]. In cases where IDH1-R132H was wildtype by immunohistochemistry in patients under 55 years, we performed pyrosequencing for both IDH1 and IDH2 mutations as previously described [29]. Cases with an IDH1 or IDH2 mutation were combined, due to the limited numbers of IDH2 mutations present to dichotomise this variable.
MRI acquisition
Diagnostic MR brain scans were performed as part of a routine clinical diagnostic investigation in each instance using a 3T scanner (Magnetom Verio/Trio TIM; Siemens Healthcare, Erlangen, Germany) equipped with an 8-channel array head coil. The protocol comprised the following axial sequences: (i) T1-weighted pre- and post-contrast axial scan: TR 600 ms, TE 82 ms, flip angle 70°, NEX = 1, slice thickness of 4 mm (ii) T2-weighted: TR 7000 ms, TE 105 ms, flip angle 150°, NEX = 1, slice thickness of 4 mm (iii) FLAIR scan: TR 9000 ms, TE 81 ms, TI 2500 ms, flip angle 150°, NEX = 1, slice thickness of 4 mm (iv) Diffusion weighted imaging (DWI): single shot spin echo planar sequence with a TE of 64 ms and flip angle of 180°, and slice thickness of 5 mm. Diffusion sensitising gradients were applied sequentially in the x, y and z directions with b values of 0 and 1000 s/mm2, and the corresponding ADC maps were generated by Syngo Software (Siemens Software, Erlangen, Germany).
MRI analysis
To evaluate the status quo, in our centre, we firstly obtained the final neuroradiological diagnosis of each newly diagnosed glioma and noted the predicted WHO grade. The final radiological diagnosis was obtained from the scan report, which was performed by a neuroradiologist with over 10 years’ experience, and dedicated training in neuro-oncology.
We subsequently performed an analysis of the same images; one reader evaluated all images, blinded to the clinical status of the patient and the pathological diagnosis. To evaluate the reproducibility of the measurements obtained, 18 scans were selected from the database, with six scans selected from each representative WHO grade (i.e. WHO grade II: n = 6, III: n = 6 and IV: n = 6). The same reader analysed the T2-weighted length and minimal ADC values and then repeated this analysis after an interval of 1 week, blinded to the clinical status of the case on both occasions. We performed the analysis on McKesson Radiology Manager (http://www.mckesson.com). This system is routinely used for diagnostic radiology in our centre, and no ‘offline’ processing of the images took place.
To standardise the imaging parameters recorded, we used select components of the Visually Accessible Rembrandt Images (VARSARI) template [30, 31]. We extracted from the template three features; we chose this smaller subset, known to be associated with IDH mutational status and WHO grade, in order to facilitate an efficient processing time of the images [32]:
-
(i)
the lesion size was measured on a T2-weighted axial image as the largest perpendicular (x–y) cross-sectional diameter;
-
(ii)
the percentage of non-enhancing or cystic abnormalities was measured through simultaneous review of a T1 and T2-weighted axial image. Cystic abnormalities were defined as a region within the tumour that does not enhance with T1-weighted imaging and contains central heterogeneous T1-weighted signal abnormalities in the absence of gadolinium contrast, and also has a high signal intensity on T2-weighted imaging; cystic abnormalities were determined by a visual assessment of the image only rather than by textual analysis of the image.
-
(iii)
the location of the tumour was based on the lobe of the brain that contained the geographic epicentre and did not include all areas of involvement. This was recorded on T2 and FLAIR images;
Two other components not contained on the template were also recorded, based on the same rationale as the VARSARI features, these two features were chosen based on known associations, and both features may be rapidly determined in routine clinical practice [32]:
-
(iv)
the non-enhancing border of the tumour was noted as being: sharp versus indistinct, through simultaneous review of T1 and T2-weighted axial images [23];
-
(v)
the cellularity of the glioma was estimated using the minimal ADC value in the tumour, as previously described [33].
To determine the feasibility of obtaining these measurements in routine clinical practise, we recorded the mean time to obtain these parameters in ten consecutive brain scans.
Statistical analysis
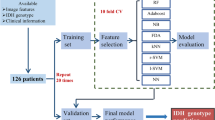
We considered the use of a machine learning algorithm, firstly as this provides an objective methodology to analyse data, and secondly its reproducibility is believed to be superior compared to an inferential approach [34]. For each required classification of the data, we used a decision forest approach for this purpose [35]. Whilst a number of available such algorithms exist, we chose a random forest model, as this has previously been demonstrated to be a robust classification algorithm for these types of data [36].
