Abstract
The goal of this study is to spatially discriminate tumor from treatment effect (TE), within the contrast-enhancing lesion, for brain tumor patients at all stages of treatment. To this end, the diagnostic accuracy of MRI-derived diffusion and perfusion parameters to distinguish pure TE from pure glioblastoma (GBM) was determined utilizing spatially-correlated biopsy samples. From July 2010 through June 2015, brain tumor patients who underwent pre-operative DWI and DSC-MRI and stereotactic image-guided biopsy were considered for inclusion in this IRB-approved study. MRI-derived parameter maps included apparent diffusion coefficient (ADC), normalized cerebral blood flow (nCBF), normalized and standardized relative cerebral blood volume (nRCBV, sRCBV), peak signal-height (PSR) and percent signal-recovery (PSR). These were co-registered to the Stealth MRI and median values extracted from the spatially-matched biopsy regions. A ROC analysis accounting for multiple subject samples was performed, and the optimal threshold for distinguishing TE from GBM determined for each parameter. Histopathologic diagnosis of pure TE (n = 10) or pure GBM (n = 34) was confirmed in tissue samples from 15 consecutive subjects with analyzable data. Perfusion thresholds of sRCBV (3575; SN/SP% = 79.4/90.0), nRCBV (1.13; SN/SP% = 82.1/90.0), and nCBF (1.05; SN/SP% = 79.4/80.0) distinguished TE from GBM (P < 0.05), whereas ADC, PSR, and PH could not (P > 0.05). The thresholds for CBF and CBV can be applied to lesions with any admixture of tumor or treatment effect, enabling the identification of true tumor burden within enhancing lesions. This approach overcomes current limitations of averaging values from both tumor and TE for quantitative assessments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A prevailing challenge in patients diagnosed with glioblastoma (GBM), throughout their treatment course, is distinguishing tumor from treatment effect (TE). Current imaging response criteria [1] rely heavily on T1w and FLAIR imaging to assess tumor burden, which may yield confounding information when treatment effects are present [2,3,4,5]. Because of this several studies have been undertaken to investigate the use of additional imaging markers for the assessment of tumor progression or treatment response [2, 3, 6, 7]. Advanced imaging parameters, derived from perfusion and diffusion MRI (pMRI, DWI), have demonstrated the potential to distinguish between GBM and TE. This information could substantially improve treatment management, while also providing further guidance for surgical biopsy or resection [5].
Although promising, the utility of pMRI and DWI-defined biomarkers for distinguishing GBM from TE has not been fully realized. This may be largely due to the use of gross regional metrics, derived from the mean, median or maximum values extracted from the entire enhancing T1w or hyperintense FLAIR areas [2, 3] to define thresholds for distinction. Though these metrics have shown value, they do not take into account the admixture of both GBM and TE within the entire abnormality, and to what degree either is present. Therefore, an accurate indication of residual tumor or TE is confounded by the spatial averaging and most likely explains the substantial variation in thresholds reported to distinguish these tissue types.
Another limitation of studies using published thresholds to distinguish tissue types is that, in several cases, the thresholds are used for purposes for which they were not originally defined. For example, a normalized (to a reference brain ROI) relative cerebral blood volume (nRCBV) threshold of 1.75 has been used to distinguish GBM from TE or to predict GBM progression [8,9,10,11]. Yet, this threshold was originally determined for the purpose of differentiating low-grade from high-grade glioma [12]. Although there is utility in this threshold, it was not validated for the intent to distinguish tumor from TE.
Tissue specimens obtained through stereotactic correlation with MRI not only diminish the effects of averaging, but also allow for a more direct correlation of imaging parameters and histopathology, with reduced sampling error and increased precision [13, 14]. A novel factor of this study, is the utilization of tissue specimens of pure histological diagnosis, which contain no admixture of tissue types. This factor enables the determination of a threshold that should be repeatable and easily validated by many sites, as it will not depend on the tissue admixture particular to each site’s study population. Also, newly explored in this study, is the evaluation of diffusion and standardized perfusion parameters for distinguishing TE from GBM. In summary, the aim of this study was to determine the diagnostic accuracy of several pMRI and DWI parameters for distinguishing TE from GBM, by using spatially correlated tissue specimens that contain only a pure histological diagnosis of TE or GBM.
