Abstract
Segmenting nuclei in histopathological images presents challenges due to variable sizes and overlapping regions, compounded by inter-class heterogeneity in shape and function. To overcome these, we proposed a robust deep learning architecture integrated with a novel edge detection technique based on local standard deviation. This method efficiently identifies nuclei edges, even at Multiscale levels, by exploiting the relationship between local standard deviation and image features. Our CNN architecture consists of stable residual, bottleneck, and attention decoding blocks, enabling effective extraction of high-level semantics and precise image localization. Evaluation on H&E stained liver disease histopathological images and a multi-organ database showcases the effectiveness of our approach, comparable to modern deep neural networks. Three performance indicators are computed to statistically assess segmentation efficacy, consistently demonstrating the superiority of our method over others. For multi-organ dataset segmentation using proposed methods attains the Jaccard index (89.99%), Precision (91.5%), Recall (91.1%), F1-measure (91.27%), and Accuracy (95.42%). Similarly for liver dataset attains Jaccard index (65.72%), Precision (88.74%), Recall (89.53%), F1-measure (90.12%), and Accuracy (94.85%). By addressing the limitations of previous unsupervised methods, our approach offers a robust solution for nuclei segmentation across diverse datasets. The significance of our work lies in its potential to enhance automated nuclei recognition in medical diagnosis and research, facilitating more accurate analysis of histopathological images and ultimately contributing to improved patient care and understanding of disease pathology.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Tumour which begins in the cells of the liver is known as liver cancer. The liver is a football-sized gland situated in the right upper quadrant of the belly, below the esophagus, and over the abdomen. The liver contains blood vessels that flow throughout the body continually, turning nutrition including medications received from the digestive system into ready-to-use molecules. Other key activities of the liver include eliminating poisons as well as other chemical organic waste from the blood as well as preparing it for excretion. The liver is especially susceptible to cancer cells migrating through the bloodstream since all of the blood throughout the body should flow through it [1]. The liver could grow various types of cancers. Hepatocellular carcinoma is by far the most common type of liver cancer which, starts in the main type of liver cell(hepatocyte). Certain kinds of liver cancer were significantly less prevalent, including intrahepatic cholangiocarcinoma and hepatoblastoma. Primary liver cancer, which starts in the liver, also a tumor that begins somewhere else in the body as well as moves to the liver, could damage the liver. Primary liver cancer was already linked to several cancer-causing agents, particularly herbicides including chemicals like vinyl chloride as well as arsenic. Smoking, particularly when paired with alcohol addiction, raises your risk. Aflatoxins, cancer-causing chemicals produced by a species of plant mold, also were linked to the outbreak [2].
The typical liver cancers are secondary or metastasis says they start somewhere else in the body. Since the liver is thought up of a variety of cell types, it could develop a variety of tumors. Many are benign (noncancerous), while others might be cancerous and could have the possibility to expand to other parts of the body (metastasize). Possible factors and treatments are used for these tumors. The growth of malignancy in the liver is much more frequent than cancer, which develops in the liver cells. Metastatic cancer, instead of liver cancer, is cancer that starts in some other part of the body, including the intestine, lungs, or breast, and afterward grows to the liver [3]. Metastatic colon cancer, for example, is cancer that forms in the intestine and develops in the liver. Liver cancer develops whenever the DNA of liver cells transforms (mutates). The substance that gives directions to every chemical reaction in the system is DNA. Variations in such directives might be the cause of gene mutations. As a result, cells might grow out of proportion and subsequently form a tumor, which is a collection of cancerous cells. In certain circumstances, including chronic hepatitis diseases, the causation of liver cancer is recognized. Moreover, liver cancer could happen in persons who have no chronic illnesses, and the etiology is still unidentified. Because of its substantial morbidity and mortality, liver cancer is a primary cause of the disease fatalities [4]. Early identification and accurate liver cancer diagnosis could significantly lower mortality and improve survival rates.
