Abstract
Background
Talaromyces marneffei (T. marneffei) is a thermally dimorphic fungus causing systemic mycosis. Due to the atypical symptoms and diverse imaging findings, T. marneffei-infected patients may be misdiagnosed thus preventing timely antifungal therapy. Moreover, HIV-negative patients with T. marneffei infection may be congenitally immunocompromised because of the mutation of immune-related genes.
Case presentation
We describe a case of an HIV-negative child who developed disseminated T. marneffei infection in a nonendemic area. Chest CT showed similar imaging changes of miliary pulmonary tuberculosis, while there was no other evidence of tuberculosis infection, and empirical antituberculosis treatment was not effective. Lymphocyte subset analysis showed reduced natural killer cells, and the immunoglobulin profile showed low levels of IgM, C3 and C4. A bone marrow smear revealed T. marneffei infection, and ascites culture also proved T. marneffei infection. Despite antifungal treatment, the child died of multiple organ failure. Two gene mutations in caspase recruitment domain-containing protein 9 (CARD9) were detected, which had not been reported previously in T. marneffei-infected patients.
Conclusions
HIV-negative patients with CARD9 mutations may be potential hosts of T. marneffei. Abnormalities in the immunoglobin profile and lymphocyte subset may provide clues for immunocompromised patients, and further genetic testing is advised to identify gene mutations in HIV-negative patients with T. marneffei infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Talaromyces marneffei (T. marneffei), previously named Penicillium marneffei, is an emerging pathogenic fungus causing systemic mycosis in Southeast Asia. It is a common but serious opportunistic infection among human immunodeficiency virus (HIV)-infected patients [1]. With the better control of HIV infection, an increasing number of cases have been reported in HIV-negative but otherwise immunocompromised individuals, such as those with various primary immunodeficiencies (PIDs), hematological malignancies, mixed connective tissue disease, transplant rejection, and diabetes mellitus and those taking corticosteroids or immunosuppressive agents [2, 3].
The clinical manifestations of T. marneffei infection are nonspecific, including recurrent fever, cough, weight loss, hepatosplenomegaly, lymphadenopathy, and gastrointestinal abnormalities [4]. T. marneffei infection can disseminate into many organs, run a rapid progressive course and can be life-threatening without timely antifungal therapy [5, 6].
Genetic susceptibility to fungal diseases in specific patients has gained increasing interest in recent years. Caspase recruitment domain-containing protein 9 (CARD9) is a human adaptor protein positioned downstream of multiple fungal sensors expressed on myeloid cells and plays a role in host defense against fungi [7]. CARD9 mutations are associated with superficial and deep fungal infections. However, to date, there have been no reports of CARD9 mutations and T. marneffei infection.
Here, we present a rare case of disseminated T. marneffei infection in an HIV-negative child in a nonendemic area with a gene mutation in CARD9 that has not been reported previously in T. marneffei infection. Moreover, a literature review was performed to further analyze the characteristics of T. marneffei-infected patients with immune-related gene mutations.
Case presentation
A 5-year-old boy was admitted to our hospital on 2 January 2020 for fever and cough lasting for half a month. Initially, he presented with a low- to moderate-grade fever and dry cough. He was treated with intravenous cephalosporin in a local hospital, but there was no significant relief of the above symptoms. Three days before admission, he had febrile symptoms with chills and a peak body temperature up to 40 °C. His coughing symptoms worsened. The boy was born in Chongqing city, Southwestern China, where the climate was warm, humid and rainy in spring and summer. It was reported by his parents that he had experienced recurrent respiratory tract infections since infancy. He had a Bacillus Calmette-Guerin Vaccine (BCG) scar on his left shoulder. His grandfather suffered from tuberculosis ten years ago, but he had been cured after antituberculosis treatment.
Upon admission to our hospital, a physical examination revealed a body temperature of 38.9 °C and poor spirit. Palpable superficial lymph nodes could be touched in the neck (approximately 5 mm in diameter), 2 of which were fused (18 mm in diameter). Inspiratory wheeze could be heard in both lungs. The liver was palpable 2 cm below the right costal margin and 4 cm below the xiphoid, and the spleen was found 6.5 cm below the ribs.
