Abstract
Huntington's disease (HD) stands as a formidable challenge in modern medicine, characterized by progressive neurodegeneration and cognitive decline. A hallmark feature of HD pathology is the aggregation of mutant huntingtin protein (mHTT), leading to cellular dysfunction and eventual neuronal demise. Despite extensive research, therapeutic interventions for HD remain elusive. This article delves into the intricate thermodynamics underlying protein aggregation in HD, exploring key molecular mechanisms and potential therapeutic avenues. By comprehensively elucidating the thermodynamic principles governing mHTT aggregation, novel insights can be garnered to inform the development of effective therapeutic strategies.
Clinical Trial: No clinical trial.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Huntington's disease (HD), a progressive neurodegenerative disorder, has opened intensive research aimed at exploring its molecular complexities. Recent advancements in understanding HD pathogenesis, with a specific focus on mutant huntingtin protein aggregation, have ignited a paradigm shift in therapeutic strategies. This comprehensive review delves into multifaceted aspects, providing a detailed exploration of recent explorations and their implications for HD research. The molecular basis of HD pathology form the cornerstone of investigation, with recent studies elucidating detail interactions governing protein aggregation [1]. A key feature of Huntington's Disease is the formation of protein aggregates, known as inclusion bodies, within affected neurons. These aggregates primarily consist of the mutant huntingtin protein, but they also incorporate other cellular components, including chaperones, ubiquitin, and various signaling molecules. The aggregation process is highly complex, governed by a delicate balance of protein–protein interactions, post-translational modifications, and cellular environments. The precise mechanisms underlying protein aggregation in HD remain incompletely understood, but it is clear that multiple factors contribute to this pathological process [2,3,4,5,6]. Protein aggregation in Huntington's disease not only disrupts cellular homeostasis but also leads to a loss of function in critical cellular processes. The sequestration of essential proteins within inclusion bodies impairs their normal physiological roles, compromising the integrity of intracellular signaling pathways, protein degradation systems, and synaptic function. Furthermore, the accumulation of misfolded proteins activates cellular stress responses, triggering a cascade of cytotoxic events that contribute to neuronal dysfunction and cell death [7,8,9,10].
In recent years, the role of thermodynamics in protein aggregation has drawn increasing attention within the field of neurodegenerative diseases. Thermodynamics, the study of energy and its transformations, provides a powerful framework for understanding the behavior of biological macromolecules in health and disease. In the context of protein aggregation, thermodynamic analyses offer insights into the stability, kinetics, and structural characteristics of protein aggregates, shedding light on the factors that drive their formation and propagation. By understanding the thermodynamic forces underlying protein aggregation, researchers can develop predictive models to assess the impact of environmental variables and therapeutic interventions on disease progression [11,12,13,14,15,16]. The study of thermodynamics in Huntington's Disease has revealed intriguing insights into the mechanisms underlying protein aggregation and neuronal dysfunction. One key finding is the role of protein conformational entropy in driving the aggregation of mutant huntingtin protein. Conformational entropy refers to the degree of disorder or flexibility within a protein structure and plays a critical role in determining its stability and folding kinetics. In the case of mutant huntingtin protein, the expansion of the polyglutamine tract leads to increased conformational entropy, destabilizing the protein structure and promoting its aggregation. This insight has profound implications for the development of therapeutics targeting protein aggregation in HD, as it suggests that interventions aimed at reducing conformational entropy may inhibit the formation of toxic protein aggregates [17,18,19,20].
Another important aspect of thermodynamics in Huntington's Disease is the influence of environmental factors on protein aggregation kinetics. Studies have shown that changes in temperature, pH, and ionic strength can significantly alter the rate and extent of protein aggregation, suggesting that environmental conditions play a crucial role in modulating disease progression. For example, elevated temperatures can accelerate protein aggregation by increasing the rate of protein unfolding and misfolding, while changes in pH can alter the electrostatic interactions between protein molecules, affecting their propensity to aggregate. By elucidating the effects of environmental variables on protein aggregation, researchers can identify potential targets for therapeutic intervention and develop strategies to mitigate the progression of HD [21,22,23,24,25].
In addition to its role in protein aggregation, thermodynamics also plays a central role in understanding the energetics of cellular processes disrupted in Huntington's Disease. For example, thermodynamic analyses have provided insights into the ATP-dependent processes involved in protein folding, degradation, and synaptic transmission, all of which are disrupted in HD. By elucidating the thermodynamic principles underlying these processes, researchers can identify new therapeutic targets and develop strategies to restore cellular homeostasis in HD [26,27,28,29].
Huntington's Disease represents a complex interplay of genetic, molecular, and environmental factors, driving a cascade of cellular dysfunctions that ultimately culminate in neuronal degeneration. Through a thermodynamic exploration of protein aggregation and molecular pathogenesis, we gain a deeper understanding of the intricacies of Huntington's Disease and uncover new avenues for therapeutic intervention. By elucidating the thermodynamic forces underlying protein aggregation and cellular dysfunction in HD, we hope to develop novel treatments that can halt or even reverse the progression of this devastating disease, bringing hope to the millions of individuals and families affected by Huntington's Disease worldwide.
Molecular basis of protein aggregation
Structure and function of huntingtin protein
The Huntingtin protein (HTT) stands as a multifaceted player in the realm of cellular biology, its complexities expanding through various cellular processes with precision and significance. At the heart of its complexity lies the HTT gene, directing the production of a protein adorned with distinct domains, each contributing uniquely to its functionality.
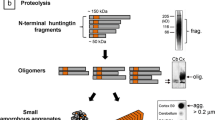
HTT, a large protein encoded by the HTT gene, opens a structural diversity characterized by several domains. Among these, the N-terminal domain commands attention with its polyglutamine (polyQ) stretch, a region whose length stands implicated in the onset and progression of HD. This domain serves as a nexus for interactions with an array of proteins pivotal in transcriptional regulation, vesicular transport, and the phenomenon of apoptosis, leading at the multifaceted role HTT plays within the cellular landscape. Moving along the protein's landscape, the central proline-rich domain emerges as a mediator of protein–protein interactions, facilitating HTT's engagement in the elaborate process of intracellular signaling pathways. Meanwhile, the C-terminal domain presents a homology with HEAT (Huntingtin, elongation factor 3, PR65/A, TOR) repeat motifs, known bastions of protein–protein interactions and guardians of protein folding integrity (Fig. 1) [30,31,32].
Ribosome occupancy was assessed on the mRNA transcript of cGAS in control, HD-het, and HD-homo cells. A Graphs depict the overlay of ribosome profiling (RPF) and mRNA readings for the cGAS (cyclic GMP-AMP synthase) transcript, obtained from the UCSC browser. Ribosome occupancy locations are indicated by arrows. B Inset highlights exon (Ex) 1 of the cGAS transcript, with arrows indicating ribosome occupancy. C Graphs illustrate the codon pause of cGAS transcripts predicted by the PausePred program, with arrows marking halted codon locations. D–G The graphs display the combination of RPF/mRNA readings for Aim2 (D), hnRNP-A2b1 (E), Sting (F), and Tbk1 (G) mRNA transcripts, obtained from the UCSC browser. Arrows indicate the expected interruption at exon 3 of Sting mRNA due to the introduction of a signal peptide into the endoplasmic reticulum (ER). The ribosome footprints shown in the figure represent data combined from all three replicates of control, HD-het, and HD-homo cells [30]
Functionally, HTT emerges as an initiator in the biochemistry of cellular processes. From the earliest stages of embryonic development to the detail intricacies of synaptic function and the directional movements of vesicular trafficking, HTT stands as a significant player, its presence shaping the landscape of cellular dynamics. Its interactions with an expansive repertoire of proteins regulate transcriptional cascades, navigate the pathways of intracellular transport, and fine-tune the mechanism of cell signaling pathways. Moreover, HTT's involvement in the regulation of autophagy, a vital cellular process governing protein degradation and maintaining homeostasis, underscores its indispensability in cellular equilibrium. Dysregulation of HTT, particularly originating from the expansion of the polyQ tract, emerges as an indicator of HD, a relentless threat to neural integrity [33,34,35].
Mutant huntingtin protein: conformational dynamics
The huntingtin protein is a large, multifunctional protein that plays essential roles in various cellular processes, including vesicle trafficking, transcriptional regulation, and synaptic function. In its normal form, huntingtin contains a polyglutamine (polyQ) tract with 6–35 glutamine residues. However, in individuals with HD, an abnormal expansion of the polyQ tract beyond 36 glutamine repeats leads to the production of mHTT [35,36,37].