Several treatment options including individual conditions are available depending on the level of cancerization. As a result, determining the level of cancerization is important. Ultrasonography, nuclear magnetic resonance imaging, as well as pathological biopsies analysis, are some of the procedures used to diagnose liver cancer. Pathological test assessment is one of them, and it could get the most reliable information about liver cancer. The nuclear diameter, nucleocytoplasmic ratio, nuclear irregularities, hyperchromatism, and anisonucleosis, as well as the nuclear texture, are all being used by pathologists to diagnose liver biopsies as well as classify the cancerization into five classes (ranging from 0 to 4) based on six parameters [5]. On the other hand, because it is qualitative as well as non-quantitative, the interpretations of unusual cases provided by pathologists are almost always different. As a consequence of this, it would have been advantageous to have an automatic rating solution that provides pathologists with a diagnosis standard. The coverage of tests can be improved by using automated testing since the automated execution of test cases is far faster than the manual execution of test cases, and it requires significantly fewer resources to execute when compared to manual testing. The nuclei segmentation using biopsy pictures is the most significant aspect of the automated rating method. The effectiveness of the computerized grading scale would be influenced by the segmented images. Cancer identification could be aided by imaging technologies that include computed tomography, magnetic resonance imaging, and ultrasound; nonetheless, histopathology image analysis (HIA) is the gold standard for oncology diagnoses [6]. Cancer is now a big public health issue. It is the second major cause of death in the US. The major cause of this phenomenon is lifestyle factors including tobacco addiction, lack of exercise, and overconsumption of unhealthy foods and processed food, so on. Recent advances in science and technology have paved the road for early cancer identification and treatment. However, there are numerous hurdles in developing an automatic technique for detecting cancer cells.
Fluorescent markers [e.g., 4′,6′-diamidino-2-phenylindole (DAPI)] or histochemical staining can be used to stain the nucleus of cells (e.g., hematoxylin). It’s critical in such systems to be able to adequately recognize the appropriate number of neurons as well as define these with the highest efficiency and lowest manual work. It’s also important to modify sophisticated algorithms quickly to samples of various tissues taken under various imaging settings. Although when nuclear shapes and sizes were perfectly partitioned, there remains inherent diversity. Nuclei in pathological specimens might have unusual forms and diameters. Another type of segmentation mistake is varying chromatin texture—highly patterned nuclei seem to be more difficult to segment, particularly whenever they are tightly crowded. Throughout this field, separating densely crowded cell nuclei has long been a concern. Because there are so many nuclei in the area (particularly in whole-slide photos), approaches that are both technically manageable and efficient are required. Finally, additional mistakes are caused by imaging distortion within the background areas, particularly because of the fluorescence data, as well as the existence of spectrum un-mixing errors in processing multi-spectral images. The technique of finding a series of image features, typically one per nucleus of the cell and closer to its center, which is frequently termed as “markers” or “seeds,” might be the most crucial part of nuclear segmentation techniques. The following methods employ these sites to define the geographic range of every nucleus of the cell. Indeed, the precision and consistency of the first seed points are important to segmentation performance [7].