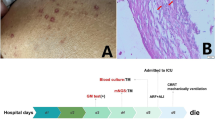
The laboratory examinations at admission reported a white blood cell count of 8.68*10^9/L, a red blood cell count of 3.25*10^12/L, hemoglobin of 92 g/L, a platelet level of 61*10^6/L and albumin of 24.7 g/L. Inflammation biomarkers, including C-reactive protein (CRP), procalcitonin (PCT), and erythrocyte sedimentation rate (ESR), were significantly elevated on admission (CRP: 47 mg/L, PCT: 66.32 mg/L, ESR: 22 mm/hr). The patient’s HIV test was negative. A chest computed tomography (CT) scan (Figure 1) showed extensive fine-grained shadows in both lungs, with cavitation in the upper lobe of both lungs and hilar enlargement, indicating a diagnosis of miliary tuberculosis. Antituberculosis therapy was empirically started on admission, including ethambutol, linezolid, isoniazid, and levofloxacin. However, no evidence of tuberculosis infection was found in the following days since the PPD test, the interferon-γ release assay, the tuberculosis gene X-pert test and repeated acid-fast staining of sputum were all negative. The blood and sputum culture were also negative. With antituberculosis and antibiotic therapy, the boy still presented with recurrent fever, and the alanine aminotransferase (ALT) level was elevated to 97.5 U/L, indicating the impairment of liver function. Isoniazid and levofloxacin were withdrawn, and the anti-infection treatment was upgraded to imipenem on 6 January. However, his clinical condition further deteriorated. He developed respiratory failure and required mechanical ventilation. He was transferred to the intensive care unit on 6 January. On 6 January, his total white blood cell count had fallen to 3.82*10^9/L, red blood cell count was 2.85*10^12/L, hemoglobin was 82 g/L, platelet level was 11*10^6/L and albumin was 18.2 g/L.
Considering the poor response to antituberculosis and antibiotic therapy, we further searched for potential evidence of infection. The serum galactomannan antigen (GM) level was 245.3 pg/mL (reference <60 pg/mL). The lymphocyte subset showed reduced natural killer (NK) cells (1.19%, reference 4–26%). The immunoglobulin profile showed low levels of IgM (0.331 g/L, reference 0.48–2.26 g/L), C3 (0.09 g/L, reference 0.7–2.06 g/L) and C4 (0.03 g/L, reference 0.11–0.61 g/L). Cytokines revealed significant elevations in IL-6 (24347.18, reference 0–16.60 pg/mL), IL-10 (319.32, reference 0–4.9 pg/mL), TNF-α (57.02, reference 0–5.2 pg/mL), and INF-γ (42.94, reference 0–17.30 pg/mL) (Table 1). Finally, microscopic examination of the bone marrow smear on 9 January identified numerous round to oval, elongated, thin-walled yeast cells with central septation distributed in the intracellular and extracellular space, which indicated T. marneffei infection (Figure 2). Subsequently, use of amphotericin B and voriconazole commenced as antifungal treatments. Unfortunately, the patient died of multiple organ failure 8 days after hospitalization. Observation of the ascites culture proved T. marneffei infection on 15 January. The abnormal serum immunoglobulin profile and lymphocyte subset encouraged us to perform further genetic tests to identify his immune status.