Materials and methods
Participants & histology
All imaging and histopathology data was obtained in accordance with HIPAA guidelines and after obtaining informed, written consent according to the guidelines approved by our Institutional Review Board (IRB). Consecutive individuals with brain lesions who had undergone image-guided biopsy utilizing pre-surgical Stealth from July 2010 through June 2015 were considered for this study. Subjects underwent surgery for their tumor based on clinical warrant, which included clinical presentation, imaging findings, and performance status [1]. Of note, co-localized histological specimens are routinely collected using a specialized protocol for deposit to an institutional bank for future correlative-image analysis. Sampling sites were guided using the post-contrast spoiled gradient recalled (SPGR) that was uploaded to the neuro-navigation unit, and determined based on surgical approach, with considerations for lesion size, location, and patient safety to best obtain a robust representation of the histopathology for diagnostic and clinical purposes. Additionally, only those patients with multiple tissue samples having a histopathologic diagnosis of pure TE or pure GBM tissue with available pre-surgical diffusion and perfusion imaging were retrospectively identified and subsequently included in this study. A neuropathologist (EC) with 27 years of experience reviewed all tissue samples. Diagnosis of TE was based upon histologic changes as previously described [15] and for GBM was based upon World Health Organization (WHO) classification criteria [16]. Specifically, tissue samples diagnosed as TE contained coagulative necrosis in combination with hyalinized and/or telangiectatic vessels, fibrinoid necrosis of vessel walls, dystrophic calcifications, or cytologic atypia with bizarre nuclei and abundant cytoplasm [15]. Mitoses should be rare [15]. Tissue samples diagnosed as GBM contained all of the following features according to the classification scheme: highly cellular neoplasm composed of astrocytic appearing cells with nuclear pleomorphism and hyperchromatism, increased mitoses, necrosis with or without pseudopalisading and/or microvascular proliferation [16]. In this study, tissue samples classified as GBM were astrocytic and did not contain any identifiable portions of TE. More specifically, tissue samples with any admixture of tumor cells and TE were purposefully not included in this study. (While some studies categorize tissue as tumor when any percentage of tumor cells are present, this can confound determination of an accurate and consistent threshold for all study cohorts.) However, one case with tissue samples of mixed histopathology, containing both GBM and TE, is used to demonstrate the application of the determined thresholds, but is not used in the analysis performed to determine the thresholds. Subjects were excluded from analysis if they received treatment for their tumor during the interval between MR imaging and surgery. Samples were excluded from analysis if they were in regions where T1w images showed no enhancement, were obtained in a location that had poor DSC signal quality, included incomplete or missing data, inadequate contrast agent injection, or incomplete anatomic coverage. The number of participants included in this study was based on availability, and not based on a power analysis.
MR imaging
Two pre-surgical MRI exams were performed on either a 1.5T or 3T MRI system (GE Healthcare, Milwaukee, WI or Siemens, Munich, Germany) prior to surgical biopsy. Data obtained in the initial MRI exam included SPGR, diffusion-weighted imaging (DWI), and dynamic susceptibility contrast imaging (DSC). The SPGR was collected with TR = 5.8–17 ms and TE = 1.8–4.8 ms, flip-angle = 10°–30°, 96–160 slices with slice thickness = 1.0–1.5 mm, inter-slice gap = 0 mm, acquisition-matrix = 256–320 × 192–320, and field of view = 220–320 × 156–320 mm2. Isotropic DWI was collected with TR = 4100–10,000 ms, TE = 46–129 ms, flip-angle = 90°, 22–32 slices with slice thickness of 5, inter-slice gap = 0–1.5 mm, acquisition-matrix = 128–352 × 128–224, and field of view = 110–240 × 105–240 mm2. The DSC gradient-echo echo-planar imaging (GRE-EPI) was collected as follows: TR = 1100–1250 ms, TE = 30 ms, flip-angle = 70°–80°, 13 slices with slice-thickness = 5 mm, inter-slice gap = 0–1.5 mm, acquisition-matrix = 96 × 96, and field-of-view = 165–220 × 165–220 mm2, using a 0.1 mmol/kg dose of gadobenate dimeglumine (Multihance: Bracco Diagnostics Inc., Princeton, NJ) contrast agent. In total, a 0.05–0.1 mmol/kg preload injection was administered before the post-contrast anatomic images were collected and a 0.1 mmol/kg dose administered during DSC data collection [17, 18]. The DSC contrast agent injection occurred at approximately 60 s into the DSC study at a rate of 3 ml/s using a power-injector. The second MRI exam was performed within 1 day of surgery and consisted of a pre-surgical Stealth exam that included SPGR, which was used to co-localize tissue samples to the MRI via a surgical navigation unit (StealthStation S7: Medtronic, Minneapolis, MN) utilized for image-guided biopsy. Since DSC is routinely acquired for diagnostic purposes and throughout patient follow-up at our institution, it is not typically acquired during the second pre-surgical exam, which is acquired foremost for surgical planning. Using the pre-surgical Stealth SPGR, all biopsy locations were visually recorded through individually labeled image captures from the navigation unit. The Stealth SPGR was collected within the same settings as the initial SPGR.