Structural studies using advanced biophysical techniques have provided insights into the structural features of mHTT. X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR) spectroscopy have revealed that mHTT adopts a predominantly disordered structure, with regions of local secondary structure elements, including α-helices and β-strands (Fig. 2). The expanded polyQ tract in mHTT confers conformational instability, promoting the formation of β-sheet-rich structures, which are characteristic of amyloid fibrils observed in HD brains. One of the defining features of mHTT is its conformational flexibility, which allows it to sample a diverse range of structural states. Molecular dynamics simulations and biophysical experiments have demonstrated that mHTT undergoes rapid conformational fluctuations, transitioning between different structural ensembles. These conformational dynamics are influenced by factors such as temperature, pH, and the presence of co-factors and ligands (Fig. 2) [38, 39].
The CR-I-TASSER pipeline operates by utilizing a query sequence and a cryo-EM density map to produce atomic models through three sequential stages: (1) In the initial step, the original data is processed to form a 3D-CNN Cα conformation. This process involves LOMETS threading and ResPRE contact-map prediction, (2) The second stage involves template selection and trace generation based on the density map, (3) Finally, in the third step, fragment reassembly simulations are guided by the density map to refine and improve the model [39]
The expanded polyQ tract in mHTT plays a critical role in modulating its conformational dynamics. The polyQ region acts as a dynamic "molecular spring," undergoing transient interactions with neighboring regions within the protein. These interactions can stabilize or destabilize specific structural motifs, leading to the formation of transiently populated intermediate states. The conformational landscape of mHTT is further influenced by post-translational modifications (PTMs), such as phosphorylation, acetylation, and ubiquitination, which regulate its stability and aggregation propensity. The conformational dynamics of mHTT have profound implications for its propensity to misfold and aggregate. Misfolding occurs when mHTT adopts non-native conformations that expose hydrophobic residues, promoting aberrant protein–protein interactions. The formation of oligomeric species, characterized by the assembly of multiple mHTT molecules, represents a crucial step in the pathogenesis of HD. These oligomers serve as nucleation sites for the subsequent formation of larger aggregates, including soluble protofibrils and insoluble amyloid fibrils, which are hallmarks of HD pathology [40, 41]. The toxicity of mHTT oligomers is attributed to their ability to disrupt cellular homeostasis through various mechanisms. Oligomers can perturb membrane integrity, leading to membrane leakage and cellular dysfunction. Additionally, oligomers can sequester essential cellular components, including molecular chaperones and signaling proteins, impairing their normal function. Furthermore, oligomers can induce cellular stress responses, such as oxidative stress and endoplasmic reticulum (ER) stress, exacerbating neuronal damage and cell death.
Targeting the conformational dynamics of mHTT represents a promising therapeutic approach for HD. By stabilizing the native conformation of huntingtin or inhibiting the formation of toxic oligomers, it may be possible to mitigate the progression of HD pathology. Small molecules, peptides, and antibodies that selectively bind to specific conformational states of mHTT have been investigated as potential therapeutic agents. One strategy involves targeting the expanded polyQ tract in mHTT to prevent its conformational transition to β-sheet-rich structures. Small molecules, such as peptidomimetics and small interfering RNAs (siRNAs), have been designed to specifically interact with the polyQ region, stabilizing its native conformation and inhibiting aggregation. Additionally, molecular chaperones, such as heat shock proteins (HSPs) and protein disulfide isomerases (PDIs), have been shown to modulate the conformational dynamics of mHTT, promoting its refolding and clearance [42, 43].
Another therapeutic approach focuses on disrupting protein–protein interactions involved in mHTT aggregation. Peptides and small molecules that target specific regions within mHTT, such as the N-terminal domain or the polyproline-rich region, have been developed to inhibit oligomerization and fibrillogenesis. Furthermore, antibodies that recognize conformational epitopes on mHTT have shown promise in selectively targeting toxic oligomers for clearance by the immune system.
Pathological implications of mHTT aggregation
The aggregation of mutant huntingtin (mHTT) protein is a hallmark pathological feature of Huntington's disease (HD). These aggregated species, including soluble oligomers and insoluble fibrils, contribute to cellular dysfunction and neurodegeneration in HD through various mechanisms. Firstly, mHTT aggregates disrupt cellular proteostasis by sequestering essential cellular components, including molecular chaperones and proteasomal machinery. This impairs protein quality control mechanisms, leading to the accumulation of misfolded proteins and further exacerbating cellular dysfunction. Moreover, mHTT aggregates induce cellular toxicity through a variety of mechanisms, including mitochondrial dysfunction, oxidative stress, and excitotoxicity. These aggregates can directly interact with cellular membranes and organelles, disrupting their structure and function. Additionally, mHTT aggregates impair axonal transport and synaptic function, leading to synaptic dysfunction and neuronal degeneration (Fig. 3) [44,45,46,47].
Reducing mutant huntingtin (mHtt) levels enhances protein synthesis and accelerates ribosomal movement along the mRNA strand. Immunoblots were conducted on mouse striatal cells lacking Htt using CRISPR/Cas9, following puromycin metabolic labeling (Experiment A). Quantification of blots from Experiment A determined the levels of Htt (B) and puromycin incorporation (C) in control (Ctrl) and Htt-depleted control, HD-homo striatal cells. Data represent the mean ± standard error of the mean (SEM) from nine different experiments. Statistical analysis was conducted using one-way ANOVA followed by Bonferroni's multiple comparisons test, with significance levels indicated as ***P < 0.001 and ****P < 0.0001. Polysome profiles (D-F) were obtained from mouse striatal cells lacking wild-type Htt. Profiles were acquired before (basal, E) and after performing a ribosome run-off test using harringtonine (2 μg/ml, 2 min, F). Corresponding quantification of polysome to monosome (PS/MS) ratios (area under the curve) was also determined. Data are expressed as mean ± SEM from three different experiments. Statistical analysis was performed using a two-tailed Student's t-test, with significance indicated by *P < 0.05. Polysome profiles (G-I) of mouse striatal cells lacking mHtt were acquired before (basal) and after performing a ribosome run-off test using harringtonine for 2 min. Corresponding quantification of polysome to monosome ratios (PS/MS) was determined. Data represent the mean ± SEM and were obtained from three separate experiments for condition H and four independent experiments for condition I. Statistical analysis was performed using a two-tailed Student's t-test, with significance set at P < 0.01. "n.s" indicates nonsignificance. Polysome profiles (J) of mouse striatal cells depleted of both wtHtt and mHtt were obtained using a ribosome run-off test with harringtonine (2 min). Associated polysome to monosome (PS/MS) ratios (K) were quantified. Data represent the mean ± SEM for two groups: wtHtt-depleted (n = 5) and mHtt-depleted (n = 6) from separate experiments. Statistical analysis was performed using a two-tailed Student's t-test, with significance set at P < 0.01. The value of n is not statistically significant [44]
Furthermore, mHTT aggregates propagate pathology through a prion-like mechanism, where misfolded protein species induce the conversion of native proteins into pathological conformations. This leads to the spread of pathology throughout the brain, contributing to disease progression and the widespread neuronal dysfunction observed in HD. Understanding the pathological implications of mHTT aggregation is essential for developing therapeutic strategies aimed at halting or reversing disease progression in HD. Targeting mHTT aggregation represents a promising therapeutic approach, with several strategies under investigation, including small molecule inhibitors, immunotherapies, and gene silencing techniques. By elucidating the mechanisms underlying mHTT aggregation and its pathological consequences, researchers can identify novel therapeutic targets and develop interventions to effectively mitigate the devastating effects of HD on patients' lives [48, 49].
Thermodynamic principles of protein aggregation
Gibbs free energy and protein folding
Understanding the dynamics of life at the molecular level requires a profound exploration of two fundamental concepts: Gibbs free energy and Protein Folding. These concepts, deeply rooted in the principles of thermodynamics and molecular biology, underpin the structural integrity, functionality, and stability of living organisms.
Gibbs free energy, symbolized as ∆G, stands as a cornerstone in the realm of thermodynamics, reflecting the energy available in a system to perform work under constant temperature and pressure. Named after the eminent physicist Josiah Willard Gibbs, this metric serves as a critical gauge for the spontaneity of chemical reactions within biological systems (Fig. 4). A negative ∆G signifies a reaction that occurs spontaneously, releasing energy into its surroundings, while a positive ∆G indicates a reaction that requires an input of energy to proceed. The relationship between Gibbs free energy (∆G), enthalpy (∆H), and entropy (∆S) elucidates the delicate balance between the enthalpic and entropic contributions to the overall spontaneity of a reaction [50,51,52].