With his parents’ consent, DNA sample from the boy was submitted to determine the genetic cause of the disease (performed at the AmCare Genomics Lab, GuangZhou, China). Medical exome sequencing (MES) was performed using custom-designed NimbleGen SeqCap probes (Roche NimbleGen, Madison, WI, USA) for in-solution hybridization to enrich coding exons for about 5000 clinically relevant disease causing genes followed by an Illumina HiSeq sequencer. Sequencing data were matched with the human genome assembly hg19 (GRCh37) as a reference and identified possible pathogenic mutations. For each variant, minor allele frequencies (> 0.1%) were filtered out in variant databases including Exome Aggregation Consortium (ExAC) database and Genome Aggregation Database (gnomAD). Considering that the proband developed disseminated T. marneffei infection and empirical antituberculosis treatment was not effective, we identified CARD9 heterozygous mutations (c.440T>C (p. L147P) and c.586A>G (p. K196E)) as the most common pathogenic variant by MES and Sanger sequencing (Figure 3). Since the boy’s parents did not undergo genetic testing, we cannot trace the source of variations. The c.440T>C (p. L147P) mutation is novel, as it is not reported in our reference gene database and is less frequent in our reference population genetic database (PM2). The frequency of c.586A>G (p. K196E) variation of CARD9 in our reference gene database is extremely low. We used the VarCards database for silico evaluation of the variants. Among the 23 silico missense predictions, 13 algorithms predicted that mutation c.440T>C (p. L147P) was harmful, and 15 algorithms predicted that mutation c.586A>G (p. K196E) was likely to affect protein structure/function. The damaging scores of the variants (c.440T>C (p. L147P) and c.586A>G (p. K196E)) were 0.57 and 0.65, respectively. From this, we conclude that heterozygote mutations c.440T>C (p. L147P) and c.586A>G (p. K196E) in CARD9 may be genetic cause of the disease.
Systematic Review
A literature search was performed in PubMed on 1 December 2020 to further analyze the characteristics of T. marneffei-infected patients who were diagnosed with immunodeficiency with documented gene mutations using the key words “Talaromyces marneffei” or “Penicillium marneffei” or “Penicilliosis” or “Talaromycosis”. Articles reporting T. marneffei-infected patients with immune-related gene mutations were included. HIV-positive cases were excluded. Disseminated T. marneffei infection was defined as infections involving more than two organs or systems, such as the respiratory system, the lymph nodes, the skin, and/or the intestine. As a result, a total of 21 cases with confirmed PID from twelve articles were analyzed, four of which died. The characteristics of the included patients are summarized in Table 2.
The age of the included patients ranged from 5 months to 34 years, and 95% (20/21) of them were pediatric patients (age < 16 years). Eighty-six percent of them were Chinese (18/21), while the rest were from Thailand. The clinical symptoms mainly presented as disseminated changes involving the lung, intestine, skin, lymph nodes, liver and spleen. Of the 21 patients, T. marneffei infection was mainly diagnosed by cultures of blood (38%, 8/21), bone marrow (24%, 5/21), bronchoalveolar lavage fluid (BALF) (14%, 3/21), throat swabs and sputum (5%, 1/21), biopsy of lymph nodes (24%, 5/21), tissue from skin lesions (5%, 1/21), tissue mass (5%, 1/21), endobronchial (10%, 2/21), liver (5%, 1/21), postoperative pathology (5%, 1/21) and next-generation sequencing of BALF (5%, 1/21). Forty-three percent (9/21) of the patients had abnormalities in the lymphocyte subset, 43% (9/21) had a normal lymphocyte subset, and 14% (3/21) had no details of the lymphocyte subset examination. In addition, 76% (16/21) of patients had abnormal immunoglobulin tests, including IgA, IgG, IgM and IgE. The reported mutations included STAT1, STAT3, CD40L and IFNGR1, and the details of the nucleotide changes and amino acid changes are shown in Table 2.
Discussion
T. marneffei is a thermally dimorphic fungus that was first isolated from a bamboo rat in Vietnam in 1956 [17]. T. marneffei infection has largely been restricted to Southeast Asia, especially in Thailand, Vietnam, northeastern India, South China, Hong Kong, Taiwan, Laos, and Malaysia [3]. Travel-related infection of T. marneffei is being increasingly recognized in nonendemic regions such as Australia, Belgium, France, Germany, Japan, the Netherlands, Oman, Sweden, Switzerland, Togo, the United Kingdom and the USA [4]. A review of 668 cases of T. marneffei infection in mainland China [6] revealed that 99.4% of the cases were reported in the southern part of China, including Guangdong and Guangxi provinces, where the climate is very humid and the temperature is similar to that in Thailand. In the present study, the boy lived in Southwestern China, where the climate was kind of similar to that of Southern China with high incidence of Talaromyces infection, such as Guangdong, Guangxi and Hongkong. However, relatively few cases of Talaromyces marneffei infection have been reported in Chongqing. And the boy in our study had never visited endemic areas. This suggested that children living in Chongqing may also be infected with T. marneffei.
