Abstract
The number of individuals working in agriculture is decreasing due to a labor shortage. When switching from manual to automation mode, image mapping must survey soil testing and plant vegetation development. The existing studies met several shortcomings in terms of higher cost remote sensing tools, higher execution time, higher computational complexity, and so on. To tackle these issues, we proposed a multispectral images feature extraction with vegetation indices for potato crops monitoring. This article discusses the many characteristics that are used to identify plants, such as plant count, plant height estimation, plant area evaluation, plant distance, crop vegetation growth detection, damaged area identification, and higher-lower vegetation. Calculating Crop growth days and minimizing leaf or root damage are provided by QGIS and Pix4Dmapper software. It measured all soil testing parameters as well as plant vegetative development. We provided a raster function for aggregation based on a low-resolution image for estimating plant area evaluation and parameters in a growing zone for higher cultivation plants. This research revealed 98% vegetation indices as well as plant characteristics. Various key parameters such as leaf area density, plants coordinates, plant height, the proportion of diffuse light for incident sunlight, and solar zenith angle are examined. The Quality Report of Vegetation Indices Value resulted in an accuracy of 96% when good or terrible weather conditions exist. All Vegetation indices values are compared with other existing technique results in terms of maximum and minimum vegetation to verify its superiority.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The Internet of Things platform (Gandhi et al., 2015) is used to interpret agricultural data collected through sensors. When IoT remote sensing data was used, HTTP updated agricultural data. HTTP agriculture identified the plant population when a robot or drone was employed in GIS. Many trails of plant structure, height, leaf indices, and nutritional data were broken by atmospheric light pictures. QGIS software is used to detect vegetation indices and the possibility for remote sensing. Pix4D mapper can display 3D models of the agricultural area after collecting overlap images.
Many trails of plant structure, height, leaf indices, and nutritional data were disrupted by atmospherically bright images (Fan & Liu, 2017). For recognizing vegetation indicators and remote sensing potential, QGIS software is utilized. Pix4D mapper can display 3D models of the agricultural area after collecting overlap images. Using QGIS software, pix4D displayed multi-spectral and hyper-spectral image information obtained during the automated drone installation process. Many scientists relaying on Remote sensing scripts using QGIS. For working in a harsh environment, various characteristics such as high efficiency, simple structure and operation, cleanliness, and energy-saving properties need to be present for developing robot-driven systems and industrial automation (Filipovic et al., 2011; Tao et al., 2021; Wei et al., 2021; Sundararaj & Selvi 2021; Jose et al., 2021).
Precision agriculture is a rapidly expanding use for unmanned aerial vehicles (UAVs). Cameras and multispectral sensors that can capture both infrared and visible light are used to monitor the health and vitality of crops in this way. Using these two wavelengths, various band-to-band ratios or discrepancies are employed to create information outputs that may be used for precision agriculture. The Normalized Difference Vegetation Index is one of the most often used vegetation indexes (NDVI). The difference between near-infrared light and red light reflected by plants is measured by this index. By dividing the difference by the total of the two wavelengths, the difference is balanced.
This normalization enables us to compare results from different datasets collected at different times or in different locations. This index is based on the notion that two essential plant components, pigments (namely chlorophyll) and leaf structure, impact those two wavelengths (red and near-infrared). Because chlorophyll absorbs light at that wavelength, healthy plants reflect little red light (chlorophyll also absorbs blue wavelengths). Because a healthy leaf structure reflects near-infrared wavelengths, healthy plants have a high near-infrared reflectance. Plants with higher chlorophyll have lower blue and red wavelength reflectance. In addition, healthier plants have a more robust leaf structure, which results in higher near-infrared reflectance. The Difference Vegetation Index (DVI) can offer a wide measure of vegetation health since visible light is not reflected nearly as much as near-infrared light. However, utilizing any of these two wavelengths is less precise than using the usual NDVI. Therefore in this work, we are utilizing different vegetation indices to yield more precision.
Farmers should be aware that dispersed light has a substantial influence on this wavelength if blue light is used. Blue light scatters more effectively in our surroundings, and this scattered light contributes to the signal we get from plant reflectance at that wavelength. Adding dispersed light to plant reflectance isn't optimal since focusing just on plant reflectance allows us to more accurately assess plant health. Figure 1 shows a drone image acquired from a DJI multi-spectral camera by MIT researchers in Chennai, at a height of 50 m above ground and a speed of 20 km per hour, with 70% overlapping. Topographic surveys and ortho-images may be generated with an RGB—multispectral camera and a NIR camera. They have various limits in terms of spatial resolution and remote sensing picture temporal value. The system is mainly developed to overcome the above shortcomings by designing a low-cost, adaptable unmanned system that may be used for a variety of purposes.