Adjusts to changes in the unfolding free energy for medium and large PEGs (a, b). Concentration charts depicting the relationship between enthalpy and entropy for PEG400 and PEG8000 are displayed in (c). Similarly, the corresponding plot for enthalpy and entropy is presented in (d). Experimental results obtained at a temperature of 298 K are represented by triangular shapes [50]
Protein Folding, on the other hand, represents the intricate process through which a linear chain of amino acids assumes its functional three-dimensional structure, crucial for its biological activity. Proteins, the workhorses of biological systems, exhibit an astonishing array of functions, ranging from catalyzing chemical reactions to providing structural support. Achieving the native conformation, characterized by precise folding patterns, is essential for the protein's functionality. This process, guided by various forces including hydrogen bonding, van der Waals interactions, hydrophobic effect, and electrostatic interactions, navigates through a vast conformational space to reach the global energy minimum, representing the native state.
The profound intersection between Gibbs free energy and Protein Folding becomes evident as thermodynamic principles govern the folding behavior of proteins. The minimization of free energy drives the folding process towards the native state, wherein the protein adopts a conformation corresponding to the lowest free energy state. The hydrophobic effect, a key player in protein folding, drives nonpolar amino acid residues to bury themselves in the protein's interior, contributing significantly to the favorable entropy of the system. Additionally, hydrogen bonding and other non-covalent interactions stabilize specific secondary structures, such as alpha helices and beta sheets, further enhancing protein stability [53,54,55].
However, protein folding is not always a smooth process, as proteins can often misfold or aggregate, leading to the formation of non-functional or toxic species. Factors such as mutations, environmental conditions, and cellular stress can perturb the folding landscape, altering the delicate balance of forces that govern protein stability.
Entropy and disorder in aggregation
Entropy, a measure of disorder or randomness in a system, is a central concept in understanding protein aggregation. When proteins are in their native state, they are highly ordered structures with specific three-dimensional conformations that allow them to perform their biological functions. However, external factors such as changes in temperature, pH, or the presence of denaturing agents can disrupt these structures, leading to the exposure of hydrophobic regions that are normally buried in the protein core.
The exposure of hydrophobic regions is a key step in protein aggregation. Hydrophobic interactions, which are driven by entropy, play a major role in the aggregation process. When hydrophobic regions are exposed, water molecules in the surrounding environment become ordered around these regions, reducing the entropy of the system. To counteract this decrease in entropy, proteins may undergo conformational changes or associate with other proteins to form aggregates, where the hydrophobic regions are sequestered away from the water molecules. The formation of protein aggregates represents a trade-off between enthalpy and entropy. Enthalpy, a measure of the internal energy of a system, tends to favor the formation of stable, ordered structures. However, the increase in entropy associated with the dispersal of hydrophobic regions can drive the aggregation process, even if it leads to the formation of less ordered structures. This balance between enthalpy and entropy determines the thermodynamic stability of protein aggregates [56,57,58].
The thermodynamic stability of protein aggregates can be further influenced by factors such as the concentration of proteins, the presence of chaperone proteins, and the specific amino acid sequences of the proteins involved. For example, proteins with highly amyloidogenic sequences, which are prone to forming β-sheet structures, are more likely to aggregate than proteins with non-amyloidogenic sequences. Thus, the thermodynamic principles of protein aggregation are governed by entropy and disorder. The exposure of hydrophobic regions, driven by entropy, plays a crucial role in initiating the aggregation process. The balance between enthalpy and entropy determines the stability of protein aggregates, with factors such as protein concentration and amino acid sequence also influencing the aggregation propensity (Fig. 5). Understanding these thermodynamic principles is essential for elucidating the mechanisms of protein aggregation and developing strategies to prevent or treat diseases associated with protein misfolding and aggregation [59,60,61].
A case of thermal hysteresis is observed for TBA-PhA12 (CT = 370 μM). Thermal hystereses are evident at various concentrations at the bottom [59]
Enthalpic contributions to protein aggregation
Enthalpy is a thermodynamic quantity that represents the internal energy of a system. In the context of proteins, enthalpy plays a crucial role in determining the stability of protein structures. Proteins are folded into specific three-dimensional structures, known as native conformations, which are stabilized by a variety of interactions, including hydrogen bonding, electrostatic interactions, and van der Waals forces. These interactions contribute to the enthalpy of the protein and help maintain its folded state [50].
During the process of protein aggregation, proteins may undergo conformational changes that lead to the exposure of hydrophobic regions, which are normally buried in the protein core. These conformational changes can be driven by external factors such as changes in pH, temperature, or the presence of denaturing agents. The exposure of hydrophobic regions can lead to the formation of intermediate structures, such as molten globules, which are partially folded states with increased solvent accessibility of hydrophobic residues [62, 63].
Hydrophobic interactions, which are driven by the tendency of nonpolar molecules to minimize their contact with water, also play a significant role in enthalpic contributions to protein aggregation. When hydrophobic regions are exposed due to conformational changes or unfolding of proteins, these regions tend to interact with other exposed hydrophobic regions on neighboring proteins, leading to the formation of hydrophobic clusters. The formation of these clusters is enthalpically favorable, as it allows the hydrophobic residues to minimize their contact with water [64]. The enthalpic contributions to protein aggregation can influence the stability and structure of protein aggregates. In some cases, the interactions between exposed hydrophobic regions can lead to the formation of highly ordered structures, such as β-sheet-rich aggregates, which are stabilized by a network of hydrogen bonds between neighboring strands. These structures are often observed in amyloid fibrils, which are associated with several neurodegenerative diseases.
Several factors can influence the enthalpic contributions to protein aggregation. For example, the specific amino acid sequence of a protein can affect its propensity to form aggregates. Proteins with sequences that promote the formation of stable secondary structures, such as β-sheets, are more likely to form aggregates than proteins with sequences that favor the formation of α-helices. Additionally, environmental factors such as pH and temperature can also influence the enthalpic contributions to protein aggregation [64,65,66].
Kinetics of protein aggregation: nucleation and growth
Nucleation is the initial step in protein aggregation and involves the formation of small, stable aggregates known as nuclei (Fig. 6). Nucleation can occur through two main pathways: primary nucleation, which involves the spontaneous aggregation of individual proteins, and secondary nucleation, which involves the aggregation of proteins on the surface of existing aggregates. Primary nucleation is typically a slow process, as it involves the formation of stable nuclei from monomeric proteins. Secondary nucleation, on the other hand, can occur more rapidly, as it involves the growth of existing aggregates through the addition of monomeric proteins [57, 67].
Graphs displaying aggregation process rates reveal the widespread occurrence of self-replication. The rate of secondary route aggregate production is plotted against the rate of main pathway aggregate production. A dashed line indicates points where the rates of the two processes are equal. The diagram distinguishes between systems dominated by primary nucleation (bottom right corner) and those where secondary processes predominate (top left corner). In systems with dominant primary nucleation and slow secondary processes, only an upper limit for secondary route rates can be determined. Similarly, if primary nucleation is slow and requires seeding, only an upper limit for the primary rate can be established, illustrated by elongated points. Proteins are categorized into three classes: pathogenic amyloids (bottom left), functioning amyloids (bottom right), and non-amyloid forming proteins under normal physiological circumstances (top right). Labels are positioned either above or to the right of the associated data point [57]
The kinetics of nucleation can be described by the nucleation rate, which is the rate at which nuclei are formed, and the critical nucleus size, which is the minimum number of proteins required to form a stable nucleus. The nucleation rate is influenced by factors such as protein concentration, temperature, and the presence of other molecules that can interact with the proteins. The critical nucleus size is determined by the stability of the nucleus, which is affected by the interactions between the proteins in the nucleus and the surrounding environment. Once nuclei are formed, they can grow through the addition of monomeric proteins. The kinetics of growth depend on the rate at which monomeric proteins can associate with the nuclei and the rate at which the nuclei can incorporate these proteins into their structure. The growth rate is influenced by factors such as protein concentration, temperature, and the stability of the nuclei. In some cases, growth can be limited by the diffusion of monomeric proteins to the surface of the nuclei, particularly in crowded environments where the movement of proteins is restricted [68, 69].
The kinetics of protein aggregation can be described by mathematical models that take into account the processes of nucleation and growth. One commonly used model is the classical nucleation and growth model, which describes the kinetics of aggregation as a two-step process involving the formation of nuclei followed by their growth. The rate of aggregation is determined by the rates of nucleation and growth, as well as the stability of the nuclei [70,71,72].
Thermodynamic drivers of MHTT aggregation
Role of hydrophobic interactions
Protein aggregation is a nucleation-dependent process involving the self-assembly of proteins into insoluble fibrillar structures. Hydrophobic interactions, along with other forces such as electrostatic interactions, hydrogen bonding, and van der Waals forces, play a crucial role in protein folding and stability (Table 1). In the context of protein aggregation, hydrophobic interactions are particularly important, as they drive the association of hydrophobic regions of proteins, leading to the formation of aggregates [73, 34, 74].
The expanded polyQ tract in mHTT is highly hydrophobic, contributing to the protein's propensity to aggregate. The hydrophobic nature of the polyQ tract promotes the exposure of hydrophobic patches on mHTT, facilitating the formation of intermolecular hydrophobic interactions between adjacent protein molecules. These interactions drive the initial nucleation events in mHTT aggregation, leading to the formation of oligomeric species [17].