Processing
Perfusion metrics of relative cerebral blood volume (rCBV) and cerebral blood flow (CBF) were calculated from DSC data using IB Neuro™ (Imaging Biometrics, Elm Grove, WI), which incorporates a rCBV leakage-correction algorithm [18, 19]. Of note, preload dosing, a mid-range flip angle and post-processing leakage correction were used to diminish unwanted changes in both T1 and T2* that can result from contrast agent leakage, which is dependent on not only contrast agent dosing but also parameter settings and field strength. Standardized rCBV (sRCBV), nRCBV, and normalized CBF (nCBF) were created with built-in standardization files, [20] or normalized to manually drawn reference regions of interest (ROIs) in normal appearing white matter, respectively. Peak signal height (PH) and percent signal recovery (PSR) were calculated using in-house scripts from the DSC time course data as previously described [21]. Using in-house scripts the apparent diffusion coefficient (ADC) was calculated from DWI data acquired at b-values of 0 and 1000 s/mm2. All in-house scripts utilized functions available within Analysis of Functional NeuroImages (AFNI) software [22].
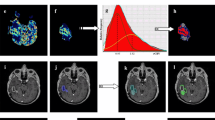
Parameter maps were then rigidly co-registered with a normalized mutual information cost function using FMRIB Software Library (FSL) [23]. Briefly, all perfusion and diffusion metrics were co-registered with the pre-surgical SPGR, where the DSC series was co-registered via a T1w reference series acquired in the exact same slice prescription as the DSC series. The pre-surgical SPGR was subsequently co-registered to the Stealth SPGR, and the same transformation matrix was applied to the perfusion and diffusion images. Spherical ROIs with a radius of 1.5 mm were manually drawn on the Stealth SPGR and visually matched in axial, sagittal, and coronal planes to the Stealth SPGR image of the biopsy coordinates captured during surgery. Actual sizes of the pure tissue samples were larger than the manually drawn spherical ROIs to mitigate potential error resulting from ROI placement or brain shift or swelling during surgery [24]. An example of this ROI placement, along with rCBV, ADC and hematoxylin and eosin (H&E) histology staining is shown in Fig. 1. Positive, non-zero median values were extracted from the ROIs for each co-registered parameter.
3D co-localization of imaging and tissue sample shown in a 31-year-old female with TE. As outlined in red, a spherical ROI was manually drawn on the pre-surgical post-contrast Stealth SPGR (d–f) by visually matching it to the post-contrast Stealth SPGR images (a–c) captured during surgery of the tissue sample location in the sagittal (a, d), coronal (b, e), and axial (c, f) planes. Also shown in the axial plane are the corresponding ADC (h), rCBV (i), and H&E histology staining (g) showing pure TE
Statistical analysis
Generalized estimating equations (GEE) analysis using a robust estimator for the variance, to account for multiple samples from the same subject, was performed twice for each parameter: initially to determine if pure GBM samples from treatment naïve and previously treated participants could be statistically combined (P > 0.05), and foremost to determine if pure GBM samples could be statistically differentiated from those with pure TE (P < 0.05). A receiver operator curve (ROC) analysis was then performed and area under the curve (AUC) determined for the parameters that showed statistically significant differences. Using this analysis, the threshold that provided the best sensitivity (SN) and specificity (SP) for distinguishing TE from GBM was then determined. In addition, a logistic regression analysis was performed to determine if combing ADC with either rCBV or CBF would improve diagnostic ability. The binary logistic output derived from the GEE analysis was used as input for the ROC analysis, AUC determination, and logistic regression analysis. All statistical analyses were performed using SPSS Statistics version 22 (IBM Corp, Armonk, NY).