More emergency analyses were performed using remote sensing camera sensors (Munyati & Mboweni, 2013; Seong et al., 2020) with increasing spectral bands and estimated the nitrogen and biomass content. The remote sensing image-focused the temperature fluorescence, vegetation indices, and water stress. Vegetation reflectance varies in the blue and red regions of the band. Vegetation indices enhanced spectral bands and signatures for shape, vigor, and size. All vegetation images were covered using NDVI, SAVI, and GNDVI. NDVI was calculated from the reflectance of the NIR image and red band. It varies in LAF and biomass structure that depends on Drone mapping areas. GNDVI is equal to the NDVI when red_band is low and Green_band is high. It shows the absorption of radiation, correlation with biomass and LAI.
Data collection and preprocessing are one major task for Drone analysis (Sari et al., 2020; Guha & Govil, 2021). Before implementing the vegetation indices, image classification is required to homogeneity images in the crop fields. The result of the classification map is displayed after crops farm verified and the number of image classification is reduced by detecting noise signals and merging drone maps. The blue color is mentioned as bare soil with a small roughness. Lighter Green color mentioned legumes of olive trees. Dark green color mentioned as normal soil with rough vines. The orange color mentioned the production of the trees.
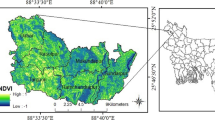
In Fig. 2, Vegetation indices Calculation is based on NDVI for detecting the Greenness of plants. The NDVI value of crops was calculated to provide maps of vigor growth and verification of uniform growth that were calculated by using Pix4D mapper. Spectral bands Green, Red, Red Eye, and NIR were stored in mapping when calculated the vegetation indices were. Pix4D software performed reflectance images and preprocessing of Crops—vegetation indices. Masks are defined as 1.6 ha of olive rees 3.6 ha of vines. The NDVI, NDRE, GNDVI, and SAVI defined the extraction of olive trees. Sometimes SAVI has L = 0.2 or 0.9, but we fixed L = 0.5 for the soil parameter.
In Fig. 3, map values of Vegetation indices estimated olive areas and estimated different values. We preferred analyzing linear correlation with VIs consideration of the Whole set of pixels. R2 correlation obtained when applying RGB image correlation Vegetation Indices with NDVI and NDRE for plotting the Crops or Olive Trees growth. The vine crops have a calculation of NDVI, GNDVI, NDRE, and SAVI set in L = 0.5 for extraction of preprocessing image. We aim to develop a user-friendly operation, R-based pipeline code to the task, build existing packages using raster, and provide analysis reports to better quality data of Agriculture in Data-set using QGIS. All data sets and packages are available from IIT and MIT Researchers. Changes in the external environment and artificial manipulations may, however, impact the results because there is no apparent difference in accuracy across various vegetation indices. Furthermore, only a few research have used vegetation indices to investigate the optimal planting methods and their viability in order to demonstrate the use of prediction models. The major objective of this study is to monitor the potato crops depending upon the vegetation indices with image feature extraction. The major contribution of this paper is presented as follows:
-
According to the literature analysis, increased computational complexity, high precision vegetation indices, longer execution time, and more expensive remote sensing techniques were used for vegetation index computation. To solve these issues, we proposed a novel methodology using different vegetations indices to precisely extract the chlorophyll content from the potato leaf multispectral images.
-
For the calculation of minimum and maximum vegetation indices, multispectral images from drone cameras are used to monitor potato fields.
-
Plant metrics such as plant count, plant height, plant area, the distance between plants, crop vegetation growth, damaged area identification, and higher-lower vegetation are all measured and analyzed in this study.
-
When good or terrible weather conditions exist, the Quality Report of Vegetation Indices Value resulted in an accuracy of 96%.
The rest of this paper is arranged accordingly. Section 2 provides a summary of different research work conducted in this field. Section 3 presents the various parameters used for computing the vegetation indexes from multispectral images. The experimental results obtained in the hyperspectral images collected from the drone are presented in Sect. 4 and the paper is concluded in Sect. 5.