The driving force behind hydrophobic interactions is the hydrophobic effect, which arises from the tendency of water molecules to exclude nonpolar molecules from their vicinity. When hydrophobic regions of proteins are exposed to water, water molecules reorganize to minimize their contact with these regions, leading to a decrease in entropy. The formation of hydrophobic aggregates allows water molecules to regain entropy by minimizing their contact with the hydrophobic surfaces, thereby driving the aggregation process.
The thermodynamics of mHTT aggregation can be understood in the context of the interplay between enthalpy and entropy changes. The association of mHTT molecules through hydrophobic interactions is enthalpically favorable, as it allows for the formation of energetically stable aggregates. However, this association is accompanied by an entropy loss due to the ordering of water molecules around the hydrophobic regions of mHTT. The net effect of these opposing forces determines the overall thermodynamic feasibility of mHTT aggregation [75, 76].
Understanding the role of hydrophobic interactions in mHTT aggregation is crucial for the development of therapeutics for HD. Various approaches have been explored to modulate hydrophobic interactions and inhibit mHTT aggregation, including the use of small molecules and peptides that target the hydrophobic regions of mHTT. By disrupting hydrophobic interactions, these compounds can prevent the formation of toxic mHTT aggregates, offering a potential therapeutic strategy for HD.
Electrostatic forces in protein aggregation
Proteins are complex molecules with both positive and negative charges distributed throughout their structure. Electrostatic forces, which arise from the interaction between these charges, can significantly impact protein behavior, including aggregation. Electrostatic interactions can be attractive or repulsive, depending on the charges involved and the distance between them. In protein aggregation, electrostatic forces primarily arise from the charged amino acid residues present on the protein surface. These forces can drive protein–protein interactions, leading to the formation of aggregates. The strength of electrostatic interactions depends on factors such as the charge distribution, the dielectric constant of the medium, and the ionic strength of the solution.
Electrostatic forces can influence protein aggregation in several ways. Attractive electrostatic interactions between proteins can promote the formation of aggregates by bringing individual protein molecules into close proximity. On the other hand, repulsive electrostatic interactions can prevent aggregation by destabilizing protein–protein interactions. In neurodegenerative diseases such as Alzheimer's and Parkinson's, protein aggregation is a hallmark pathology. Electrostatic forces are thought to play a role in the aggregation of proteins such as amyloid-beta and alpha-synuclein, which are associated with these diseases. Understanding the role of electrostatic forces in protein aggregation could lead to new therapeutic strategies for these conditions [77, 78].
Several strategies can be employed to modulate electrostatic forces and mitigate protein aggregation. For example, altering the pH or ionic strength of the solution can affect the strength of electrostatic interactions (Fig. 7). Additionally, introducing mutations that change the charge distribution on the protein surface can alter the propensity for aggregation. Various factors influence the strength and nature of electrostatic forces in protein aggregation. The distribution of charged residues on the protein surface, the dielectric constant of the medium, and the ionic strength of the solution all play crucial roles. Proteins with a high density of charged residues are more likely to experience strong electrostatic interactions, leading to increased aggregation propensity. In a high dielectric constant medium like water, electrostatic interactions are weakened, making it easier for proteins to aggregate. Conversely, in a low dielectric constant medium, such as an organic solvent, electrostatic interactions are stronger, promoting protein aggregation. The ionic strength of the solution also affects electrostatic interactions, with high ionic strengths screening electrostatic interactions and reducing aggregation propensity. The role of electrostatic forces extends beyond driving the formation of aggregates; they also influence the structure of the resulting aggregates. In some cases, electrostatic interactions can lead to the formation of highly ordered aggregates, such as amyloid fibrils, which are associated with several neurodegenerative diseases. The specific arrangement of charged residues within these aggregates can affect their stability and toxicity [27, 79, 80].
Water removal over time via the dewatering process was observed for pure CNF suspensions at pH 3 (A). The investigation focuses on determining the rate of dewatering for various grades of cellulose nanofibrils (CNF) under different chemical conditions (B). The dewatering rate is assessed by calculating the gradient between the points where 10% and 30% of the water has been removed [79]
Therapeutic strategies targeting protein aggregation
Small molecule inhibitors
Several theories have been proposed to explain the mechanisms of protein aggregation. The nucleation-dependent polymerization model suggests that the aggregation process is initiated by the formation of a critical nucleus, followed by the elongation of fibrils through the addition of monomeric proteins. The nucleation-elongation model proposes that the aggregation process is driven by the sequential addition of monomeric proteins to preformed oligomers, leading to the formation of fibrils. Other models, such as the template-assisted mechanism, suggest that pre-existing aggregates or fibrils serve as templates for the recruitment and aggregation of monomeric proteins [81, 82].
Small molecule inhibitors are compounds that can bind to specific target proteins and modulate their function. In the context of protein aggregation, small molecule inhibitors can target various stages of the aggregation process, including protein misfolding, oligomerization, and fibril formation. These inhibitors can act by stabilizing the native conformation of proteins, preventing their aggregation into toxic oligomers and fibrils, or promoting the clearance of aggregates by enhancing cellular proteostasis mechanisms.
The development of small molecule inhibitors for protein aggregation is a challenging task due to the complex nature of the aggregation process and the structural diversity of target proteins. However, advances in computational modeling, high-throughput screening, and structure-based drug design have enabled the identification of promising lead compounds for further optimization [83]. One approach to developing small molecule inhibitors is to screen libraries of chemical compounds for their ability to inhibit protein aggregation in vitro. Compounds that show promising inhibitory activity can then be further optimized for improved potency, selectivity, and pharmacokinetic properties. Structure–activity relationship (SAR) studies can help identify key structural features required for inhibitory activity and guide the design of more potent inhibitors [84, 85]. Another approach is to use computational methods, such as molecular docking and virtual screening, to identify potential small molecule inhibitors that can bind to specific target proteins involved in the aggregation process. These computational approaches can rapidly screen large libraries of chemical compounds and prioritize lead compounds for experimental validation.
Small molecule inhibitors targeting protein aggregation hold great promise as potential therapeutics for neurodegenerative diseases. By inhibiting the formation of toxic protein aggregates, these inhibitors have the potential to slow or halt disease progression and preserve neuronal function. Several small molecule inhibitors targeting different aspects of the aggregation process have shown promising results in preclinical studies and are currently being evaluated in clinical trials. For example, small molecule inhibitors targeting the beta-amyloid protein, which forms plaques in the brains of Alzheimer's patients, have shown potential in reducing beta-amyloid aggregation and improving cognitive function in animal models of the disease. Similarly, small molecule inhibitors targeting alpha-synuclein, a protein implicated in Parkinson's disease, have shown promise in reducing alpha-synuclein aggregation and neurotoxicity in animal models [86,87,88,89].
Chaperone-mediated protein folding
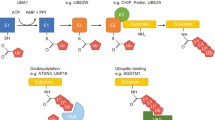
Chaperone-mediated protein folding is a critical cellular process that helps proteins fold correctly and prevents aggregation (Table 2). Chaperones are a diverse group of proteins that assist in protein folding by interacting with unfolded or misfolded proteins, stabilizing intermediates, and facilitating correct folding. There are several classes of chaperones, including Hsp70, Hsp90, and the chaperonins (GroEL/GroES in bacteria and CCT/TRiC in eukaryotes), each with specific functions in protein folding [90, 91].
Hsp70 chaperones, such as DnaK in bacteria and Hsp70 in eukaryotes, bind to exposed hydrophobic regions of unfolded or misfolded proteins, preventing them from aggregating and facilitating their folding into the correct conformation. Hsp70 chaperones work in conjunction with co-chaperones, such as Hsp40, which help deliver substrates to Hsp70 and stimulate its ATPase activity. Hsp90 chaperones, such as Hsp90 in eukaryotes, play a crucial role in the folding and stabilization of many signaling proteins and transcription factors. Hsp90 interacts with a large number of client proteins, and its activity is regulated by co-chaperones, such as Hsp70 and Hsp40, as well as by post-translational modifications. Chaperonins, such as GroEL/GroES in bacteria and CCT/TRiC in eukaryotes, provide a protected environment for protein folding, allowing them to fold correctly without the risk of aggregation. Chaperonins function by encapsulating unfolded or partially folded proteins within a barrel-like structure, providing a folding chamber where proteins can fold undisturbed. Therapeutic strategies targeting protein aggregation often aim to modulate chaperone activity to promote the correct folding of proteins and prevent aggregation. Small molecules known as pharmacological chaperones can stabilize folding intermediates and facilitate correct folding, thereby preventing aggregation. For example, the small molecule 4-phenylbutyrate has been shown to stabilize the mutant cystic fibrosis transmembrane conductance regulator (CFTR) protein and promote its correct folding and trafficking to the cell surface [92, 93].