Results
There were 18 consecutive participants that met inclusion criteria for analysis in this study. Of these, one subject was excluded due to a poor contrast agent injection. Another subject was excluded because all biopsy samples were acquired in non-enhancing regions. A third subject was excluded because they received a course of chemotherapy between imaging and surgical biopsy. Individual samples were also excluded because they were from non-enhancing regions (n = 2) or the sample was obtained within a region where no DSC signal was present or had insufficient signal (n = 2). Therefore, 44 samples with histologic diagnosis of pure TE (n = 10) or pure GBM (n = 34) were included in this analysis from the remaining 15 participants. For these patients all correlative imaging was acquired a median of 5 days prior to surgery. The average age of the 8 male (51 years, range = 31–64) and 7 female (58 years, range = 30–68) participants was 54 (range = 30–68) years. The Stealth MRI exam, used with the surgical navigation unit for co-localization of tissue samples, was performed within one day of surgery for all participants. An average of 3 samples were obtained from each participant (2–5 samples/participant). Participants were previously treated with radiation therapy (RT) alone (n = 2), RT in combination with chemotherapy (n = 4), or RT in combination with chemotherapy followed by bevacizumab with TRC105 on first recurrence (n = 1), while 8 were treatment naïve. Of the patients who received treatment, image-correlated specimens were obtained from surgery that was performed while subjects were off treatment (n = 5), during the treatment course of adjuvant chemotherapy (n = 1), or during treatment with bevacizumab and TRC105 (n = 1). The pure GBM samples came from both treatment naïve (n = 25 samples) and treated (n = 9 samples) patients. The pure TE samples came from treated patients only (n = 10 samples).
Representative H&E histology staining is seen in Fig. 2, showing specimens of pure GBM from both a treated and treatment naïve patient, a specimen of containing pure TE, and lastly a specimen containing GBM admixed with TE. The admixed sample was not used in analysis for calculation of a threshold, but is presented to draw a distinction between samples that were included in this study from those that were not. Representative diffusion and perfusion image parameter maps are shown in Fig. 3. GEE analysis revealed no statistically significant differences in parameter values between treated (n = 9) and untreated (n = 25) GBM samples (P > 0.05) across all parameters as displayed in Table 1. Therefore, GBM samples were analyzed collectively irrespective of prior treatment. The ADC (P = 0.149), PH (P = 0.325), and PSR (P = 0.742) could not statistically distinguish between TE and GBM (P > 0.05). However, the sRCBV (<3575 a.u., P = 0.001), nRCBV (< 1.13 a.u.; P = 0.0004), and nCBF (1.05 a.u.; P = 0.002) were able to distinguish TE and GBM. These results are also shown in Table 1. Both sRCBV (SN/SP%=79.4/90.0; AUC = 0.853) and nRCBV (SN/SP%=82.4/90.0; AUC = 0.888) had better specificity than did nCBF (SN/SP%=79.4/80.0; AUC = 0.859). Applying thresholds of sRCBV, nRCBV, and nCBF enable the creation of fractional tumor burden (FTB) maps [25] as shown in Fig. 4. The FTB maps in Fig. 4 were generated for a subject whose data was not used in the calculation of any thresholds, but is presented for prospective illustration only. As shown, the FTB map can be used to distinguish areas of TE from GBM within T1w enhancement. The tissue samples obtained from this patient were confirmed as being a mixture of GBM and TE, where the degree of TE and GBM present in the FTB maps reflect the histology of the biopsy specimens. Logistic regression analysis revealed that ADC was not significant when combined with nCBF (χ2(ADC) = 0.113, df = 1, P = 0.737), nRCBV (χ2(ADC) = 0.374, df = 1, P = 0.541), or sRCBV (χ2(ADC) = 0.000, df = 1, P = 0.987).