2 Review of related works
Tucker et al. (2005) proposed a method that determines the spectral vegetation indices, processing the data, estimation of radiometric values, and compared the data that were obtained from 1982 to 1991 remote sensing. This method estimates the SPOT vegetation, GNDVI, and NDVI and acquired a maximum NDVI value of 0.65 and 0.1 as minimum NDVI. The Satellite NDVI image of agriculture areas was utilized by Gandhi et al. (2015). They proposed a novel change detection and land cover classification for various NDVI values. The NDVI values obtained by this method are 0.2 (maximum) and 0.1 (minimum).
Fan and Liu (2017) proposed a method that can be used to derive the statistical indices, data collection, and preprocessing. This method utilized the NN approach to detect the number of hidden nodes and NDVI inter-calibration outcomes. The resultant NDVI vegetation values are 0.35 (maximum) and 0.17 (minimum). Munyati and Mboweni (2013) proposed a method to analyze the SPOT HRG image and NDVI images. This method was also used to estimate the NDVI vegetation value and obtained maximum vegetation of 0.29 and minimum vegetation of 0.14.
NDVI crop coverage registration system was stated by Sari et al (2020). To find the crop coverage the author designed apiary schedule models. The performances were analyzed over MODIS dataset images. The NDVI and CCRS were merged for further enhancement and obtained NDVI values are 0.89 (maximum) and 0.45 (minimum vegetation). Cui et al. (2020) delineated a novel method based on Neural networks. The NDVI forecasting was analyzed by the adoption of Data and CNN algorithms. NDVI values obtained by this method range from 0.57 to 0.27.
Zaitunah et al (2018) proposed a method to analyze NDVI for landcover of Landsat8 images and covered Indonesia watershed places. The maximum and minimum values of NDVI obtained by this method are 0.876 and 0.15 respectively. Seong et al. (2020) stated a novel method to determine the NDVI estimation and BRDF correction through Himawari-8/AHI. The author explained the method of Atmospheric Correction for TOC Reflectance and BRDF for Normalized Reflectance. The maximum vegetation NDVI value obtained is 0.6 and the minimum vegetation of 0.3.
Guha and Govil (2021) delineated the Land surface temperature (LST) and NDVI values. The paper explained the methodology of Retrieving LST from Landsat data, Generation of NDVI, validation of LST-NDVI, and analyzed in between 2002 and 2018 NDVI data. NDVI values obtained by this method are 0.5 (maximum vegetation) and 0.2 (minimum vegetation). Cucho-Padin et al. (2020) proposed novel low-cost remote sensing tools to NDVI values. This can be estimated by SNR images from UAV platforms. This paper ensures high NDVI values of about 0.8.
Tsouros et al.(2019) proposed a method that was based on UAVs. This paper described IoT precision Agricultural using UAV and enabled the acquisition of real-time environmental data. IoT devices help to capture high spatial and temporal resolution images. Budiharto et al. (2019) stated a novel method in which precision agriculture is obtained by using Drone. It solved the pesticide sprayers problem using vegetation indices.
Panda et al. (2010) proposed a Neural network-based agriculture crop yield prediction. It is described to investigate the strength of vegetation indices using neural network techniques. The author also investigated NIR, NDVI, GVI, SAVI, and PVI vegetation indices. Jorge et al. (2019)proposed a novel method to detect the irrigation inhomogeneities in olive grove cultivation. This paper described the photogrammetric method to find heterogeneous areas of crops using a Drone camera. NDRE value is used to identify the growth of inhomogeneities in crops. NDRE, NDVI, GNDVI, and SAVI vegetation indices were used to find the irrigation irregularities in olive Crops.
Morlin Carneiro et al. (2020) proposed an approach to detect the spatial and temporal variabilities in soybean crops using canopy sensors. The canopy was used to find the vegetation indices values in the variability of crop growths. Robinson et al. (2017)proposed a novel Dynamic Landsat Derived Normalized Difference Vegetation Index (NDVI) Product for the Conterminous United States. The atmospheric contamination, data processing failures, and instrument errors were analyzed by the Remote sensing datasets.
3 Proposed approach
Images stored in Aerial SD-CARD using DJI phantom4 V3 Pro and Sentra Multispectral camera sensor with spectral bands RGB, Red edge, and NIR. The Pix4D mapper makes the orthomosaic 3D model using the Aerial image manually. This flight altitude parameter set is based on Heritability.