Another approach is to enhance the expression or activity of endogenous chaperones using genetic or pharmacological methods. For example, overexpression of Hsp70 has been shown to reduce aggregation of mutant huntingtin protein, the causative agent of Huntington's disease, in cell and animal models. Similarly, upregulation of Hsp90 has been shown to promote the degradation of misfolded proteins associated with neurodegenerative diseases. Thus, chaperone-mediated protein folding is a crucial cellular process that helps proteins fold correctly and prevents aggregation. Therapeutic strategies targeting protein aggregation often aim to modulate chaperone activity to promote correct folding and prevent aggregation. Further research into the mechanisms of chaperone-mediated protein folding and the development of novel therapeutic approaches targeting protein aggregation are warranted to combat protein misfolding diseases.
Immunotherapy approaches
Protein misfolding and aggregation are hallmark features of several neurodegenerative diseases, including Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD), as well as other proteinopathies such as amyotrophic lateral sclerosis (ALS) and prion diseases. These diseases are characterized by the accumulation of misfolded proteins in the brain, leading to the formation of insoluble aggregates and the progressive loss of neuronal function. Traditional therapeutic strategies for these diseases have focused on targeting the symptoms or downstream effects of protein aggregation, with limited success in halting or reversing disease progression. Immunotherapy has emerged as a promising therapeutic approach for targeting protein aggregation in neurodegenerative diseases. The basic premise of immunotherapy is to harness the body's immune system to target and clear pathological proteins, such as amyloid-beta (Aβ) in AD, alpha-synuclein (α-syn) in PD, and mutant huntingtin (mHTT) in HD, thereby slowing or halting disease progression. Several immunotherapy approaches have been explored, including passive immunization with monoclonal antibodies (mAbs), active immunization with vaccines, and modulation of the immune response.
Passive immunization involves the administration of pre-formed antibodies that specifically target pathological proteins. In the case of AD, several monoclonal antibodies targeting Aβ have been developed and tested in clinical trials. For example, aducanumab, a human monoclonal antibody that targets aggregated forms of Aβ, showed promising results in early clinical trials, leading to its approval by the FDA for the treatment of AD. Similarly, monoclonal antibodies targeting α-syn and mHTT are also being developed and tested for their efficacy in PD and HD, respectively. Active immunization, or vaccination, involves the administration of antigens that stimulate the immune system to produce antibodies against pathological proteins. In AD, active immunization with Aβ peptides has been investigated as a potential therapeutic approach. However, clinical trials of Aβ vaccines, such as AN1792, have shown mixed results, with some trials being halted due to safety concerns. Despite these challenges, research in this area continues, with efforts focused on improving vaccine design and delivery to enhance efficacy and safety [94, 95].
Modulation of the immune response is another approach to immunotherapy that has been explored in the context of protein aggregation diseases. One strategy is to use small molecules or biologics to modulate the activity of immune cells, such as microglia, which play a key role in clearing misfolded proteins from the brain. For example, drugs that target the TREM2 pathway, which is involved in microglial activation and phagocytosis, are being investigated as potential therapeutics for AD and other neurodegenerative diseases. Hence, immunotherapy holds great promise as a therapeutic approach for targeting protein aggregation in neurodegenerative diseases. Passive immunization with monoclonal antibodies, active immunization with vaccines, and modulation of the immune response are all viable strategies for targeting pathological proteins and slowing disease progression. Further research is needed to optimize these approaches and develop safe and effective immunotherapies for neurodegenerative diseases.
Gene silencing techniques
Gene silencing techniques offer a promising approach for targeting protein aggregation by reducing the production of the toxic proteins involved in disease pathogenesis. Several gene silencing approaches have been developed, including RNA interference (RNAi), antisense oligonucleotides (ASOs), and CRISPR-Cas9 gene editing, each with unique mechanisms and applications in neurodegenerative disease therapy.
RNA interference (RNAi) is a natural cellular process for regulating gene expression. In RNAi, double-stranded RNA molecules called small interfering RNAs (siRNAs) bind to target messenger RNA (mRNA) molecules, leading to their degradation and subsequent reduction in protein expression. RNAi has been widely used to silence genes associated with neurodegenerative diseases, including genes encoding amyloid-beta (Aβ) in AD, alpha-synuclein (α-syn) in PD, and mutant huntingtin (mHTT) in HD. Antisense oligonucleotides (ASOs) are synthetic single-stranded nucleic acid molecules that bind to target mRNA molecules through Watson–Crick base pairing. Once bound, ASOs can induce mRNA degradation, inhibit translation, or modulate alternative splicing, depending on their design and target sequence. ASOs have been used to target genes involved in neurodegenerative diseases, such as SOD1 in ALS and tau in AD, with promising results in preclinical and clinical studies [96, 97]. CRISPR-Cas9 gene editing is a powerful tool for targeted gene silencing and modification. CRISPR-Cas9 uses a guide RNA (gRNA) to direct the Cas9 nuclease to specific DNA sequences, where it induces double-strand breaks (DSBs) that can lead to gene knockout or modification through error-prone repair mechanisms. CRISPR-Cas9 has been used to silence genes associated with neurodegenerative diseases, including APP and BACE1 in AD, and has shown promise in preclinical studies. One of the main challenges in using gene silencing techniques for therapeutic purposes is the delivery of nucleic acid molecules to the target cells in the brain. Several delivery strategies have been developed to overcome this challenge, including viral vectors, lipid nanoparticles, and cell-penetrating peptides, which can efficiently deliver siRNAs, ASOs, and CRISPR-Cas9 components to the brain [98,99,100].
Hence, gene silencing techniques offer a promising approach for targeting protein aggregation in neurodegenerative diseases. RNA interference, antisense oligonucleotides, and CRISPR-Cas9 gene editing can be used to reduce the production of toxic proteins involved in disease pathogenesis, potentially slowing or halting disease progression. Further research is needed to optimize these approaches and develop safe and effective gene silencing therapies for neurodegenerative diseases.
Challenges and future directions
Understanding the thermodynamics of protein aggregation in Huntington's disease is a complex and challenging endeavor. Many proteins implicated in Huntington's disease undergo post-translational modifications or require co-factors for their aggregation. Investigating how these factors influence the thermodynamics of aggregation could provide new insights into potential therapeutic targets.
Current methods for studying protein aggregation kinetics and thermodynamics are often time-consuming and labor-intensive. Developing high-throughput screening methods could accelerate the discovery of small molecules or peptides that modulate protein aggregation. Computational models play a crucial role in understanding the thermodynamics of protein aggregation. Future efforts should focus on improving the accuracy of these models by incorporating more detailed molecular interactions and experimental data. Given the complexity of protein aggregation in Huntington's disease, targeting multiple pathways simultaneously may be more effective than single-target approaches. Future research should explore the thermodynamic implications of multi-target therapeutic strategies.
While significant progress has been made in understanding the thermodynamics of protein aggregation, translating these findings into clinically effective therapeutics remains a challenge. Future research should focus on developing strategies to translate basic research into clinical applications. Developing therapeutics for Huntington's disease requires consideration of safety and pharmacokinetic profiles. Future research should focus on developing compounds with favorable safety profiles and pharmacokinetics for clinical use. Immunotherapy approaches targeting protein aggregation have shown promise in preclinical studies. Future research should focus on further exploring the thermodynamic aspects of these approaches and translating them into clinical applications. Gene silencing techniques, such as RNA interference, have the potential to modulate protein aggregation in Huntington's disease. Future research should explore the thermodynamic implications of these techniques and their potential as therapeutic strategies.
Hence, addressing the challenges and exploring future directions in understanding the thermodynamics of protein aggregation in Huntington's disease could lead to the development of novel therapeutics that target protein aggregation, ultimately improving the lives of patients with Huntington's disease.
Conclusion
In conclusion, the thermodynamics of protein aggregation in Huntington's disease (HD) reveal a complex interplay of factors that contribute to the formation of toxic protein aggregates. The aggregation process is driven by a delicate balance of hydrophobic interactions, electrostatic forces, and other molecular forces, leading to the formation of oligomers and fibrils that are key features of HD pathology. Understanding the thermodynamic drivers of protein aggregation is crucial for the development of effective therapeutics for HD. Targeting these thermodynamic processes offers promising strategies for intervention, including the design of small molecules that stabilize the native state of huntingtin protein or inhibit the formation of toxic aggregates. Additionally, immunotherapy approaches that target protein aggregates show potential in clearing existing aggregates and preventing further aggregation.