Statistical analyses were carried out using the R software environment (R Development Core Team, R 3.3.3, 2016, http://www.R-project.org/). In order to evaluate the reproducibility of the MRI measures obtained we calculated the intra-class correlation coefficient (ICC), and subsequently 1-ICC [37]. In this context 1-ICC represents the proportion of variability which is due to measurement error rather than biological variation. Subsequently, to evaluate the accuracy of the neuroradiologist’s diagnostic report we constructed a confusion matrix using the final neuropathological diagnosis for comparison using the ‘caret’ package.
To then determine the efficacy of machine learning, we used a two-class random forest classifier with 500 trees, again using the ‘caret’ package. A fivefold cross validation was applied to the training set of data, in order to estimate the performance of the classifier, and to validate the model [18]. Each dataset was randomly split into training and testing sets (70:30 ratio) using the createDataPartition function. To avoid overfitting the model we limited the dependent variables to those being considered in each hypothesis i.e. WHO grade or genetic marker [38]. For discriminating tumour grade, a two-class discrimination and a three-class discrimination were performed. For two-class discrimination, WHO grade II/III were combined into one class and discriminated against grade IV. For three-class discrimination, two-class tests were performed on all pairwise combinations of tumour grades: grade II/III; III/IV; and II/IV.
The number of training samples from grade and mutation classes were imbalanced; therefore, synthetic samples were generated using the SMOTE (Synthetic Minority Over-sampling Technique) method (DMwR package) [39] This technique is a data sampling procedure that uses both up-sampling of the minority class and down-sampling of the majority class to help balance the training set. For each dataset, the trained random forest model was employed to obtain a classification score for the test sample. A receiver operator curve was calculated from the full set of classification scores, the area under the receiver operator curve (AUC), along with specificity and sensitivity, was employed as a measure of classification performance.
Results
Patient demographics
We identified 381 patients with a de novo glioma diagnosed within the past 2 years in our institution. Following neurosurgical intervention, 57 patients were classified pathologically as having a WHO grade II glioma, grade III: n = 63 and grade IV: n = 261. In total there were: 76 IDH mutant gliomas, and 305 IDH wildtype gliomas. All patients included had a diagnostic MRI brain scan available, and no cases were excluded from analysis. A summary of the demographics, WHO grade and IDH status identified is displayed in Table 1.
Reproducibility analysis of MRI metrics analysed
In determination of the reproducibility of measurement of MRI metrics incorporated into the machine learning algorithm, we observed a high level of reproducibility of both T2 lesion length: ICC = 0.989 (95% CI 0.972, 0.996), and ADC: ICC = 0.936 (95% CI 0.802, 0.981). The value of 1-ICC was low in both instances, T2 lesion: 1-ICC = 0.011 and ADC: 1-ICC = 0.0604, suggesting a very low proportion of variability due to measurement error with both variables.
MRI variables
With increasing WHO grade, as expected, we observed a trend for a higher mean lesion size on T2-weighted imaging: WHO grade II = 4.65 cm (± 2.02), III = 5.82 cm (± 2.08), IV = 6.09 cm (± 1.87); an increase in mean percentage of cystic abnormalities, WHO grade II = 14.23% (± 18.4), III = 53.12% (± 28.39), IV = 83.98% (± 16.59); and a lower mean value of ADC, WHO grade II = 1.23 × 10−3 mm2/s (± 0.24), III = 0.91 × 10−3 mm2/s (± 0.12), IV = 0.69 × 10−3 mm2/s (± 0.11). As these variables were incorporated into the machine learning algorithm, an inferential statistical comparison was therefore not performed, to avoid conflation of two differing statistical approaches. A summary of the MRI variables used as continuous predictors in the machine learning algorithm (as mean and standard deviation), based on WHO grade of glioma and in each genetic mutation analysed are presented in Table 2.
The mean time to obtain the five imaging parameters in ten consecutive brain scans was: 1 min 33 s.
WHO grade of glioma
The neuroradiology reports in these 381 newly diagnosed gliomas were found to contain the following levels of accuracy in relation to each WHO grade: II 96.49% (95% CI 0.88, 0.99); III 36.51% (95% CI 0.24, 0.50); IV 72.9% (95% CI 0.67, 0.78). In no instances did the radiologist’s report comment on the likely IDH status in the glioma, without concomitant usage of machine learning.