In Eq. (1), \(G\left( {\sigma^{2} } \right)\) is the identical twin correlation and \(E\left( {\sigma^{2} } \right)\) is the fraternal twin correlation which is used to compute the variance value for crops. Broad sense heritability (H2) is defined as the proportion of trait variance that is due to all genetic factors including dominance interaction and gene–gene interactions. The potato vegetation growth was calculated from NDVI images from 2017 up to 2019. Watershed transformation help in calculating plant stand count. The potato's higher growth is estimated by remote sensing GIS. The Image prepared for data extraction such as Fig. 1 and Fig. 3 show the orthomosaic image. The field Crop function is based on image cropping to calculate the area of interest which is marked in four corners of land using R-studio. In Fig. 3, the mask function of the field is classified on VI and specified by the no of users. The overall flow diagram with respect to the proposed model is delineated in Fig. 4.
3.1 Region of soil removed for vegetation analysis
Wet and Dry climate provided changes in soil color. Sometimes Vegetation Index could not detect from the NDVI image. In Fig. 5, Soil removal is applied for finding vegetation of plants. The soil color is hidden Fungal: bacterial ratio in plant image. Data extraction requires for detecting soil presence and absence. This is important for Remote sensing image analysis. We involved filed mask file to removing soil, where we apply Red_band is1, Green_band is 2, Blue_band is 3, and index_value is HUE.
3.2 Detection of plants count
Seed quality is measured from the number of count plants per land. The early vigor was measured from plant count for adjusting yield calculation and less water usage. The field mask created the binary maks using distance mapping transformation. IT generated an NDVI image from a combination of NIR and grayscale images. Boundaries are identified in between from watershed basins Which are interrupted in various plants. They evaluated distinct plants when touching the plants with 95% accuracy. When the differences early vigor in the emergency duration of images and genotypes, images stored in the dataset for stand count estimation and stored count of plant types, shown in Fig. 6 and values shown in Table 1.
3.3 Estimation of plant height
Plant height is measured from the correlation of life span, the mass of seeds, maturity time, and atmospherically light or solar brightness value. This measurement is calculated by increasing the color thickness of the direction of the plant per day. Image analysis CHM-Canopy height model implemented in this analysis for detecting plant height. This model covered the DSM-Digital surface model and Soil based model in the growth of plants per cycle. DSM models compared all days up to cultivation day which plants increased color in NDVI image. Tif file is shown time evaluation for plant height. Field image Region programming estimated plant height within three or four trails of potato NDVI calculation with the estimation of MIoU value from 0.77 to 1.00.
The determination and outcome of plant height are shown in Fig. 7 produced using a shapefile with CHM at 0.3 nm wavelength.CHM measured from DSM1 and DSM0 in 0.3 nm wavelength, DSM1 is soil base in 331.4 nm wavelength and DSM1 is vegetation growth in 331.4 nm wavelength. In Fig. 3, plant height data is plotted. Height of plant measured in minimum 12.2 cm and maximum 69 cm and weight measured in minimum 2.550 g and maximum 6.790 g. Soil index value and Water index value change depending upon the volume and height of the plant.
3.4 Evaluation of plant area
Image size and brightness of the image with resolution evaluated in processing time. We proposed one raster function with a low-resolution image for aggregation and field information function, field count function, and field area function for setting the plant area evaluation and its parameters for higher cultivation plant in a growth region while improving the higher cultivation work. In feature Extraction, the Resolution is kept as 1.7 cm in 115 s, and field information kept plot averages. When reducing the resolution to 3.5 cm, time 45 s decreased with parameter changes. The processing time reduced depends upon multiple cores and when using different parameters-25 s for four cores. In Fig. 8, shows the income and outcome plot view in the Evaluation of the plant area.
3.5 Distance between plant
When eliminating soil content from a target Tif image using the field crop function, the distance between plants and plant length is measured. Using row and column lines, we selected four points from a Tif image to form a rectangle or square shape. Only when four lines were picked was the complete shape detected. Others can be removed by using the field shape—python function. When plot values are compared, Plot 13 is deleted since it is less than plots 14 and 15, as seen in Fig. 5. NGRDI is one of Normalized Red and Green Difference Index to solving vegetation detection with low-density wavelength while detection of vegetation based on
The NGRDI index Python packages assisted in selecting the object to remove, cropping the image, removing the soil, and plotting shapefiles using data extraction to determine the distance between the two objects as shown in Fig. 9. The calculation of Heritability and NGRDI is based on Eqs. (1) and (2).