The pathologist assistant’s job is to assess the tissue’s cell arrangements. As a result, this type of assessment is wholly subjective, with several variables and errors. It could be handled with the help of a computer-assisted automatic detection technique utilizing histopathological images. Because the automated diagnosis of cancer offers enhanced cancer treatment, it has piqued the interest of academics in previous times [8]. Histopathological pictures are widely utilized in a variety of potential treatments, including medical diagnosis, cancer malignancy assessment, and therapy efficacy predictions because they provide detailed information about cellular morphology as well as tissue structure. Unfortunately, a pathologist’s subjective examination of histological pictures takes patience and therefore is imprecise. The goal of a digital histopathology image study is to examine histopathological pictures automatically, which can significantly enhance diagnostic repeatability and objectivity. The duty of dissecting each nucleus in histological photographs is an especially important one. However, there are still some difficulties in segmenting overlapping, or touching nuclei, and there is also a restricted ability to apply the method to diverse types of tissues and epithelial tissue [9]. It would appear that highly patterned nuclei are more difficult to segment, particularly when there is a dense concentration of them. Because there are so many nuclei in the vicinity, there has long been a concern over the difficulty of separating densely crowded cell nuclei. As a result, automated cancer in the liver of HIA is required to increase the effectiveness and precision of diagnostics. Computer-aided automatic image assessment of liver cancer has become possible due to better advancements in pattern classification as well as visual analytical techniques. Computer-aided approaches are easier, faster, and more impartial than manual processes [10]; additionally, they could be implemented by specifying a collection of useful texture features and classification techniques. The latest automated approaches, nevertheless, have had some drawbacks. Developing feature extraction, for starters, involves specialist knowledge, which would be biased and time-consuming. Secondly, categorization effectiveness is overly reliant on the features picked. Lastly, when the database varies, the feature extraction cannot be continuously improved.
Robust unsupervised edge detection methodology is suggested for the nucleus segmentation of liver cancer H&E colored histopathological images. Variations in lighting, color, shading, and textures can produce an edge, which can be used to determine the width, height, direction, and surfacing aspects of a digital image. These qualities can be determined by using the edge. The digital analysis of the image helps in the selection of edge points by deleting data that is not necessary for the analysis. Image processing, computer vision, and machine vision are all examples of applications that make use of edge detection for image segmentation and data extraction. Noise can muddle the identification of refined modifications, which is dependent upon the pixel’s threshold of modification that identifies an edge. Detecting these boundaries is challenging and consumes more time, particularly whenever an image is distorted by noise. In the core fields of image processing, including feature detection and feature extraction, edge detection is a basic and important tool. The finding of boundaries that represent the limits and split the representation of the image from other regions or objects in a digital image is known as edge detection. When edge detection is performed, the edge that is being looked for is detected based on the brightness variations that occur inside the image pixels. It is a collection of processes for finding the points in an image where clearly and unambiguously a rise in brightness happens. This can be accomplished by looking at the image and comparing it to itself. To put it another way, ED is used to locate points in a digital image where there are abrupt changes in the level of brightness, often known as discontinuities. Edges are defined as points in an image where the brightness of the surrounding pixels varies dramatically. The sequence of actions is required to extract picture-related information such as image sharpening, augmentation, and object positioning. Input a color image initially. Filtration has been used to eliminate as much noise as possible while preserving the true edges of the object. To enhance the effectiveness of edges, use differentiating. The edge intensity threshold is being used to eliminate excessive edge pixels and keep the rest of the pixels contained. Some applications will require subpixel resolution to identify the distance of an edge and the space among pixels. After the edge exposure, get the images. Figure 1 shows the Flow diagram of edge detection.
In digital photos, the gradient-based method is also known as a mask. The digitized mask is used to compute differential approximation in either the horizontal or vertical direction of the image. Several edge detectors work well throughout uncorrupted photos, but they are extremely susceptible to distortion and therefore won’t work well on images that are blurry or won’t have “sharp” edges. Pre-filtering an image before doing edge detection is often beneficial for such purposes. When images are distorted by excessive amounts of distortion, smoothing filters are generally utilized as pre-filters. A median filter, for example, could be used to eliminate impulses. Such filters, on the other hand, incline to diminish gradients of the edge. Although linear sharpeners could improve edge gradients, but are still incapable of generating step edges and therefore are highly sensitive to interference and outliers. Furthermore, they frequently create significant ringing, which might result in false edges. The usage of certain edges with nonlinear are improving filters as pre-filters for gradient-based segmentation techniques. It is demonstrated that using nonlinear sharpeners could greatly boost the effectiveness of gradient-based edge detectors. Those filters share some characteristics that make them useful in this situation. It could, for example, transform fuzzy edges into excellent step edges while also suppressing noise. Furthermore, there is no ringing at the margins with these filters. As a result, false alarms are reduced, and gradients are broad and focused. It results in inherently narrower edge outlines and decreased threshold sensitivities. Following prefiltering, smooth edges that might go undetected were recognized. The suggested edge detection method is not as prone to noise as existing gradient-based edge detection techniques. A calculation method is also provided to demonstrate that the suggested edge detection approach produces less noise than traditional gradient-based edge detection methods.