High accuracy levels were obtained using machine learning in all grades of glioma: WHO grade II/III AUC = 98%, sensitivity = 0.82, specificity = 0.94; grade II/IV AUC = 100%, sensitivity = 1.0, specificity = 1.0; grade III/IV AUC = 97%, sensitivity = 0.83, specificity = 0.97. Furthermore, to facilitate direct comparison with a prior machine learning study radiologically determining the WHO status of a glioma [18], we also classified WHO grade based on a combination of grade II and III gliomas versus grade IV; resulting in an extremely high accuracy of 99%, sensitivity = 1.0, specificity = 0.92.
A summary of the accuracy of classification, as well as sensitivity and specificity are provided in Table 3. A graphical representation demonstrating the classification of the gliomas by WHO grade II-IV by MRI parameters recorded is shown in Fig. 1. Whilst, we included five MRI parameters in the machine learning algorithm to classify grade, the most informative metric was the ADC.
Boxplots of the MRI parameters, a lesion size (cm), b apparent diffusion coefficient (mm2/s), c degree of cystic abnormalities (%) and d the age of the patient, used to classify gliomas by WHO grade II, III and IV. The parameters combined resulted in an accuracy of over 98% in classification of WHO grade
IDH mutation status
Through the use of machine learning, we obtained moderate to high accuracy rates for the discrimination of IDH status as mutant versus wildtype, resulting in an AUC of 88%, sensitivity = 0.81, specificity = 0.77. Figure 2 demonstrates the classification of IDH status as mutant versus wildtype using three most informative MRI parameters in this regard.
Discussion
This study demonstrates the ability of a machine learning algorithm to classify gliomas on MRI scans according to both WHO grade and IDH status with a high degree of accuracy, thereby demonstrating the utility of such an approach as an adjunct for radiologists reporting MRI scans with newly diagnosed gliomas. There are three novel findings in this report. Firstly, this study using ‘real world’ data, demonstrated a high degree of diagnostic accuracy in relation to both WHO grade and IDH status of gliomas using machine learning. Secondly in contrast to prior studies, all images analysed were taken exclusively from routine clinical diagnostic scans and the analysis was performed on a radiologist’s work station, without employing any additional software or post-processing of the images (in less than two minutes per case). Finally, the levels of accuracy obtained were equivalent to prior machine learning reports in relation to both WHO grade and IDH status, without an acquisition of spectroscopy of cerebral blood volume, both of which would increase the scanning time.
A radiologist’s report has an influential role on the management of a patient with a newly diagnosed glioma. However, a number of factors may influence the accuracy of such a report, thereby leading to potential errors or misinterpretation of the radiological abnormalities noted. Firstly, physical factors, which may include the frequency of interruptions during a reporting session. Interruptions in reporting lead to an impairment in working memory, resulting in up to 13% increase in time for reporting and an increased potential for errors [40]. Fatigue is also a factor, as this adversely impacts the visual system including: worse accommodation, decreased saccadic velocity and reduced gaze volume and coverage [41]. Secondly, a number of cognitive biases may adversely affect the accuracy of a radiologists report of a glioma including: anchoring—the tendency to latch on to the first abnormality seen, such as contrast enhancement in a low grade glioma; satisfaction of search—termination of search following identification of an abnormality, such as failure to identify a second lesion in a multifocal glioma; confirmation bias—collection and interpretation of data to confirm an initial suspicion—misdiagnosis of a low grade glioma as a demyelinating plaque [42]. In order to reduce reporting time and cognitive biases, both of which may lead to reporting and diagnostic errors, machine learning offers a significant advantage [6], particularly in the context of general radiologists who may lack expertise in neuro-oncology.
The goal of machine learning is to devise a mathematical model, so that this formula may be applied to a new dataset. In the context of radiology such an approach is typically supervised, using labelled data as an endpoint [7], in the context of this study the neuropathological diagnosis guided the algorithm. Prior studies in neuro-oncology have used machine learning in a variety of capacities, including: modelling patient survival following glioma diagnosis [43], identification of EGFR amplification in a glioblastoma [44], and discrimination of a glioma from other brain lesions [45]. This study by Zacharaki et al. employed 161 features derived from MR images to analyse using both Gabor texture and image intensity characteristics, resulting in a 96% level of accuracy in differentiating between a low-grade and high-grade glioma.