3.6 Detection of crops vegetation growth
Plant growth is measured from the Vegetation index. Most VIs are designed with programming for finding spectral differences reflectance from starting day to the ending day. The field index is designed and developed when storing pixels in the image. The field information function calculates the average VI indexes for coloring or RGB files to detect NDVI images. Biomass algorithms accurately concealed the Greenness percentage by 95%. It has a tendency to saturate with a canopy of less than 90%. The normalized difference red edge index classifies NDRE as a setting (NDRE). For detecting plant status with conditions, NDRE is more sensitive than NDVI. The NDVI image Calculation-2018 is represented in Fig. 10.
In potato cultivation, after the plant gets maturity in 100 days, It cultivated on a visual rating of 2–8 of senescence. Table 1 compared Drone Survey for MIoU-H calculation with users’ cultivation weight of plants per day. The comparison of Min-Vegetation indices with previous Research articles- 2020 is described in Table 2. It is compared to NDVI and NDRE in visual rating. NDVI and NDRE reading is higher than the visual rating in-between days calculation. Both indexes are most reliable and provide mature plant reading for improving remote sensing data of Plant growth value.
4 Experimental analysis and results
A vegetation index is a single number that quantifies vegetation biomass and/or plants vigor for each pixel in a remote sensing image. The index is computed using several spectral bands that are sensitive to plant biomass and vigor. Different kinds of vegetative indices are explained in the following sub-sections.
4.1 NDVI image calculation
NDVI image Calculation is shown in Fig. 11. We computed two bands 58 and 90 for the generation of NDVI images with wavelengths NIR (666-671) nm and NIR (826-831) nm. Multispectral and Hyperspectral images analyzed using field image python program. The soil area is separated using the NDVI technique and the results obtained are shown in Fig. 10. The technique is the same as that of rotating an image, taking a mask, plotting a shapefile, and analyzing multispectral and hyperspectral images. In Figs. 12 and 13, the value of blue is set as 3 and the index is HUE.
LST Calculation depends on NDVI, ITOI, E-Land Surface Emissivity, LST-Land Surface Temperature, PV-Proportion of vegetation, BT-Brightness temperature in Celsius, Radiance Mult Band10, and Radiance Add Band10. The LST calculation measures estimation of Land surface temperature with a high range and is shown in Fig. 14.
The following steps involved detecting LST calculation.
-
Step 1 Converted digital to spectral radiance. \(L\lambda = M_{L} *\,Q_{cal} + A_{L}\)
-
Step 2 Set Radiance Mult Band10 value \(3.3420\exp^{ - 4}\)
-
Step 3 Set Radiance Add Band10 value 0.10000.
-
Step 4 Measured Brightness temperature value = \(\frac{{K_{2} }}{{In\left( {\frac{{K_{1} }}{L\lambda + 1}} \right)}} - 273.15\)
-
Step 5 Set K2 Constant Band10 = 1321.07 and K1 Constant Band10 = 774.88.
-
Step 6 Measured NDVI value \(\frac{{{\text{Re}} d\,5 - {\text{Re}} d\,4}}{{{\text{Re}} d\,5 + {\text{Re}} d\,4}}\)
-
Step 7 Measured PV—Proportion of vegetation \(\left( {\frac{{NDVI_{\max } - NDVI_{\min } }}{{NDVI_{\max } \, + NDVI_{\min } }}} \right)^{2}\)
-
Step 8 Measured E-Land Surface Emissivity \(E\, = \,0.004*PV + 0.986\)
-
Step 9 Measured LST-Land Surface Temperature = \(\frac{BT}{1} + W*\frac{BT}{{14380}}*In\left( E \right)\)
4.2 Final Drone Survey Report between [2017–2019]: 3D model
4.2.1 Drone Survey for MIoU calculation
Drone recognized a survey of Agricultural land with the help of train datasets. The agriculture vision subset has 100,000 HD Tif images from different farmlands in the US. Every image had RGB and NIR with resolutions per pixel. 10 types of a field annotated for agricultural research. It is based on the Acquisition method, NIR camera or NDVI camera and Farmland image classification, GIS location, and Color Evaluation. Acquisition method completed images capturing on Drone. The NIR camera or NDVI camera sets high-speed image processing with ground resolution. Crop-type farmland was used in Cornland for this survey. Evaluation of metric Intersection over union
All Tif image files are kept through Drone from 2017 to 2019 on the Ketty website and land located in the US. Color evaluation computed from Drone to evaluate QGIS using IoT controller. We have compared from 2017 to 2019 in 90% images of a single land. Farmland images sub-sampled in full size 512*512. The annotation region of the image is cropped into multiple sub-images. 13,000 images trained, 5000 images evaluated, and finally, 4000 images tested from valuated images. In total, seven classes were examined namely Double plant, skipping plants, water content, waterway, weed cluster, land background, and cloud color-shadow as shown in Fig. 15, and the MIoU graph obtained for these readings are plotted in Fig. 16.