Histopathology image processing, as an integral aspect of digital pathology, is becoming increasingly significant in cancer diagnosis, as it could provide direct as well as trustworthy information to prove the rating and form of cancer. The goal of nuclei segregation is to acquire precise information about every nucleus in addition to comparing the number of nuclei. Regarding nuclei identification, the shape of every nucleus is produced rather than only the positioning of its central points. As a result, it could precisely remove every nucleus from the images for additional investigation. Segmentation of the nucleus in histopathological images was studied for over a decade. To achieve reliable segmentation of photos of various categories, a wide range of interventions were presented. Most classic nuclei segmentation techniques may be broken down into two steps: first, recognizing the nuclei, and then obtaining the contours of every nucleus. The region of nuclei or the seed of every nucleus should be generated by the detection step. DL-based procedures have grown in recent years in image segmentation owing to their high effectiveness throughout several computer vision applications. These have had a substantial impact on all aspects of image processing and computer vision, including object categorization, identification, and segmentation.
This study introduces an automated edge detection method that operates independently of algorithm variables, followed by thresholding to create binary images. The proposed CNN architecture focuses on robust residual block and attention techniques for nuclei segmentation, effectively extracting deep features while eliminating unnecessary elements. Addressing the challenge of nuclei segmentation in histopathological images due to staining variations and diverse visual properties of cell types, the study leverages deep learning techniques with significant success. Overall, the main contributions lie in the development of an automated edge detection method, a CNN architecture tailored for nuclei segmentation, and the application of deep learning to address challenging segmentation problems in biological contexts.
The paper is divided into six sections. In Section 2, existing nuclei segmentation techniques are briefly discussed. The problem statement is discussed in Section 3. Section 4 covers the entire edge detection process, including the threshold operator as well as the CNN network segmentation algorithm. Section 5 contains the results and comments, as well as tables and graphs. Finally, the paper is concluded in Section 6.
2 Related works
According to the use of diverse staining, scanning, overlapped, clustered nuclei, as well as the confusing boundaries among adjacent cell nuclei, previous nuclei segmentation approaches have only yielded poor effectiveness using multi-center and multi-organ whole-slide images (WSIs). Researchers have developed a stain-aware nucleus segmentation idea of multi WSIs that is based on deep learning to address these problems. They present an effective method that utilizes stain clustering to pick several different staining patterns, in contrast to earlier research that normalized WSIs by using a single stain pattern obtained from the database. The rise in digital pathology has been accompanied by an increase in interest in automated nuclei segmentation in hematoxylin and eosin (H&E) stained histology images. Despite this significance, automated nuclei segmentation is still difficult to achieve because different types of nuclei have a variety of distinct appearances. This heterogeneity might result in the nuclei having a varying intensity of hematoxylin, which frequently harms the success of the approaches that are now being used. Different deep learning networks use another staining pattern, and afterward, the segmented masking of the specific models was combined using a Choquet integral-based aggregation algorithm. The experimental results reveal that the improved technique outperforms state-of-the-art nucleolus segmentation methodologies with aggregated Jaccard index (AJI) and F1scores of 73.23% and 89.32%, respectively, whereas accomplishing a lesser set of variables with a difficult multi-center multi-organ WSIs data frame. However, the strategy does not allow for the introduction of stain normalization techniques into the deep learning model and the use of multiple hierarchical procedures to merge the distinct nuclei segmentation models [11].