Representative H&E histology staining in tissue samples from CRT-treated (a, c, d) and treatment naïve (b) patients. Shown are samples of pure GBM (a, b), pure TE (c), and GBM admixed with TE (d). The sample of GBM admixed with TE is shown to illustrate non-pure samples which were excluded from analysis. The samples of pure GBM (a, b) contain all of the same characteristic features regardless of treatment status
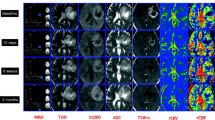
Example parameter maps in a 61-year-old male with GBM. Shown are post-contrast SPGR (a), sRCBV (b), nRCBV (c), nCBF (d), FLAIR (e), ADC (f), PSR (g), and PH (h). Perfusion parameter maps are overlaid on post-contrast SPGR. ADC is overlaid on FLAIR. Noticeable in the PSR image is the absence of tumor contrast compared to other tissue, which occurs when a preload dose of contrast agent is administered for the purpose of reducing T1-related leakage effects in the calculation of rCBV
Example fractional tumor burden (FTB) maps in a 58-year-old female, whose samples were not included in the analysis for calculation of thresholds to differentiate TE and GBM. The tissue histopathology of the region encircled in blue is TE with sparse neoplastic astrocytes, and encircled in yellow contains sections of both TE and GBM. Using the respective thresholds, GBM (red) and TE (white) are shown in areas within enhancement on post-contrast SPGR (a) using sRCBV (b), nRCBV (c), and nCBF (d) thresholds
While treated and untreated GBM samples were not statistically different, a separate sub-analysis of seven patients with treated GBM (n = 9) and TE (n = 10) samples was performed. These results also showed no statistical ability to distinguish TE from treated GBM samples for ADC (P = 0.064), PH (0.145), or PSR (P = 0.368). Additionally, TE was distinguished from treated GBM samples with equivalent specificity for sRCBV (P = 0.010; SN/SP% = 88.9/90.0; AUC = 0.900), nRCBV (P = 0.015; SN/SP% = 88.9/90.0; AUC = 0.933) and nCBF (P = 0.0003; SN/SP% = 77.8/80.0; AUC = 0.867) as for the combined analysis. Logistic regression analysis also revealed that ADC was not significant when combined with nCBF (χ2(ADC) = 1.390, df = 1, P = 0.238), nRCBV (χ2(ADC) = 0.115, df = 1, P = 0.735), or sRCBV (χ2(ADC) = 1.296, df = 1, P = 0.255).
Discussion
Thresholds for perfusion parameters sRCBV, nRCBV, and nCBF were determined to distinguish between pure GBM and pure TE, each providing an accuracy greater than 0.80. In a recent report that included nine patients with 26 biopsy samples, where nRCBV was the only parameter examined, a nRCBV threshold of 1.00 was found to differentiate GBM from TE [25]. However, in their study, the GBM samples analyzed were allowed to contain any admixture of TE, which likely forced a lower threshold in order to reliably distinguish GBM from TE in the studied population. In this study, which incorporated a larger population with a greater number of tissue samples, the nRCBV threshold of 1.13 is slightly higher, as expected since all samples were pure GBM or pure TE. However, the fact that both sites determined a threshold close in value bodes well for using rCBV to consistently distinguish tumor from treatment effect. It is highly relevant that sRCBV was found to distinguish GBM from TE with similar accuracy as nRCBV, as use of standardization provides increased repeatability and decreased variability among patients and between imaging sessions, while also eliminating the need to subjectively identify a reference region of interest for normalization [19].
Interestingly, PH and PSR demonstrated no ability to differentiate GBM from TE. However, as previously described [21], using a preload, while helpful for the creation of rCBV maps, does decrease PSR contrast. Therefore, PH and PSR may show potential for distinguishing GBM from TE if the DSC-MRI data is collected without a preload of contrast agent or with a lower flip angle, the approach used in a previous study demonstrating the utility of these parameters [6]. Use of dual-echo DSC-MRI method such as SPICE (spiral perfusion imaging with consecutive echoes) [26] would allow for the optimal determination of both parameters since a contrast agent preload is not required. This results from the fact that T1 leakage effects are automatically eliminated by virtue of the dual-echo acquisition [18, 27].
Surprisingly, ADC did not statistically distinguish GBM from TE, nor did combining ADC with rCBV or CBF improve diagnostic accuracy [3]. However, in this study, only samples within T1w enhancement were included. There may be further utility in examining ADC in areas of hyperintense FLAIR without post-contrast T1 enhancement [13, 28]. Furthermore, whereas this study was restricted to two b-values, examining ADC collected with alternative or additional b-values may offer an improved characterization of GBM and TE. For example, higher b-values have shown promise in discerning true tumor progression following chemo-radiotherapy, such as in the study by Chu et al. in which a histogram analysis of ADC calculated from b-values of 0 and 3000 s/mm2 had better diagnostic performance than ADC calculated from b-values of 0 and 1000 s/mm2 [29].
Although not examined in this study, the metabolite concentrations identified with MR spectroscopy (MRS) have also been shown to be useful in distinguishing focused regions of TE from GBM [2, 3]. Additionally, dynamic contrast enhanced (DCE) and arterial spin labeling (ASL) imaging have shown utility in regional characterization, and in the future may prove to be additional accurate biomarkers for spatially characterizing GBM and TE [2, 30, 31].