4.3 Identification of damaged area
Multiple images were evaluated using python code in QGIS. This result provided root area, leave indices, seeds count, structures of the plant, and damaged areas of the field. The filtered plant damage area and detected damaged Area is shown in Fig. 17. Vegetation Indices BGI, VARI, and SCI are involved in Identifying the plant’s damaged area. Four points set boundaries of vegetation growth in the hyperspectral image. This shapefile makes tiff file extraction from the area of damaging pests, field blocks, differences of soil, and so on. When Extract is true, the selected image file is converted to shapefile. It estimated and evaluated seeds, leaves, and diseases of the plant.
4.4 Dataset description
In Ecublens place Project in Pix4Dmapper Demo, NDVI value found maximum vegetation 0.9 and minimum vegetation 0.2. Ecublens WGS84 /UTM ZONE 32N project described drone mapping of agricultural field, ray cloud, and index calculation using pix4Dmapper. This project captured and analyzed the NDVI Tif image file of the UK smart place using Red, Green, Red edge, and NIR bands. Multispectral (Tif) images of Drone located Agricultural field in Ecublens Switzerland. The description of this dataset is provided in Table 3.
4.5 Final results using Pix4D
The Vegetation index used is computed using the formulas shown in this section and Table-1. Normalized difference vegetation index (NDVI) measured analyzes remote sensing data and live greenness vegetation with a NIR band. The NDVI result is shown in Fig. 18. Brightness Index Modified (BIM) helped to identify pollen and field count total with a C band. The formula to compute BIM is presented below:
Spectral Vegetation Indices (SVI) are formed by integrating multiple spectral bands into one. The results obtained for SVI are shown in Fig. 19. Green Leaf Index (GLI) with C band used to measure wheat cover, where red-green-DNS and applied. It is computed using the formula shown below:
Normalized Green Red difference index (NGRDI) is used for estimating the biomass of corn and found a linear correlation with C band using python in QGIS. The NGRDI result is shown in Fig. 20. Hue Index (HI) index is used for Primary colors with C band and it is computed using the formula shown below
SI-Spectral Slope Saturation Index (SI) used for estimating multispectral images in tif file with a C band and the formula to compute it is \(\frac{{\left( {R - B} \right)}}{{\left( {R + B} \right)}}\). Visible Atmospherically Resistant Index (VARI) was used for estimating the fraction of vegetation with the C band. The following formula is used to compute VARI
BGI is used for supporting archaeological investigations [1–5] and the formula used is \(\frac{B}{G}\) with C band. Plant Senescence Reflectance Index (PSRI) is used for health monitoring, stress detection of plant physiological and crop production, and yield analysis. The formula is shown in Table-1 which is applied with the RedEdge band. The PSRI result is shown in Fig. 21. Triangular Vegetation Index (TVI) was used for the Estimation of the Green leaf area index and applied formula \(\left( {0.5*\left( {120*\left( {NIR1 - G} \right) - 200*\left( {R - G} \right)} \right)} \right)\) with NIR band. The TVI result is shown in Fig. 22.
Chlorophyll Vegetation Index (CVI) used for crop development variation in different conditions with a NIR band. The CVI result is shown in Fig. 23. Green Chlorophyll Index (CIG) measured the greenness field of Chlorophyll with the NIR band and the CIG results obtained are shown in Fig. 24. Bare Soil Index (BSI) is used with a C band for identifying bare areas of soil and lands fallow. It is computed using the below formula
Chlorophyll Index Vegetation Index (CIRE) is measured using red-edge and NIR with NIR band. The CIRE result is shown in Fig. 25. Green Normalized Difference Vegetation Index (GNDVI) is used to determine water and nitrogen content with the NIR band. The GNDVI result is shown in Fig. 26. Ratio Vegetation Index (RVI) is used for estimating Vegetation Water Content and applied with the NIR band. The RVI result is shown in Fig. 27. Normalized Difference Red Edge (NDRE) index used for measuring healthy vegetation or not in the multi-spectral image with NIR band. The NDRE result is shown in Fig. 28. Difference Vegetation Index (DVI) was used for indicating plant water status with the NIR band. The DVI result is shown in Fig. 29. The enhanced vegetation index (EVI) is used for reducing the noise of background atmospheric saturation with the NIR band. The EVI value is computed using the formula shown below:
In Fig. 30, the phenological stage of Aerial-Drone view1 and view2 is shown using Vegetation Indices which cannot be calculated without the Blue band value. In Tables 1 and 3, the Maximum and Minimum Vegetation indices are compared with conventional techniques. We received a 96% Accuracy value of NDVI and other VIs using Pix4Dmapper that the value similar to like same as Ecublens Project et al. Other paper vegetation result accuracy is less than my report VIs value.