The gold standard in tumor prediction and diagnosis is histopathological tissue examination. Whole slide imaging (WSI), or the screening and digitizing including whole histological images, is increasingly have been used in pathology labs all over the world. Depending on WSI data, trained histopathologists may accurately diagnose biopsy samples. Because of the complexity of WSIs as well as the growing quantity of probable cancer cases, interpreting these images takes time. Automatic tumor tissue division improves the accuracy, efficiency, and reproducibility of investigations. Deep learning-based algorithms have recently shown cutting-edge performance in a wide range of image analysis applications, along with the interpretation of digitized slides. To resolve these concerns, researchers present a generic deep learning-based system for histopathological tissue analysis. The approach is essentially a set of separate techniques in the preprocessing training inference pipeline that, when combined, enhance the analysis’ effectiveness and generalization. Among the methodologies that are discussed are an aggregate segmentation strategy, a split of the WSI into fewer local patches despite the resolution of classification mismatches, quick reasoning approaches, and an efficient, patch-based probability estimation framework. The most significant drawback of the patch-based segmentation technique for WSI images was the fact that the smaller patch sizes were unable to take into account the broader context of the surrounding area. Using training images for which the label maps are already established, a patch database is constructed. Similar patches from the database are obtained and compared with each patch in the testing picture. After that, the labels that correspond to these individual patches are concatenated to generate an initial segmentation map for the test instance [12].
The contribution of this analysis was to create quantitative feature-based models using histopathological images to identify hepatocellular carcinoma (HCC) from neighboring healthy cells as well as estimate HCC patients’ prognosis following surgical removal. The features have been extracted from hematoxylin and eosin (H&E)-stained whole-slide images of HCC patients from The Cancer Genome Atlas as well as tissue microarray pictures from West China Hospital, in a completely automated pipeline utilizing computational methods to evaluate the quantification characteristics of histopathological slides of HCC patients. The collected characteristics have been utilized to build predictive methods that categorize tissue slides but also apply machine-learning techniques for predicting individual prognostic value. Every histopathology slide yielded a total of 1733 quantifiable image characteristics. In both the test and objective evaluation sets, the diagnostic classifier based on 31 characteristics was able to effectively discriminate HCC from neighboring normal tissues (AUC 0.886). As per their allocated risk ratings, the random-forest prognosis models with 46 characteristics were able to completely identify patients within every set into longer- or shorter-term survival categories. Furthermore, the prognosis algorithm researchers developed had an equivalent predictive accuracy of individuals’ mortality at various periods following surgery as TNM staging methods. These results suggested that image-based machine-learning systems could aid doctors in HCC diagnosis and post-hepatectomy prognosis predictions. Such highly dimensional datasets, on the other hand, necessitate professional examination or specialized systems and technology, which makes it difficult for doctors to use these procedures in everyday practice [13].
Long-term and recurrent liver damage causes hepatocellular carcinoma (HCC), a kind of primary liver cancer. The radical curative treatment of choice is usually surgical resection. Since initial HCC recurrent following resection is linked to poor overall mortality, predicting recurrence of the following resection is essential in medical care. However, the pathologic hallmarks of HCC recurrence in the initial stages are still to be determined. Relying on digitized pathologic pictures of hematoxylin and eosin-stained samples and machine learning using a support vector machine, researchers sought to forecast the rapid recurrence of HCC after resection (SVM). The study comprised 158 HCC individuals who met the Milan criteria and had surgical removal. The patients were categorized into three groups. The SVM-based prediction approach correctly differentiated the three groups by 89.9% (80/89). Group I was accurately guessed in all instances, whereas Group II was accurately predicted through one case and Group III was correctly predicted in eight cases. Digital histopathology and machine learning might be designed to estimate HCC relapse following surgical removal with good accuracy, and a better understanding of recurrence. Normal blood tests and diagnostic testing are now employed for follow-up monitoring following HCC resection in the majority of cases; nevertheless, any use of diagnostic imaging combined with computer vision provides potential as a technique for accurate postoperative follow-up monitoring [14].