There were several limitations in this study. The IDH status was not available for the subjects included in this study. It has been reported that rCBV is higher [32] and ADC lower [33] in those with IDH wildtype tumors, which is prevalent in approximately 90% of GBM cases [34]. Therefore, the results of this study likely appropriately reflect the general GBM population with a greater representation of IDH wildtype subjects. However, if possible IDH status should be accounted for in future studies. Additionally, though there were 44 tissue samples utilized, the number of participants included was based on available datasets and not a pre-determined power analysis. The number of samples utilized was also restricted since non-pure tissue types were intentionally not included, to allow for a well-defined comparison of two distinct groups. Yet, with the number of samples included, good statistical power was achieved with the use of GEE, which also accounts for multiple samples from the same subject, in differentiating GBM from TE. Still, further confirmation of the determined threshold is recommended using a greater number of correlated tissue and imaging samples.
As rCBV and CBF were co-registered to higher resolution anatomical images, the application of calculated thresholds is somewhat limited by resolution. Yet, with advances in parallel imaging, this should become less important. Similarly, brain shift and swelling or compression may also affect the accuracy of stereotactic co-localization of imaging to biopsy site [24]. To mitigate potential errors, samples were not collected for analysis when tissue swelling beyond the resection margins or sinking into the resection cavity occurred. Guided by the STEALTH navigational probe for confirmation, this was determined at the discretion of a neurosurgeon (WM) with 26 years of experience. Additionally, all tissue biopsy specimens were larger than the ROI used to extract imaging parameter values, which provides another level of compensation for any potential error due to brain shift or swelling, and diminishes potential errors caused by extracting values from lower resolution parameter maps at a higher resolution following co-registration.
Another possible limitation to this analysis is that the pure GBM samples were combined from patients with both treatment-naïve and treated GBM and therefore underlying biological differences may be present. Yet, combining data from both groups was justified by the fact that treated and untreated tumor were indiscernible based on H&E analysis and the imaging results from a sub-analysis of only treated-subject samples were equivalent to those obtained when the treated and untreated GBM populations were combined. In fact, the inclusions of both treated and untreated GBM may make the results from this study more robust in that they are more generally applicable throughout a patient’s treatment course and for a range of tumor conditions. Lastly, we also recognize that there may be inherent bias in selecting symptomatic patients, since these are the patients more likely to undergo surgery and thus have more aggressive tumor on imaging.
This study demonstrates that although thresholds for ADC, PH, and PSR could not be determined to spatially map areas of GBM and TE, thresholds for sRCBV, nRCBV, or nCBF could be determined enabling a more decisive representation of the lesion. Unlike histogram analysis or gross-regional metrics, a clear benefit of this approach is the additional visual assessment that may be provided to clinicians to determine tumor growth or response to treatment, in addition to the ability to quantitatively assess the percent of enhancing lesion that is GBM.
Conclusion
The ability to determine whether a patient with a brain tumor is responding to therapy or progressing has remained a difficult challenge. To date this assessment has been based primarily on the radiographic appearance of contrast-agent enhancing lesions on MRI, with disappointing results. Here we validate, with spatially-correlated tissue samples, that normalized and standardized cerebral blood volume (nRCBV, sRCBV) can distinguish tumor from treatment effect on a per-voxel basis, whereas ADC cannot. Knowledge of the proportion of enhancing lesion that represents tumor has the potential to impact treatment management decisions in daily clinical practice as well as for the evaluation of new therapies in clinical trials.
Funding
This study was funded by the National Institutes of Health / National Cancer Institute (R01 CA082500; U01 CA176110) and Advancing a Healthier Wisconsin.