A paradigm shift in machine learning has resulted in the application of simpler models but significantly larger datasets, leading to an increase in the effectiveness of the training stage of the algorithm [46]. In the absence of more advanced MRI derived parameters such as perfusion [8] or spectroscopy [17], our study focused on obtaining a larger dataset (n = 381). This is in contrast to prior studies where smaller cohort sizes (n = 28 [17], n = 102 [8], n = 129 [15], n = 37 [18]) have relied on complexity of image analysis to generate data for the machine learning algorithm. In this present study, we believe that the large cohort size obfuscated the requirement for significant post-processing of the images, such as textural analysis [8], to derive a large dataset for training the machine learning algorithm. The derived algorithm may then have application to clinical practice, as the reporting radiologist could potentially input five data points into the reporting software which may incorporate a machine learning algorithm, to predict features in a glioma such as WHO grade with accuracy.
Prior studies employing machine learning to predict WHO grade, have dichotomised gliomas into low grade versus high grade through the addition of spectroscopy [17], or a measure of relative oxygen extraction fraction combined with a total of 116 MRI features [18]. However, except for ADC, Guzman-De-Villoria et al. found no advantage in the use of quantitative MRI for classification of glioma grade. Our results are in agreement with these findings, where the ADC was the most predictive feature, in terms of classifying glioma grade. However, in contrast to prior studies, our intentionally parsimonious approach to image analysis, restricted to five MRI variables, resulted in a novel machine learning algorithm that classified three grades of glioma independently, rather than a dichotomised approach, as has been the case in all prior reports. Furthermore, our approach provided greater accuracy than routine radiology reporting in our centre, and a level of accuracy in discriminating WHO grade that is equivalent to prior machine learning reports [18]—without the implementation of extensive image processing or a prolonged scanning protocol. These findings suggest that the approach employed in this study may provide a time efficient useful adjunct for radiologists predicting WHO grade of a newly diagnosed glioma.
The IDH status of a glioma, has both therapeutic and prognostic implications for a newly diagnosed glioma [21], therefore, it would also be desirable for a radiologist’s report to provide a prediction of whether a tumour is likely to be IDH mutant or wildtype. Neuroimaging studies have demonstrated that the location of a glioma, in particular frontal lobe, and a well-defined border on an axial FLAIR image, may predict IDH status [22, 23, 47]. More recent studies have used texture based analysis, resulting in 42 texture features in a study by Zhou et al. and 2970 imaging features derived from T1, T2 and DWI, resulting in an AUC of 89% [26]. Using clinical imaging alone, the algorithm used in this study resulted in a similar level of accuracy of 88% for classification of IDH status. Another approach has been the use of MR spectroscopy to predict IDH status [24], however, MR spectroscopy has limited spatial resolution and may suffer from partial volume effects due to overlap of the voxel with surrounding normal tissue, thereby limiting its widespread use in routine practice [48]. In order to overcome a reduction in signal to noise ratio, high field strength, multi-channel phased array coils and efficient pulse sequences are required, although these requirements may not always be met outside of a research setting. The prediction of IDH status was also conferred through machine learning in this present study, as this approach may extract information from MRI that is not immediately apparent on visual inspection, therefore implementation of this approach in radiological reporting may significantly enhance reports with the provision of the likely IDH status of a newly diagnosed glioma.
Study limitations
A few limitations of this work must be considered. Firstly, the cross-sectional data used did not provide the ability to identify markers of survival [49]. A future longitudinal study could provide such data. Secondly, the clinical details were not available in every case studied. This restricted interpretation as to whether biopsy or resected tissue was studied. Through the use of electronic operative records in our centre, any future such studies may be able to obtain more detailed neurosurgical information. Finally, the neuroradiologist’s report was derived from visual inspection of the MRI scans, rather than measurement of the same five metrics employed in the machine learning algorithm. Therefore, direct comparison was not possible, this may also be analysed in a future similar study. Nonetheless our premise was to provide an objective data-driven adjunct for the neuradiologist, particularly those without dedicated neuro-oncology training, rather than a potential replacement.