In current history, hepatocellular carcinoma (HCC), the much more frequent kind of common malignant liver cancer, has emerged as the main cause of cancer fatalities. For tumor loading evaluation surgical planning, including postoperative evaluation and automatic segmentation of HCC lesions was crucial. Because the morphology of HCC lesions differs so much from patient to patient, detection and segmentation are time-consuming as well as an error-prone process. As a result, there is a great demand for a completely automated and dependable HCC segmentation solution. In this paper, researchers describe a unique hybrid neural network for efficient HCC recognition of hematoxylin and eosin (H&E)-stained whole slide pictures using multi-task training and ensemble learning methods (WSIs). To begin, three task-specific branchings are combined to expand the feature space, allowing the systems to learn additional basic characteristics and reducing the danger of overfitting. Secondly, to achieve feature aggregation, a supervised learning technique is used, during which selected kernel modules (SKMs) and spatially and channel-wise squeeze-and-excitation components (scSEMs) are used to capture characteristics from various regions and dimensions. On three publically accessible databases, the suggested technique exhibits state-of-the-art performance, with segmentation accuracy and reliability of 0.797, 0.923, and 0.765 in the PAIP, CRAG, & UHCMC&CWRU datasets, accordingly, demonstrating its efficacy in tackling the HCC segmentation challenge. However, this study would not look into whether a unified framework, using approaches like levels of learning and domain adaptability, could not achieve the same level of accuracy as the ensemble method [15].
A novel deep learning architecture named LiverNet is proposed for multi-class classification of HCC histopathology images, focusing on different sub-types of liver HCC tumors. The dataset comprises both a novel KMC dataset and the publicly available TCGA-LIHC dataset. By integrating convolutional block attention modules (CBAM) and atrous spatial pyramid pooling (ASPP) blocks, LiverNet effectively captures multi-scale features in H&E-stained liver histopathology data. Results demonstrate that LiverNet outperforms existing architectures in terms of classification quality and computational efficiency, achieving significant improvements in precision, recall, F1-score, intersection over union (IoU), and accuracy. LiverNet exhibits superior performance across all metrics, showcasing an approximate improvement in accuracy and F1-score on both the KMC and TCGA-LIHC datasets [16].
For advancing digital pathology applications such as cell phenotype determination, cancer grading, and prognosis an AI-based nuclear segmentation method is introduced for segmenting nuclei in histopathological images. This technique utilizes a new nuclear segmentation network enhanced with residual skip connections to address the challenges posed by diverse nuclei types and morphologies. Experiments were conducted on two datasets: The Cancer Genome Atlas (TCGA) and Triple-Negative Breast Cancer (TNBC). The result indicates that the proposed technique achieves superior performance compared to existing methods, with an aggregated JI (67.94) and dice coefficient (80.84) on the TCGA dataset, and a JI (73.32), dice coefficient (84.41), precision (83.52), recall(83.06), and F1-measure (83.29) on the TNBC dataset [17].
A novel approach for the early detection and diagnosis of oral cancer aimed to address the significant global health burden posed by this disease. Leveraging the mouth’s exceptional sensory capabilities, the study utilizes deep neural networks, particularly automated systems, to identify intricate disease patterns. Transfer learning techniques are applied to enhance performance in target categories, with a focus on achieving optimal learning rates through comparative analyses. The study conducts a categorization analysis of reference results and presents detailed findings. Preliminary results indicate the effectiveness of deep learning in tackling this challenging problem, with the Inception-V3 algorithm demonstrating superior accuracy compared to the existing methods [18].
With the widespread availability of mobile devices and affordable sensors, data is leveraged to enhance diagnostic capabilities. With heart disease being a leading global cause of mortality and disability, the study focuses on improving cardiac prediction using sensor data. Additionally, the study aims to develop an algorithmic model for sentiment classification using EEG signals, recognizing the impact of emotions on various aspects of life. Furthermore, the investigation delves into forecasting neuromuscular disease classification methods in clinical and biological settings. The study’s framework explores potential deep learning applications across these domains to advance diagnostic capabilities and improve healthcare outcomes [19].