It is believed that infection with T. marneffei results from the inhalation of fungal conidia into the alveolar spaces within the lungs, and the ability of the fungus to survive the killing process and replicate inside macrophage cells has an important role in its pathogenicity [18, 19]. Infection of T. marneffei in those patients with immune dysfunction usually causes fungemia and disseminated disease to various organs involving the skin, lymph nodes, lung, spleen, bone and central nervous system [20, 21]. The radiological appearance is variable, including interstitial to alveolar infiltrates, or both, and reticulonodular consolidation [22].
T. marneffei-infected patients can be misdiagnosed with other pathogen infections. Tuberculosis is the most common misdiagnosis in patients with T. marneffei infection since they share similar clinical features and lung imaging findings. Qiu et al. [23] retrospectively evaluated clinical data from patients with confirmed T. marneffei infection involving the respiratory system and found that 38.1% (24/63) of them were misdiagnosed as having pulmonary tuberculosis and 7.9% (5/63) were diagnosed as having bacterial pneumonia. When empirical antibiotic therapy is not satisfactory, clinicians need to be attentive to special pathogenic infections. Studies have shown that bone marrow smear examination is a simple and rapid way to identify disseminated T. marneffei [24, 25]. This point was verified by our study. Galactomannan (GM) is the heteropolysaccharide present in the cell walls of most Aspergillus and Penicillium species. Some research has reported that GM levels in serum are significantly increased in patients with T. marneffei infection [26, 27]. Our study also found that the GM level in serum was perceptibly elevated in T. marneffei infection. Therefore, GM is a useful biomarker for the diagnosis of T. marneffei infection.
In recent years, the proportion of T. marneffei infection in HIV-negative patients has risen, and these patients usually have different degrees of immunosuppression, such as neutropenia, lymphopenia, and abnormalities in the serum immunoglobulin profile and lymphocyte subset. Genetic testing is essential in the diagnosis of underlying primary immunodeficiencies (PIDs). From our literature review, gene mutations in STAT1, STAT3, CD40L and IFNGR1 were reported in T. marneffei-infected patients that were immunocompromised. STAT1 and CD40L accounted for 33.3% (7/21) and 38.1% (8/21) of the known immune-related gene mutations in patients with T. marneffei infection, respectively. However, there are other gene mutations that predispose the host to T. marneffei infection.
Human CARD9 deficiency caused by mutations in the CARD9 gene is associated with selective defects in host defense against invasive fungal infection, often involving specific families of pathogenic fungi, including Candida species, dark-walled molds, yeast-like fungi and extrapulmonary aspergillus [28,29,30]. However, there are no reports on mutations in the CARD9 gene causing T. marneffei infection. The patient we reported started with respiratory symptoms and eventually developed multiple organ failure, which was later confirmed to be T. marneffei infection. Two compound heterozygous mutations in the CARD9 gene were found in the patient, c.440T>C (p. L147P) and c.586A>G (p. K196E), and the pathogenicity of the two loci of the gene has not yet been reported. However, his parents did not undergo a gene sequence test. Therefore, it is not clear whether these two mutations originated from his parents. Further study is required to determine the relationship between CARD9 gene mutation and T. marneffei infection.
Conclusions
This is the first patient with a CARD9 mutation and T. marneffei infection to be described, which indicates that HIV-negative patients with a CARD9 mutation may be potential hosts of T. marneffei. Abnormalities in the immunoglobin profile and lymphocyte subset may provide clues for immune defects in these T. marneffei-infected patients, and further genetic testing is advised to identify underlying immune dysfunction.
References
Kawila R, Chaiwarith R, Supparatpinyo K. Clinical and laboratory characteristics of penicilliosis marneffei among patients with and without HIV infection in Northern Thailand: a retrospective study. BMC Infect Dis. 2013;13:464.