Gene silencing techniques, such as RNA interference (RNAi), hold great promise for HD therapeutics by selectively targeting the expression of mutant huntingtin protein. These approaches can potentially prevent the accumulation of toxic aggregates and slow the progression of the disease. Overall, a comprehensive understanding of the thermodynamics of protein aggregation in HD is essential for the development of effective therapeutic strategies. Continued research in this field is crucial for advancing our understanding of HD pathology and developing novel therapies to combat this devastating disease.
Data availability
The review has no data to be provided.
References
Latoszek E, Wiweger M, Ludwiczak J, Dunin-Horkawicz S, Kuznicki J, Czeredys M. Siah-1-interacting protein regulates mutated huntingtin protein aggregation in Huntington’s disease models. SpringerE Latoszek, M Wiweger, J Ludwiczak, S Dunin-Horkawicz, J Kuznicki, M CzeredysCell & bioscience, 2022•Springer [Internet]. 2022 Dec 1 [cited 2024 May 23];12(1). Available from: https://springerlink.bibliotecabuap.elogim.com/article/10.1186/s13578-022-00755-0
Tipping WJ, Merchant AS, Fearon R, Tomkinson NCO, Faulds K, Graham D. Temporal imaging of drug dynamics in live cells using stimulated Raman scattering microscopy and a perfusion cell culture system. RSC Chem Biol [Internet]. 2022 Aug 31 [cited 2024 May 23];3(9):1154–64. Available from: https://pubs.rsc.org/en/content/articlehtml/2022/cb/d2cb00160h
Chawla S, Ahmadpour D, Schneider KL, Kumar N, Fischbach A, Molin M, et al. Calcineurin stimulation by Cnb1p overproduction mitigates protein aggregation and α-synuclein toxicity in a yeast model of synucleinopathy. Cell Communication and Signaling. 2023 Aug 24;21(1):220.
Yu D, Zarate N, White A, Coates D, Tsai W, Nanclares C, et al. CK2 alpha prime and alpha-synuclein pathogenic functional interaction mediates synaptic dysregulation in Huntington’s disease. Acta Neuropathol Commun. 2022 Dec 3;10(1):83.
Ochneva A, Zorkina Y, Abramova O, Pavlova O, Ushakova V, Morozova A, et al. Protein Misfolding and Aggregation in the Brain: Common Pathogenetic Pathways in Neurodegenerative and Mental Disorders. Int J Mol Sci. 2022 Nov 21;23(22):14498.
Díaz SA, Pascual G, Patten LK, Roy SK, Meares A, Chiriboga M, et al. Towards control of excitonic coupling in DNA-templated Cy5 aggregates: the principal role of chemical substituent hydrophobicity and steric interactions. Nanoscale. 2023;15(7):3284–99.
Clore GM. NMR spectroscopy, excited states and relevance to problems in cell biology – transient pre-nucleation tetramerization of huntingtin and insights into Huntington’s disease. J Cell Sci. 2022 Jun 15;135(12):jcs258695. https://doi.org/10.1242/jcs.258695.
Rui H, Ashton KS, Min J, Wang C, Potts PR. Protein–protein interfaces in molecular glue-induced ternary complexes: classification, characterization, and prediction. RSC Chem Biol. 2023;4(3):192–215.
Gropp MHM, Klaips CL, Hartl FU. Formation of toxic oligomers of polyQ-expanded Huntingtin by prion-mediated cross-seeding. Mol Cell. 2022 Nov;82(22):4290-4306.e11.
Boros BD, Schoch KM, Kreple CJ, Miller TM. Antisense Oligonucleotides for the Study and Treatment of ALS. Neurotherapeutics. 2022 Jul;19(4):1145–58.
Nassar A, Satarker S, Gurram PC, Upadhya D, Fayaz S, Nampoothiri M. Repressor Element-1 Binding Transcription Factor (REST) as a Possible Epigenetic Regulator of Neurodegeneration and MicroRNA-Based Therapeutic Strategies. Mol Neurobiol. 2023 Oct 16;60(10):5557–77.
Zhaliazka K, Serada V, Matveyenka M, Rizevsky S, Kurouski D. Protein-to-lipid ratio uniquely changes the rate of lysozyme aggregation but does not significantly alter toxicity of mature protein aggregates. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2023 May;1868(5):159305.
Chowdhury SR, Lu HP. Unraveling the mechanism of tau protein aggregation in presence of zinc ion: The earliest step of tau aggregation. Chemical Physics Impact. 2022 Jun;4:100060.
Fongaro B, Cappelletto E, Sosic A, Spolaore B, Polverino de Laureto P. 3,4‐Dihydroxyphenylethanol and 3,4‐dihydroxyphenylacetic acid affect the aggregation process of <scp>E46K</scp> variant of α‐synuclein at different extent: Insights into the interplay between protein dynamics and catechol effect. Protein Science. 2022 Jul 16;31(7):e4356. https://doi.org/10.1002/pro.4356.
Bertoglio D, Verhaeghe J, Miranda A, Wyffels L, Stroobants S, Mrzljak L, et al. Longitudinal preclinical evaluation of the novel radioligand [11C]CHDI-626 for PET imaging of mutant huntingtin aggregates in Huntington’s disease. Eur J Nucl Med Mol Imaging. 2022 Mar 15;49(4):1166–75.
Sonar K, Mancera RL. Characterization of the Conformations of Amyloid Beta 42 in Solution That May Mediate Its Initial Hydrophobic Aggregation. J Phys Chem B. 2022 Oct 13;126(40):7916–33.
Wankhede NL, Kale MB, Upaganlawar AB, Taksande BG, Umekar MJ, Behl T, et al. Involvement of molecular chaperone in protein-misfolding brain diseases. Biomedicine & Pharmacotherapy. 2022 Mar;147:112647.
Pereira CA de S, Medaglia N de C, Ureshino RP, Bincoletto C, Antonioli M, Fimia GM, et al. NAADP-Evoked Ca2+ Signaling Leads to Mutant Huntingtin Aggregation and Autophagy Impairment in Murine Astrocytes. Int J Mol Sci. 2023 Mar 15;24(6):5593.
Nazarov S, Chiki A, Boudeffa D, Lashuel HA. Structural Basis of Huntingtin Fibril Polymorphism Revealed by Cryogenic Electron Microscopy of Exon 1 HTT Fibrils. J Am Chem Soc. 2022 Jun 22;144(24):10723–35.
Shing K, Sapp E, Boudi A, Liu S, Seeley C, Marchionini D, et al. Early whole-body mutant huntingtin lowering averts changes in proteins and lipids important for synapse function and white matter maintenance in the LacQ140 mouse model. Neurobiol Dis. 2023 Oct;187:106313.
Escobedo A, Piccirillo J, Aranda J, Diercks T, Mateos B, Garcia-Cabau C, et al. A glutamine-based single α-helix scaffold to target globular proteins. Nat Commun. 2022 Nov 18;13(1):7073.
Felício D, du Mérac TR, Amorim A, Martins S. Functional implications of paralog genes in polyglutamine spinocerebellar ataxias. Hum Genet. 2023 Dec 16;142(12):1651–76.
Wilbertz JH, Frappier J, Muller S, Gratzer S, Englaro W, Stanek LM, et al. Time-resolved FRET screening identifies small molecular modifiers of mutant Huntingtin conformational inflexibility in patient-derived cells. SLAS Discovery. 2022 Jun;27(4):219–28.
Sap KA, Geijtenbeek KW, Schipper-Krom S, Guler AT, Reits EA. Ubiquitin-modifying enzymes in Huntington’s disease. Front Mol Biosci. 2023 Feb 8;10.
Song H, Wang C, Zhu C, Wang Z, Yang H, Wu P, et al. Suppression of toxicity of the mutant huntingtin protein by its interacting compound, desonide. Proceedings of the National Academy of Sciences. 2022 Mar 8;119(10). e2114303119. https://doi.org/10.1073/pnas.2114303119.
Makeeva VS, Dyrkheeva NS, Lavrik OI, Zakian SM, Malakhova AA. Mutant-Huntingtin Molecular Pathways Elucidate New Targets for Drug Repurposing. Int J Mol Sci. 2023 Nov 27;24(23):16798.
Buell AK. Stability matters, too – the thermodynamics of amyloid fibril formation. Chem Sci. 2022;13(35):10177–92.
Abelein A. Metal Binding of Alzheimer’s Amyloid-β and Its Effect on Peptide Self-Assembly. Acc Chem Res. 2023 Oct 3;56(19):2653–63.
Abelein A, Johansson J. Amyloid inhibition by molecular chaperones in vitro can be translated to Alzheimer’s pathology in vivo. RSC Med Chem. 2023;14(5):848–57.
Sharma M, Rajendrarao S, Shahani N, Ramírez-Jarquín UN, Subramaniam S. Cyclic GMP-AMP synthase promotes the inflammatory and autophagy responses in Huntington disease. Proceedings of the National Academy of Sciences. 2020 Jul 7;117(27):15989–99.