Several key parameters such as leaf area density, plants coordinates, plant height, the proportion of diffuse light for incident sunlight, and solar zenith angle based on Forest Light Environmental Simulator (FLiES) are examined. The canopy structure on canopy reflectance and solar radiation conditions are characterized by evaluating the effect of these parameters. Table 4 depicts the FLiES parameter sets used in this study.
4.6 Computational complexity
The computational complexity of the proposed methodology is explained in this section. The complexity associated with the training process of the proposed method is \(O(X)\). From this, \(O(XHtF)\) is the computational complexity of this technique to process the spectral bands and \(O(XHtFD)\) is the overall computational complexity for a total of D iterations.
5 Conclusion
The Tif file information was extracted and calculated using Pix4D mapper and RStudio Software. Remote sensing data was segregated and rotated for NDVI and NDRE calculation in particular parts of Multispectral and hyperspectral image (Tif). Canopy data and NDVI—NDRE data are measured from field area function, field polygon function and field plot function, spatial visualization function, and non-grid shape function. The Plant parameters are computed like the detection of plants Count, Estimation of plant height, Evaluation of plant area, the distance between plants, detection of crops vegetation growth, identification of damaged area, and higher-Lower vegetation. Several key parameters such as leaf area density, plants coordinates, plant height, the proportion of diffuse light for incident sunlight, and solar zenith angle are examined. Crops growth days of LST and reducing the damage of leaf or roots calculated using QGIS software. All parameters of soil testing and plant vegetation growth were tested and tabulated. This paper reported vegetation indices of NDVI and NDRE 98% and plant parameters. The vegetation indices NDVI, SVI, NGRDI, PSRI, GNDVI, RVI, NDRE, TVI, CVI, CIG, CIRE, and DVI were generated using the image calculation using pix4dmapper with the Green band, red band, red edge band, and NIR band. The pix4dmapper software generated a Quality report of Drone mapping images. The Quality Report of Vegetation indices value Tabulated in result accuracy 96%, when Good or bad climate conditions. All Benchmarking results of Vegetation indices compared with other paper results.
Availability of data and materials
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
Budiharto, W., Chowanda, A., Gunawan, A. A., Irwansyah, E., & Suroso, J. S. (2019). A review and progress of research on autonomous drone in agriculture, delivering items and geographical information systems (GIS). In 2nd World symposium on communication engineering (WSCE) (pp. 205–209). IEEE. https://doi.org/10.1109/WSCE49000.2019.9041004
Cucho-Padin, G., Loayza, H., Palacios, S., Balcazar, M., Carbajal, M., & Quiroz, R. (2020). Development of low-cost remote sensing tools and methods for supporting smallholder agriculture. Applied Geomatics, 12(3), 247–263. https://doi.org/10.1007/s12518-019-00292
Cui, C., Zhang, W., Hong, Z., & Meng, L. (2020). Forecasting NDVI in multiple complex areas using neural network techniques combined feature engineering. International Journal of Digital Earth, 3(12), 1733–1749. https://doi.org/10.1080/17538947.2020.1808718
Fan, X., & Liu, Y. (2017). A comparison of NDVI intercalibration methods. International Journal of Remote Sensing, 38(19), 5273–5290. https://doi.org/10.1080/01431161.2017.1338
Filipovic, V., Nedic, N., & Stojanovic, V. (2011). Robust identification of pneumatic servo actuators in the real situations. Forschung Im Ingenieurwesen, 75(4), 183–196.
Gandhi, G. M., Parthiban, S., Thummalu, N., & Christy, A. (2015). NDVI: Vegetation change detection using remote sensing and gis—A case study of Vellore District. Procedia Computer Science, 57, 1199–1210.