Conclusions
In conclusion, this study demonstrates the use of a machine learning algorithm, derived from a large dataset of ‘real word’ MRI scans, that can accurately classify WHO grade and IDH status in newly diagnosed gliomas. The minimal image processing performed in this study may facilitate translation of such an approach into clinical practice as an adjunct to a neuroradiologist to provide accurate and rapid objective reports in de novo gliomas.
References
Ostrom QT, Gittleman H, Fulop J et al (2015) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol 17:iv1–iv62. https://doi.org/10.1093/neuonc/nov189
Gittleman H, Kromer C, Ostrom QT et al (2017) Is mortality due to primary malignant brain and other central nervous system tumors decreasing? J Neurooncol 133:265–275. https://doi.org/10.1007/s11060-017-2449-1
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 Mutations in Gliomas. N Engl J Med 360:765–773. https://doi.org/10.1056/NEJMoa0808710
Metellus P, Coulibaly B, Colin C et al (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 120:719–729. https://doi.org/10.1007/s00401-010-0777-8
Thrall JH, Li X, Li Q et al (2018) Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol 15:504–508. https://doi.org/10.1016/j.jacr.2017.12.026
Kohli M, Prevedello LM, Filice RW, Geis JR (2017) Implementing machine learning in radiology practice and research. Am J Roentgenol 208:754–760
Zacharaki EI, Wang S, Chawla S, Soo D (2010) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62:1609–1618. https://doi.org/10.1002/mrm.22147.Classification
Zacharaki EI, Morita N, Bhatt P et al (2012) Survival analysis of patients with high-grade gliomas based on data mining of imaging variables. AJNR Am J Neuroradiol 33:1065–1071. https://doi.org/10.3174/ajnr.A2939
Svolos P, Tsolaki E, Kapsalaki E et al (2013) Investigating brain tumor differentiation with diffusion and perfusion metrics at 3T MRI using pattern recognition techniques. Magn Reson Imaging 31:1567–1577. https://doi.org/10.1016/j.mri.2013.06.010
Wen PY, Reardon DA (2016) Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol 12:2015–2016. https://doi.org/10.1038/nrneurol.2015.242
Kondziolka D, Lunsford LD, Martinez a J (1993) Unreliability of contemporary neurodiagnostic imaging in evaluating suspected adult supratentorial (low-grade) astrocytoma. J Neurosurg 79:533–536. https://doi.org/10.3171/jns.1993.79.4.0533
Aronen HJ, Gazit IE, Louis DN et al (1994) Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology 191:41–51. https://doi.org/10.1148/radiology.191.1.8134596
Knopp EA, Cha S, Johnson G et al (1999) Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology 211:791–798. https://doi.org/10.1148/radiology.211.3.r99jn46791
Guzman-De-Villoria JA, Mateos-Perez JM, Fernandez-Garcia P et al (2014) Added value of advanced over conventional magnetic resonance imaging in grading gliomas and other primary brain tumors. Cancer Imaging 14:1–10. https://doi.org/10.1186/s40644-014-0035-8
Miloushev VZ, Chow DS, Filippi CG (2015) Meta-analysis of diffusion metrics for the prediction of tumor grade in gliomas. AJNR Am J Neuroradiol 36:302–308. https://doi.org/10.3174/ajnr.A4097
Ranjith G, Parvathy R, Vikas V et al (2015) Machine learning methods for the classification of gliomas: initial results using features extracted from MR spectroscopy. Neuroradiol J 28:106–111. https://doi.org/10.1177/1971400915576637
Wiestler B, Kluge A, Lukas M et al (2016) Multiparametric MRI-based differentiation of WHO grade II/III glioma and WHO grade IV glioblastoma. Sci Rep 6:35142. https://doi.org/10.1038/srep35142
Flavahan WA, Drier Y, Liau BB et al (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529:110–114. https://doi.org/10.1038/nature16490
Sanson M, Marie Y, Paris S et al (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27:4150–4154. https://doi.org/10.1200/JCO.2009.21.9832
Beiko J, Suki D, Hess KR et al (2014) IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol 16:81–91. https://doi.org/10.1093/neuonc/not159
Qi S, Yu L, Li H et al (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 7:1895–1902. https://doi.org/10.3892/ol.2014.2013
Delfanti RL, Piccioni DE, Handwerker J et al (2017) Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol 135:601–609. https://doi.org/10.1007/s11060-017-2613-7
Branzoli F, Di Stefano AL, Capelle L et al (2017) Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro Oncol. https://doi.org/10.1093/neuonc/nox214