Rook ME, Southwell AL. Antisense Oligonucleotide Therapy: From Design to the Huntington Disease Clinic. BioDrugs. 2022 Mar 7;36(2):105–19.
Naeli P, Winter T, Hackett AP, Alboushi L, Jafarnejad SM. The intricate balance between microRNA‐induced mRNA decay and translational repression. FEBS J. 2023 May 11;290(10):2508–24.
Pan H, Liu Z, Ma J, Li Y, Zhao Y, Zhou X, et al. Genome-wide association study using whole-genome sequencing identifies risk loci for Parkinson’s disease in Chinese population. NPJ Parkinsons Dis. 2023 Feb 9;9(1):22.
Gu X, Richman J, Langfelder P, Wang N, Zhang S, Bañez-Coronel M, et al. Uninterrupted CAG repeat drives striatum-selective transcriptionopathy and nuclear pathogenesis in human Huntingtin BAC mice. Neuron. 2022 Apr;110(7):1173-1192.e7.
Costa MD, Maciel P. Modifier pathways in polyglutamine (PolyQ) diseases: from genetic screens to drug targets. Cellular and Molecular Life Sciences. 2022 May 3;79(5):274.
Childs-Disney JL, Yang X, Gibaut QMR, Tong Y, Batey RT, Disney MD. Targeting RNA structures with small molecules. Nat Rev Drug Discov. 2022 Oct 8;21(10):736–62.
Kotowska-Zimmer A, Przybyl L, Pewinska M, Suszynska-Zajczyk J, Wronka D, Figiel M, et al. A CAG repeat-targeting artificial miRNA lowers the mutant huntingtin level in the YAC128 model of Huntington’s disease. Mol Ther Nucleic Acids. 2022 Jun;28:702–15.
Tants J, Schlundt A. Advances, Applications, and Perspectives in Small‐Angle X‐ray Scattering of RNA. ChemBioChem. 2023 Sep 19;24(17). e202300110. https://doi.org/10.1002/cbic.202300110.
Zhang X, Zhang B, Freddolino PL, Zhang Y. CR-I-TASSER: assemble protein structures from cryo-EM density maps using deep convolutional neural networks. Nat Methods. 2022 Feb 7;19(2):195–204.
Wu GH, Smith-Geater C, Galaz-Montoya JG, Gu Y, Gupte SR, Aviner R, et al. CryoET reveals organelle phenotypes in huntington disease patient iPSC-derived and mouse primary neurons. Nat Commun. 2023 Feb 8;14(1):692.
Gupta H, Sahi S. High-throughput virtual screening of potential inhibitors of GPR52 using docking and biased sampling method for Huntington’s disease therapy. Mol Divers. 2023 Dec 1. https://doi.org/10.1007/s11030-023-10763-y
Wells C, Brennan S, Keon M, Ooi L. The role of amyloid oligomers in neurodegenerative pathologies. Int J Biol Macromol. 2021 Jun;181:582–604.
Galyan SM, Ewald CY, Jalencas X, Masrani S, Meral S, Mestres J. Fragment-based virtual screening identifies a first-in-class preclinical drug candidate for Huntington’s disease. Sci Rep. 2022 Nov 16;12(1):19642.
Eshraghi M, Karunadharma PP, Blin J, Shahani N, Ricci EP, Michel A, et al. Mutant Huntingtin stalls ribosomes and represses protein synthesis in a cellular model of Huntington disease. Nat Commun. 2021 Mar 5;12(1):1461.
Caron NS, Banos R, Yanick C, Aly AE, Byrne LM, Smith ED, et al. Mutant Huntingtin Is Cleared from the Brain via Active Mechanisms in Huntington Disease. The Journal of Neuroscience. 2021 Jan 27;41(4):780–96.
Lemarié FL, Caron NS, Sanders SS, Schmidt ME, Nguyen YTN, Ko S, et al. Rescue of aberrant huntingtin palmitoylation ameliorates mutant huntingtin-induced toxicity. Neurobiol Dis. 2021 Oct;158:105479.
Caron NS, Banos R, Aly AE, Xie Y, Ko S, Potluri N, et al. Cerebrospinal fluid mutant huntingtin is a biomarker for huntingtin lowering in the striatum of Huntington disease mice. Neurobiol Dis. 2022 May;166:105652.
Oura S, Noda T, Morimura N, Hitoshi S, Nishimasu H, Nagai Y, et al. Precise CAG repeat contraction in a Huntington’s Disease mouse model is enabled by gene editing with SpCas9-NG. Commun Biol. 2021 Jun 23;4(1):771.
Han JY, Seo J, Choi Y, Im W, Ban JJ, Sung JJ. CRISPR-Cas9 mediated genome editing of Huntington’s disease neurospheres. Mol Biol Rep. 2023 Mar 23;50(3):2127–36.
Stewart CJ, Olgenblum GI, Propst A, Harries D, Pielak GJ. Resolving the enthalpy of protein stabilization by macromolecular crowding. Protein Science. 2023 Mar 21;32(3):e4573. https://doi.org/10.1002/pro.4573
Galano-Frutos JJ, Nerín-Fonz F, Sancho J. Calculation of Protein Folding Thermodynamics Using Molecular Dynamics Simulations. J Chem Inf Model. 2023 Dec 25;63(24):7791–806.
Vila JA. Protein folding rate evolution upon mutations. Biophys Rev. 2023 Aug 15;15(4):661–9.
Heinz LP, Grubmüller H. Spatially resolved free-energy contributions of native fold and molten-globule-like Crambin. Biophys J. 2021 Aug;120(16):3470–82.
Doyle LA, Takushi B, Kibler RD, Milles LF, Orozco CT, Jones JD, et al. De novo design of knotted tandem repeat proteins. Nat Commun. 2023 Oct 24;14(1):6746.
Wesołowski PA, Wales DJ, Pracht P. Multilevel Framework for Analysis of Protein Folding Involving Disulfide Bond Formation. J Phys Chem B. 2024 Apr 4;128(13):3145–56.
Fassler JS, Skuodas S, Weeks DL, Phillips BT. Protein Aggregation and Disaggregation in Cells and Development. J Mol Biol. 2021 Oct;433(21):167215.
Meisl G, Xu CK, Taylor JD, Michaels TCT, Levin A, Otzen D, et al. Uncovering the universality of self-replication in protein aggregation and its link to disease. Sci Adv. 2022 Aug 12;8(32):eabn6831. https://doi.org/10.1126/sciadv.abn6831.
Prabakaran R, Rawat P, Thangakani AM, Kumar S, Gromiha MM. Protein aggregation: in silico algorithms and applications. Biophys Rev. 2021 Feb 17;13(1):71–89.
Gallonde WT, Poidevin C, Houard F, Caytan E, Dorcet V, Fihey A, et al. Kinetic Delay in Cooperative Supramolecular Polymerization by Redefining the Trade‐Off Relationship between H‐Bonds and Van der Waals/π–π Stacking Interactions. Angewandte Chemie. 2023 Dec 4;135(49).
Lundahl MLE, Fogli S, Colavita PE, Scanlan EM. Aggregation of protein therapeutics enhances their immunogenicity: causes and mitigation strategies. RSC Chem Biol. 2021;2(4):1004–20.
Brudar S, Hribar-Lee B. Effect of Buffer on Protein Stability in Aqueous Solutions: A Simple Protein Aggregation Model. J Phys Chem B. 2021 Mar 18;125(10):2504–12.
Verma AK, Khan E, Mishra SK, Kumar A. Small Molecule Screening Discovers Compounds that Reduce FMRpolyG Protein Aggregates and Splicing Defect Toxicity in Fragile X-Associated Tremor/Ataxia Syndrome. Mol Neurobiol. 2022 Mar 18;59(3):1992–2007.
Shmool TA, Martin LK, Matthews RP, Hallett JP. Ionic Liquid-Based Strategy for Predicting Protein Aggregation Propensity and Thermodynamic Stability. JACS Au. 2022 Sep 26;2(9):2068–80.
Ferenczy GG, Kellermayer M. Contribution of hydrophobic interactions to protein mechanical stability. Comput Struct Biotechnol J. 2022;20:1946–56.
Liu Y, Li K, Tian J, Gao A, Tian L, Su H, et al. Synthesis of robust underwater glues from common proteins via unfolding-aggregating strategy. Nat Commun. 2023 Aug 24;14(1):5145.
Ventura S, Cristina Vega M, Lacroix E, Angrand I, Spagnolo L, Serrano L. Conformational strain in the hydrophobic core and its implications for protein folding and design. Nat Struct Biol. 2002 Jun 1;9(6):485–93.
Das TK, Chou DK, Jiskoot W, Arosio P. Nucleation in Protein Aggregation in Biotherapeutic Development: A look into the Heart of the Event. J Pharm Sci. 2022 Apr;111(4):951–9.