Qiu Y, Liao H, Zhang J, Zhong X, Tan C, Lu D. Differences in clinical characteristics and prognosis of Penicilliosis among HIV-negative patients with or without underlying disease in Southern China: a retrospective study. BMC Infect Dis. 2015;15:525.
Chan JF, Lau SK, Yuen KY, Woo PC. Talaromyces (Penicillium) marneffei infection in non-HIV-infected patients. Emerg Microbes Infect. 2016;5(3): e19.
Cao C, Xi L, Chaturvedi V. Talaromycosis (Penicilliosis) due to Talaromyces (Penicillium) marneffei: Insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. Mycopathologi. 2019;184(6):709–20.
Lee PP, Chan KW, Lee TL, Ho MH, Chen XY, Li CH, et al. Penicilliosis in children without HIV infection–are they immunodeficient? Clin Infect Dis. 2012;54(2):e8–19.
Hu Y, Zhang J, Li X, Yang Y, Zhang Y, Ma J, et al. Penicillium marneffei infection: an emerging disease in mainland China. Mycopathologia. 2013;175(1–2):57–67.
Drummond RA, Lionakis MS. Mechanistic insights into the role of C-Type lectin receptor/CARD9 signaling in human antifungal immunity. Front Cell Infect Microbiol. 2016;6:39.
Lee PP, Lao-Araya M, Yang J, Chan KW, Ma H, Pei LC, et al. Application of flow cytometry in the diagnostics pipeline of primary immunodeficiencies underlying disseminated talaromyces marneffei infection in HIV-negative children. Front Immunol. 2019;10:2189.
Lee PP, Mao H, Yang W, Chan KW, Ho MH, Lee TL, et al. Penicillium marneffei infection and impaired IFN-γ immunity in humans with autosomal-dominant gain-of-phosphorylation STAT1 mutations. J Allergy Clin Immunol. 2014;133(3):894-6. e5.
Chen X, Xu Q, Li X, Wang L, Yang L, Chen Z, et al. Molecular and phenotypic characterization of nine patients with STAT1 GOF mutations in China. J Clin Immunol. 2020;40(1):82–95.
Fan H, Huang L, Yang D, Lin Y, Lu G, Xie Y, et al. Pediatric hyperimmunoglobulin E syndrome: A case series of 4 children in China. Medicine (Baltimore). 2018;97(14): e0215.
Pan M, Qiu Y, Zeng W, Tang S, Wei X, Zhang J. Disseminated Talaromyces hepatic granulomatous inflammation in an infant with STAT3 mutation: a case report. BMC Infect Dis. 2020;20(1):394.
Zhang W, Ye J, Qiu C, Wang L, Jin W, Jiang C, et al. Rapid and precise diagnosis of T. marneffei pulmonary infection in a HIV-negative patient with autosomal-dominant STAT3 mutation: a case report. Ther Adv Respir Dis. 2020; 14:1753466620929225.
Kamchaisatian W, Kosalaraksa P, Benjaponpitak S, Hongeng S, Direkwattanachai C, Lumbiganon P, et al. Penicillosis in patients with X-linked Hyperimmunoglobulin M syndrome (XHIGM), case reports from Thailand. J Allergy Clin Immunol. 2006;117(2):S282.
Liu D, Zhong LL, Li Y, Chen MJZddekzzCjocp. Recurrent fever, hepatosplenomegaly and eosinophilia in a boy. Zhongguo Dang Dai Er Ke Za Zhi. 2016;18(11):1145-9.
Du X, Tang W, Chen X, Zeng T, Wang Y, Chen Z, et al. Clinical, genetic and immunological characteristics of 40 Chinese patients with CD40 ligand deficiency. Scand J Immunol. 2019;90(4):e12798.
Cao C, Xi L, Chaturvedi VJM. Talaromycosis (Penicilliosis) Due to Talaromyces (Penicillium) marneffei: Insights into the Clinical Trends of a Major Fungal Disease 60 Years After the Discovery of the Pathogen. Mycopathologia. 2019;184(1).
Pongpom M, Vanittanakom P, Nimmanee P, Cooper CR, Jr., Vanittanakom N. Adaptation to macrophage killing by Talaromyces marneffei. Future Sci OA. 2017;3(3): Fso215.