Guha, S., & Govil, H. (2021). An assessment on the relationship between land surface temperature and normalized difference vegetation index. Environment, Development and Sustainability: A Multidisciplinary Approach to the Theory and Practice of Sustainable Development, 23(2), 1944–1963.
Jorge, J., Vallbé, M., & Soler, J. A. (2019). Detection of irrigation inhomogeneities in an olive grove using the NDRE vegetation index obtained from UAV images. European Journal of Remote Sensing, 52(1), 169–177. https://doi.org/10.1080/22797254.2019.1572459
Jose, J., Gautam, N., Tiwari, M., Tiwari, T., Suresh, A., Sundararaj, V. & Rejeesh, M.R. (2021). An image quality enhancement scheme employing adolescent identity search algorithm in the NSST domain for multimodal medical image fusion. Biomedical Signal Processing and Control, 66, 102480
MorlinCarneiro, F., AngeliFurlani, C. E., Zerbato, C., Candida de Menezes, P., da Silva Gírio, L. A., & Freire de Oliveira, M. (2020). Comparison between vegetation indices for detecting spatial and temporal variabilities in soybean crop using canopy sensors. Precision Agriculture, 21, 979–1007. https://doi.org/10.1007/s11119-019-09704-3
Munyati, C., & Mboweni, G. (2013). Variation in NDVI values with change in spatial resolution for semi-arid Savanna vegetation: A case study in northwestern South Africa. International Journal of Remote Sensing, 34(7), 2253–2267.
Panda, S. S., Ames, D. P., & Panigrahi, S. (2010). Application of vegetation indices for agricultural crop yield prediction using neural network techniques. Remote Sensing, 2(3), 673–96. https://doi.org/10.3390/rs2030673
Robinson, N. P., Allred, B. W., Jones, M. O., Moreno, A., Kimball, J. S., Naugle, D. E., Erickson, T. A., & Richardson, A. D. (2017). A dynamic Landsat derived normalized difference vegetation index (NDVI) product for the conterminous United States. Remote Sensing, 9(8), 863. https://doi.org/10.3390/rs9080863
Sari, F., Kandemir, İ, & Ceylan, D. A. (2020). Integration of NDVI imagery and crop coverage registration system for apiary schedule. Journal of Apicultural Science, 64(1), 105–121.
Seong, N. H., Jung, D., Kim, J., & Han, K. S. (2020). Evaluation of NDVI estimation considering atmospheric and BRDF correction through Himawari-8/AHI. Asia-Pacific Journal of Atmospheric Sciences, 56, 265–274.
Sundararaj, V., Selvi, M. Opposition grasshopper optimizer based multimedia data distribution using user evaluation strategy. Multimed Tools Appl, 80, 29875–29891. https://doi.org/10.1007/s11042-021-11123-4 (2021).
Tao, H., Li, X., Paszke, W., Stojanovic, V., & Yang, H. (2021). Robust PD-type iterative learning control for discrete systems with multiple time-delays subjected to polytopic uncertainty and restricted frequency-domain. Multidimensional Systems and Signal Processing, 32(2), 671–692.
Tsouros, D. C., Bibi, S., & Sarigiannidis, P. G. (2019). A review on UAV-based applications for precision agriculture. Information, 10(11), 349. https://doi.org/10.3390/info10110349
Tucker, C. J., Pinzon, J. E., Brown, M. E., Slayback, D. A., Pak, E. W., Mahoney, R., Vermote, E. F., & El Saleous, N. (2005). An extended AVHRR 8-km NDVI dataset compatible with MODIS and SPOT vegetation NDVI data. International Journal of Remote Sensing, 26(20), 4485–4498. https://doi.org/10.1080/01431160500168686
Wei, T., Li, X., & Stojanovic, V. (2021). Input-to-state stability of impulsive reaction–diffusion neural networks with infinite distributed delays. Nonlinear Dynamics, 103(2), 1733–1755.
Zaitunah, A., Ahmad, A. G., & Safitri, R. A. (2018). Normalized difference vegetation index (Ndvi) analysis for land cover types using landsat 8 oli in besitang watershed, Indonesia. In InIOP Conference Series: Earth and Environmental Science (Vol. 126, No. 1, p. 012112). IOP Publishing.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Meivel, S., Maheswari, S. Monitoring of potato crops based on multispectral image feature extraction with vegetation indices. Multidim Syst Sign Process 33, 683–709 (2022). https://doi.org/10.1007/s11045-021-00809-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11045-021-00809-5