Nashed NT, Aniana A, Ghirlando R, Chiliveri SC, Louis JM. Modulation of the monomer-dimer equilibrium and catalytic activity of SARS-CoV-2 main protease by a transition-state analog inhibitor. Commun Biol. 2022 Mar 1;5(1):160.
Cascella R, Bigi A, Cremades N, Cecchi C. Effects of oligomer toxicity, fibril toxicity and fibril spreading in synucleinopathies. Cellular and Molecular Life Sciences. 2022 Mar 4;79(3):174.
Navarro S, Ventura S. Computational methods to predict protein aggregation. Curr Opin Struct Biol. 2022 Apr;73:102343.
Vasilenko EO, Kozin SA, Mitkevich VA, Buchelnikov AS, Nechipurenko YuD. A Thermodynamic Model of the Formation of Protein Aggregates on a Matrix. Biophysics (Oxf). 2023 Dec 18;68(6):934–44.
Delacour J, Doumic M, Martens S, Schmeiser C, Zaffagnini G. A mathematical model of p62-ubiquitin aggregates in autophagy. J Math Biol. 2022 Jan 14;84(1–2):3.
Farkas A, Zsindely N, Nagy G, Kovács L, Deák P, Bodai L. The ubiquitin thioesterase YOD1 ameliorates mutant Huntingtin induced pathology in Drosophila. Sci Rep. 2023 Dec 11;13(1):21951.
Miguez A, Gomis C, Vila C, Monguió-Tortajada M, Fernández-García S, Bombau G, et al. Soluble mutant huntingtin drives early human pathogenesis in Huntington’s disease. Cellular and Molecular Life Sciences. 2023 Aug 3;80(8):238.
Ramadan A, Mohammed A, Elnour AA, Sadeq A, Al Mazrouei N, Alkaabi M, et al. The flavonoid luteolin reduces mutant huntingtin aggregation and cytotoxicity in huntingtin-mutated neuroblastoma cells. Saudi Pharmaceutical Journal. 2023 Dec;31(12):101871.
Alkanli SS, Alkanli N, Ay A, Albeniz I. CRISPR/Cas9 Mediated Therapeutic Approach in Huntington’s Disease. Mol Neurobiol. 2023 Mar 9;60(3):1486–98.
Zhaliazka K, Kurouski D. Nanoscale imaging of individual amyloid aggregates extracted from brains of Alzheimer and Parkinson patients reveals presence of lipids in α‐synuclein but not in amyloid β 1–42 fibrils. Protein Science. 2023 Apr 16;32(4):e4598. https://doi.org/10.1002/pro.4598.
Evans CG, O’Brien J, Winfree E, Murugan A. Pattern recognition in the nucleation kinetics of non-equilibrium self-assembly. Nature. 2024 Jan 18;625(7995):500–7.
Fall A, Henriksson M, Karppinen A, Opstad A, Heggset EB, Syverud K. The effect of ionic strength and pH on the dewatering rate of cellulose nanofibril dispersions. Cellulose. 2022 Sep 20;29(14):7649–62.
Duan C, Wang R. A Unified Description of Salt Effects on the Liquid–Liquid Phase Separation of Proteins. ACS Cent Sci. 2024 Feb 28;10(2):460–8.
Liu Y, Feng W, Wang Y, Wu B. Crosstalk between protein post-translational modifications and phase separation. Cell Communication and Signaling. 2024 Feb 12;22(1):110.
Elenbaas BOW, Khemtemourian L, Killian JA, Sinnige T. Membrane-Catalyzed Aggregation of Islet Amyloid Polypeptide Is Dominated by Secondary Nucleation. Biochemistry. 2022 Jul 19;61(14):1465–72.
Itabashi H, Tashiro K, Koshikawa S, Datta S, Yagai S. Distinct seed topologies enable comparison of elongation and secondary nucleation pathways in seeded supramolecular polymerization. Chemical Communications. 2023;59(48):7375–8.
Moghadam Farid S, Noori M, Nazari Montazer M, Khalili Ghomi M, Mollazadeh M, Dastyafteh N, et al. Synthesis and structure–activity relationship studies of benzimidazole-thioquinoline derivatives as α-glucosidase inhibitors. Sci Rep. 2023 Mar 16;13(1):4392.
Forozan R, Ghomi MK, Iraji A, Montazer MN, Noori M, Dastyafteh N, et al. Synthesis, in vitro inhibitor screening, structure–activity relationship, and molecular dynamic simulation studies of novel thioquinoline derivatives as potent α-glucosidase inhibitors. Sci Rep. 2023 May 15;13(1):7819.
Mukherjee S, Abdalla M, Yadav M, Madhavi M, Bhrdwaj A, Khandelwal R, et al. Structure-Based Virtual Screening, Molecular Docking, and Molecular Dynamics Simulation of VEGF inhibitors for the clinical treatment of Ovarian Cancer. J Mol Model. 2022 Apr 24;28(4):100.
Nwabufo CK, Aigbogun OP. Diagnostic and therapeutic agents that target alpha-synuclein in Parkinson’s disease. J Neurol. 2022 Nov 13;269(11):5762–86.
Fleming SM, Davis A, Simons E. Targeting alpha-synuclein via the immune system in Parkinson’s disease: Current vaccine therapies. Neuropharmacology. 2022 Jan;202:108870.
Calabresi P, Mechelli A, Natale G, Volpicelli-Daley L, Di Lazzaro G, Ghiglieri V. Alpha-synuclein in Parkinson’s disease and other synucleinopathies: from overt neurodegeneration back to early synaptic dysfunction. Cell Death Dis. 2023 Mar 1;14(3):176.
Liu Y, Tan L, Tan MS. Chaperone-mediated autophagy in neurodegenerative diseases: mechanisms and therapy. Mol Cell Biochem. 2023 Oct 25;478(10):2173–90.
Rossi MA, Pozhidaeva AK, Clerico EM, Petridis C, Gierasch LM. New insights into the structure and function of the complex between the Escherichia coli Hsp70, DnaK, and its nucleotide-exchange factor, GrpE. Journal of Biological Chemistry. 2024 Jan;300(1):105574.
Tiwari S, Fauvet B, Assenza S, De Los Rios P, Goloubinoff P. A fluorescent multi-domain protein reveals the unfolding mechanism of Hsp70. Nat Chem Biol. 2023 Feb 20;19(2):198–205.
Kim H, Park J, Roh SH. The structural basis of eukaryotic chaperonin TRiC/CCT: Action and folding. Mol Cells. 2024 Mar;47(3):100012.
Sengupta U, Kayed R. Amyloid β, Tau, and α-Synuclein aggregates in the pathogenesis, prognosis, and therapeutics for neurodegenerative diseases. Prog Neurobiol. 2022 Jul;214:102270.
Song C, Shi J, Zhang P, Zhang Y, Xu J, Zhao L, et al. Immunotherapy for Alzheimer’s disease: targeting β-amyloid and beyond. Transl Neurodegener. 2022 Dec 18;11(1):18.
Katti A, Diaz BJ, Caragine CM, Sanjana NE, Dow LE. CRISPR in cancer biology and therapy. Nat Rev Cancer. 2022 May 22;22(5):259–79.
Terada C, Oh K, Tsubaki R, Chan B, Aibara N, Ohyama K, et al. Dynamic and static control of the off-target interactions of antisense oligonucleotides using toehold chemistry. Nat Commun. 2023 Dec 2;14(1):7972.
Gosset P, Maxan A, Alpaugh M, Breger L, Dehay B, Tao Z, et al. Evidence for the spread of human-derived mutant huntingtin protein in mice and non-human primates. Neurobiol Dis. 2020 Jul;141:104941.
Southwell AL, Skotte NH, Bennett CF, Hayden MR. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol Med. 2012 Nov;18(11):634–43.
Luo M, Lee LKC, Peng B, Choi CHJ, Tong WY, Voelcker NH. Delivering the Promise of Gene Therapy with Nanomedicines in Treating Central Nervous System Diseases. Advanced Science. 2022 Sep 18;9(26):e2201740. https://doi.org/10.1002/advs.202201740.
Acknowledgements
Authors acknowledge Lovely Professional University to provide opportunity to work.
Funding
There is no funding for this manuscript.
Author information
Authors and Affiliations
Contributions
Tejasvi Paney drafted the manuscript.
Rajinder Singh Kaundal formatted figures.
Vivek Pandey finalized the manuscript with proper details.
Corresponding author
Ethics declarations
Ethics approval
Not Applicable.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaundal, R.S., Pandey, T. & Pandey, V. Exploring the thermodynamics of protein aggregation: an insight to Huntington's disease therapeutics. Neurosci Behav Physi (2024). https://doi.org/10.1007/s11055-024-01639-1
Published:
DOI: https://doi.org/10.1007/s11055-024-01639-1