Hamilton A. Penicillium marneffei: penicilliosis and the red peril in the east. Mycologist. 2003;17(2):84–5.
Wongkamhla T, Chongtrakool P, Jitmuang A. A case report of Talaromyces marneffei Oro-pharyngo-laryngitis: a rare manifestation of Talaromycosis. BMC Infect Dis. 2019;19(1):1034.
Ye F, Luo Q, Zhou Y, Xie J, Zeng Q, Chen G, et al. Disseminated penicilliosis marneffei in immunocompetent patients: a report of two cases. Indian J Med Microbiol. 2015;33(1):161–5.
Salzer HJF, Burchard G, Cornely OA, Lange C, Rolling T, Schmiedel S, et al. Diagnosis and Management of Systemic Endemic Mycoses Causing Pulmonary Disease. Respiration. 2018;96(3):283–301.
Qiu Y, Zhang JQ, Pan ML, Zeng W, Tan CMJCmj. Determinants of prognosis in Talaromyces marneffei infections with respiratory system lesions. Chin Med J (Engl). 2019;132(16):1.
Mo W, Deng Z, Li S. Clinical blood routine and bone marrow smear manifestations of disseminated penicilliosis marneffei. Chin Med J (Engl). 2002;115(12):1892–4.
Qin L, Zhao L, Tan C, Chen XU, Yang Z, Mo W. A novel method of combining Periodic Acid Schiff staining with Wright-Giemsa staining to identify the pathogens Penicillium marneffei, Histoplasma capsulatum, Mucor and Leishmania donovani in bone marrow smears. Exp Ther Me. 2015;9(5):1950–4.
Han XJ, Su DH, Yi JY, Zou YW, Shi YL. A Literature Review of Blood-Disseminated P. marneffei Infection and a Case Study of this Infection in an HIV-Negative Child with Comorbid Eosinophilia. Mycopathologia. 2019 Feb;184(1):129-139.
Huang YT, Hung CC, Liao C, Sun HY, Chang SC, Chen YC. Detection of circulating galactomannan in serum samples for diagnosis of Penicillium marneffei infection and Cryptococcosis among patients infected with human immunodeficiency virus. J Clin Microbiol. 2007;45:2858–62.
Drewniak A, Gazendam RP, Tool AT, van Houdt M, Jansen MH, van Hamme JL, van Leeuwen EM, Roos D, Scalais E, de Beaufort C, Janssen H, van den Berg TK, Kuijpers TW. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121(13):2385–92.
Drummond RA, Franco LM, Lionakis MS. Human CARD9: A Critical Molecule of Fungal Immune Surveillance. Front Immunol. 2018;9:1836.
Rieber N, Gazendam RP, Freeman AF, Hsu AP, Collar AL, Sugui JA, et al. Extrapulmonary Aspergillus infection in patients with CARD9 deficiency. JCI Insight. 2016;1(17): e89890.
Acknowledgements
We thank the patients and their families and all staff in the department for their support.
Funding
No funding was obtained for this study
Author information
Authors and Affiliations
Contributions
All authors contribute to this work. YF and CL conceived the study and coordinated and finalized the manuscript. CY participated in the design, statistical analysis and preparation of the manuscript draft. FH performed data collection. SL, DP, YF and CL were involved in the direct clinical care of the reported patient. FX participated in the coordination. All authors read and approved the final manuscript. YF and CL are both corresponding authors of this work and take responsibility for the article as a whole.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This case report and literature review were approved by Ethics Committee of Children’s Hospital, Chongqing Medical University.
Informed consent
The patient’s parents have provided written informed consent for the publication of the case report and accompanying images.
Additional information
Handling editor: Patrick CY Woo
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
You, Cy., Hu, F., Lu, Sw. et al. Talaromyces Marneffei Infection in an HIV-Negative Child with a CARD9 Mutation in China: A Case Report and Review of the Literature. Mycopathologia 186, 553–561 (2021). https://doi.org/10.1007/s11046-021-00576-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-021-00576-8