References
Eisele SC, Wen PY, Lee EQ (2016) Assessment of brain tumor response: RANO and its offspring. Curr Treat Options Oncol 17:35. doi:10.1007/s11864-016-0413-5
Raimbault A, Cazals X, Lauvin MA, Destrieux C, Chapet S, Cottier JP (2014) Radionecrosis of malignant glioma and cerebral metastasis: a diagnostic challenge in MRI. Diagn Interv Imaging 95:985–1000. doi:10.1016/j.diii.2014.06.013
Verma N, Cowperthwaite MC, Burnett MG, Markey MK (2013) Differentiating tumor recurrence from treatment necrosis: a review of neuro-oncologic imaging strategies. Neuro Oncol 15:515–534. doi:10.1093/neuonc/nos307
Brandes AA, Tosoni A, Spagnolli F, Frezza G, Leonardi M, Calbucci F, Franceschi E (2008) Disease progression or pseudoprogression after concomitant radiochemotherapy treatment: pitfalls in neurooncology. Neuro Oncol 10:361–367. doi:10.1215/15228517-2008-008
Parvez K, Parvez A, Zadeh G (2014) The diagnosis and treatment of pseudoprogression, radiation necrosis and brain tumor recurrence. Int J Mol Sci 15:11832–11846. doi:10.3390/ijms150711832
Barajas RF Jr, Chang JS, Segal MR, Parsa AT, McDermott MW, Berger MS, Cha S (2009) Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 253:486–496. doi:10.1148/radiol.2532090007
Gahramanov S, Muldoon LL, Varallyay CG, Li X, Kraemer DF, Fu R, Hamilton BE, Rooney WD, Neuwelt EA (2013) Pseudoprogression of glioblastoma after chemo- and radiation therapy: diagnosis by using dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging with ferumoxytol versus gadoteridol and correlation with survival. Radiology 266:842–852. doi:10.1148/radiol.12111472
Nasseri M, Gahramanov S, Netto JP, Fu R, Muldoon LL, Varallyay C, Hamilton BE, Neuwelt EA (2014) Evaluation of pseudoprogression in patients with glioblastoma multiforme using dynamic magnetic resonance imaging with ferumoxytol calls RANO criteria into question. Neuro Oncol 16:1146–1154. doi:10.1093/neuonc/not328
Law M, Young RJ, Babb JS, Peccerelli N, Chheang S, Gruber ML, Miller DC, Golfinos JG, Zagzag D, Johnson G (2008) Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 247:490–498. doi:10.1148/radiol.2472070898
Al-Okaili RN, Krejza J, Woo JH, Wolf RL, O’Rourke DM, Judy KD, Poptani H, Melhem ER (2007) Intraaxial brain masses: MR imaging-based diagnostic strategy–initial experience. Radiology 243:539–550. doi:10.1148/radiol.2432060493
Khalifa J, Tensaouti F, Chaltiel L, Lotterie JA, Catalaa I, Sunyach MP, Ibarrola D, Noel G, Truc G, Walker P, Magne N, Charissoux M, Ken S, Peran P, Berry I, Moyal EC, Laprie A (2016) Identification of a candidate biomarker from perfusion MRI to anticipate glioblastoma progression after chemoradiation. Eur Radiol. doi:10.1007/s00330-016-4234-5
Law M, Yang S, Wang H, Babb JS, Johnson G, Cha S, Knopp EA, Zagzag D (2003) Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 24:1989–1998
Barajas RF Jr, Phillips JJ, Parvataneni R, Molinaro A, Essock-Burns E, Bourne G, Parsa AT, Aghi MK, McDermott MW, Berger MS, Cha S, Chang SM, Nelson SJ (2012) Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro Oncol 14:942–954. doi:10.1093/neuonc/nos128
Jain R, Gutierrez J, Narang J, Scarpace L, Schultz LR, Lemke N, Patel SC, Mikkelsen T, Rock JP (2011) In vivo correlation of tumor blood volume and permeability with histologic and molecular angiogenic markers in gliomas. AJNR Am J Neuroradiol 32:388–394. doi:10.3174/ajnr.A2280
Perry A, Schmidt RE (2006) Cancer therapy-associated CNS neuropathology: an update and review of the literature. Acta Neuropathol 111:197–212. doi:10.1007/s00401-005-0023-y
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A, Reifenberger G, von Deimling A (2016) World Health Organization classification of tumours of the central nervous system, 4th edn, chap 1. International Agency for Research on Cancer, pp 28–45
Boxerman JL, Prah DE, Paulson ES, Machan JT, Bedekar D, Schmainda KM (2012) The role of preload and leakage correction in gadolinium-based cerebral blood volume estimation determined by comparison with MION as a criterion standard. AJNR Am J Neuroradiol 33:1081–1087. doi:10.3174/ajnr.A2934
Paulson ES, Schmainda KM (2008) Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology 249:601–613 doi:10.1148/radiol.2492071659
Prah MA, Stufflebeam SM, Paulson ES, Kalpathy-Cramer J, Gerstner ER, Batchelor TT, Barboriak DP, Rosen BR, Schmainda KM (2015) Repeatability of standardized and normalized relative CBV in patients with newly diagnosed glioblastoma. AJNR Am J Neuroradiol 36:1654–1661. doi:10.3174/ajnr.A4374