Zhou H, Vallières M, Bai HX et al (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19:862–870. https://doi.org/10.1093/neuonc/now256
Zhang B, Chang K, Ramkissoon S et al (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19:109–117. https://doi.org/10.1093/neuonc/now121
Elkhaled A, Jalbert LE, Phillips JJ et al (2012) Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med 4:116ra5–116ra5. https://doi.org/10.1126/scitranslmed.3002796
Reuss DE, Sahm F, Schrimpf D et al (2015) ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol 129:133–146. https://doi.org/10.1007/s00401-014-1370-3
Arita H, Narita Y, Matsushita Y et al (2015) Development of a robust and sensitive pyrosequencing assay for the detection of IDH1/2 mutations in gliomas. Brain Tumor Pathol 32:22–30. https://doi.org/10.1007/s10014-014-0186-0
Bakas S, Akbari H, Sotiras A et al (2017) Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci data 4:170117. https://doi.org/10.1038/sdata.2017.117
Clark K, Vendt B, Smith K et al (2013) The Cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057. https://doi.org/10.1007/s10278-013-9622-7
Smits M, van den Bent MJ (2017) Imaging correlates of adult glioma genotypes. Radiology 284:316–331. https://doi.org/10.1148/radiol.2017151930
Lee EJ, terBrugge K, Mikulis D et al (2011) Diagnostic value of peritumoral minimum apparent diffusion coefficient for differentiation of glioblastoma multiforme from solitary metastatic lesions. Am J Roentgenol 196:71–76. https://doi.org/10.2214/AJR.10.4752
Breiman L (2001) Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat Sci 16:199–231. https://doi.org/10.1214/ss/1009213726
Zikic D, Glocker B, Konukoglu E et al (2012) Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. Med Image Comput Comput Assist Interv 15:369–376
Fernández-Delgado M, Cernadas E, Barro S et al (2014) Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res 15:3133–3181
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310
Srivastava N, Hinton G, Krizhevsky A et al (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Williams LH, Drew T (2017) Distraction in diagnostic radiology: how is search through volumetric medical images affected by interruptions? Cogn Res Princ Implic 2:12. https://doi.org/10.1186/s41235-017-0050-y
Waite S, Kolla S, Jeudy J et al (2017) Tired in the reading room: the influence of fatigue in radiology. J Am Coll Radiol 14:191–197. https://doi.org/10.1016/j.jacr.2016.10.009
Lee CS, Nagy PG, Weaver SJ, Newman-Toker DE (2013) Cognitive and system factors contributing to diagnostic errors in radiology. Am J Roentgenol 201:611–617
Emblem KE, Pinho MC, Zöllner FG et al (2015) A generic support vector machine model for preoperative glioma survival associations. Radiology 275:228–234. https://doi.org/10.1148/radiol.14140770
Kickingereder P, Bonekamp D, Nowosielski M et al (2016) Radiogenomics of Glioblastoma: Machine Learning-based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology 281:907–918. https://doi.org/10.1148/radiol.2016161382
Zacharaki EI, Wang S, Chawla S et al (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62:1609–1618. https://doi.org/10.1002/mrm.22147
Halevy A, Norvig P, Pereira F (2009) The Unreasonable effectiveness of data. IEEE Intell Syst 24:8–12. https://doi.org/10.1109/MIS.2009.36
Wang Y, Zhang T, Li S et al (2015) Anatomical localization of isocitrate dehydrogenase 1 mutation: a voxel-based radiographic study of 146 low-grade gliomas. Eur J Neurol 22:348–354. https://doi.org/10.1111/ene.12578
Horská A, Barker PB (2010) Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin N Am 20:293–310. https://doi.org/10.1016/j.nic.2010.04.003
Jain R, Poisson LM, Gutman D et al (2014) Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 272:484–493. https://doi.org/10.1148/radiol.14131691
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors of this study have any conflict of interest in relation to this work.
Human and animal participants
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all participants in this study.
Rights and permissions
About this article
Cite this article
De Looze, C., Beausang, A., Cryan, J. et al. Machine learning: a useful radiological adjunct in determination of a newly diagnosed glioma’s grade and IDH status. J Neurooncol 139, 491–499 (2018). https://doi.org/10.1007/s11060-018-2895-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-2895-4