Bedekar D, Jensen T, Schmainda KM (2010) Standardization of relative cerebral blood volume (rCBV) image maps for ease of both inter- and intrapatient comparisons. Magn Reson Med 64:907–913. doi:10.1002/mrm.22445
Boxerman JL, Paulson ES, Prah MA, Schmainda KM (2013) The effect of pulse sequence parameters and contrast agent dose on percentage signal recovery in DSC-MRI: implications for clinical applications. AJNR Am J Neuroradiol 34:1364–1369. doi:10.3174/ajnr.A3477
Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173
Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM (2012) FSL. Neuroimage 62:782–790. doi:10.1016/j.neuroimage.2011.09.015
LaViolette PS, Rand SD, Ellingson BM, Raghavan M, Lew SM, Schmainda KM, Mueller W (2011) 3D visualization of subdural electrode shift as measured at craniotomy reopening. Epilepsy Res 94:102–109. doi:10.1016/j.eplepsyres.2011.01.011
Hu LS, Eschbacher JM, Heiserman JE, Dueck AC, Shapiro WR, Liu S, Karis JP, Smith KA, Coons SW, Nakaji P, Spetzler RF, Feuerstein BG, Debbins J, Baxter LC (2012) Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol 14:919–930. doi:10.1093/neuonc/nos112
Paulson ES, Prah DE, Schmainda KM (2016) Spiral perfusion imaging with consecutive echoes (SPICE) for the simultaneous mapping of DSC- and DCE-MRI parameters in brain tumor patients: theory and initial feasibility. Tomography 2:295–307. doi:10.18383/j.tom.2016.00217
Schmainda KM, Prah M, Baxter LC, Paulson ES, Maze S, Pipe J, Wang D, Debbins J, Hu LS (2016) Simultaneous measurement of DSC- and DCE-MRI parameters using dual-echo spiral with a standard dose of gadolinium in comparison to single-echo GRE-EPI methods in brain tumors. Proceedings of the Twenty-Third Meeting of the International Society for Magnetic Resonance in Medicine #487. Mira Smart Conferencing, Toronto, Ontario, Canada
Eidel O, Burth S, Neumann JO, Kieslich PJ, Sahm F, Jungk C, Kickingereder P, Bickelhaupt S, Mundiyanapurath S, Baumer P, Wick W, Schlemmer HP, Kiening K, Unterberg A, Bendszus M, Radbruch A (2017) Tumor infiltration in enhancing and non-enhancing parts of glioblastoma: a correlation with histopathology. PLoS ONE 12:e0169292. doi:10.1371/journal.pone.0169292
Chu HH, Choi SH, Ryoo I, Kim SC, Yeom JA, Shin H, Jung SC, Lee AL, Yoon TJ, Kim TM, Lee SH, Park CK, Kim JH, Sohn CH, Park SH, Kim IH (2013) Differentiation of true progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide: comparison study of standard and high-b-value diffusion-weighted imaging. Radiology 269:831–840. doi:10.1148/radiol.13122024
Ye J, Bhagat SK, Li H, Luo X, Wang B, Liu L, Yang G (2016) Differentiation between recurrent gliomas and radiation necrosis using arterial spin labeling perfusion imaging. Exp Ther Med 11:2432–2436. doi:10.3892/etm.2016.3225
Larsen VA, Simonsen HJ, Law I, Larsson HB, Hansen AE (2013) Evaluation of dynamic contrast-enhanced T1-weighted perfusion MRI in the differentiation of tumor recurrence from radiation necrosis. Neuroradiology 55:361–369. doi:10.1007/s00234-012-1127-4
Kickingereder P, Sahm F, Radbruch A, Wick W, Heiland S, Deimling A, Bendszus M, Wiestler B (2015) IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 5:16238. doi:10.1038/srep16238
Xing Z, Yang X, She D, Lin Y, Zhang Y, Cao D (2017) Noninvasive assessment of IDH mutational status in World Health Organization grade II and III astrocytomas using DWI and DSC-PWI combined with conventional MR imaging. AJNR Am J Neuroradiol 38:1138–1144. doi:10.3174/ajnr.A5171
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. doi:10.1007/s00401-016-1545-1
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Ownership interest in Imaging Biometrics LLC (KMS). No conflict of interest to report (MAP, MMA, WMM, EJC, RGH, JMC).
Human Subjects Research
All procedures performed in this study were in accordance with the ethical standards of the Institution’s IRB and with the 1964 Helsinki declaration and its later amendments.
Animal Research
This article does not contain any studies with animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Prah, M.A., Al-Gizawiy, M.M., Mueller, W.M. et al. Spatial discrimination of glioblastoma and treatment effect with histologically-validated perfusion and diffusion magnetic resonance imaging metrics. J Neurooncol 136, 13–21 (2018). https://doi.org/10.1007/s11060-017-2617-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-017-2617-3