A novel spatial-channel attention-based modified U-Net architecture with ResNet blocks to address nuclei segmentation challenges in histology images under variable conditions. It aims to simultaneously handle issues like poor staining quality, stain variability, tissue variability, and morphological variability common in clinical settings. Leveraging both channel and spatial features, the method provides solutions for staining and morphological variability. Utilizing three benchmark datasets, the proposed model demonstrates superior segmentation performance compared to existing methods. It also exhibits generalization across 20 cancer sites, surpassing previous literature. Moreover, the model’s simplicity makes it more accessible for practical implementation. Its impact extends to improving nuclei segmentation, classification, and cancer grading in histology analysis [20].
3 Problem statement
In histological image analysis, nuclei segmentation is a basic but difficult issue. One of the major issues is the presence of overlapping regions that make it more complicated to segment single nuclei. Segmentation of various-sized, contacting nuclei is another important problem during nuclei segmentation. Furthermore, automated nuclei identification is complicated by nucleus size and shape diversity among classes. Two key challenges in effectively segmenting and evaluating histopathology images are color variability in the images with varied nucleus architecture. Numerous machine learning algorithms depend significantly on hand-crafted features that are limited by manual thresholding. Deep learning techniques, on the other hand, were lately been applied to challenging segmentation problems in biology with significant performance. The nuclei areas inside the liver cancer Hematoxylin and Eosin (H&E) stained histopathological pictures are segmented using an unsupervised edge detection technique utilizing a thresholding operator with the CNN approach suggested in this paper. The method can recover nuclei edges quickly, even at multiscale, because local standard deviation values are connected with picture edge information. A thresholding procedure is used to turn the edge-detected images into binary images. Three blocks are included in the CNN architecture. The suggested deep-learning architecture outperformed other nuclei segmentation approaches while applying to nuclei segmentation challenges.
4 Proposed methodology
Sample images are imported. Image pre-processing is a method for enhancing the quality of an image. After being captured, the images were enhanced and pre-processed. The fundamental purpose of edge detection is to eliminate the images’ irrelevant noise. Furthermore, using the three blocks of CNN architecture, the necessary features of the images are retrieved, as well as the nuclei to identify liver cancer. Figure 2 depicts the segmentation procedure.
4.1 Dataset
Pathologists reportedly acquired a database of 80 H&E stained histopathological photographs for a specific organ, the liver. Initially, tissue collecting, fixing, implantation, subdividing, and coloring are performed on histopathological slides. Following that, an Olympus scanning visualizes every one of the colored images at the similar 40 microscopic zooms, and those are then turned into digital photos and transferred to a system. Each image is 8.31 MB in size and 1920 × 1440 pixels in size. The optimal image for cancer diagnosis, as per the pathologists, is a 40 microscopic zoom. Adobe Photoshop 2019 was used to generate a series of ground certainties for stained H&E liver cancer histopathological pictures. However these processes are manually done, and the ground certainties are generated using an automated selection feature found in Photoshop 2019. As per the visualization, operator error is quite low with this strategy. Experts have identified the nuclei (or pathologists). The nuclei looked blue in H&E stained histopathological pictures in the majority of instances. It is to explain that such ground implications were created solely to compare them to the ultimate segmented image as well as to measure the quality metrics. The Kumar dataset, created by Dr.Amit Sethi’s group at the Indian Institute of Technology, Guwahati, is another freely available multi-organ database. Every image within that collection is made up of 1000 × 1000 patches recovered from a single Whole Slide Imaging (WSI). The collection also includes cells with 7 distinct organisms: the stomach, the bladder, the breast, the prostate, the colorectal, the kidney, and the liver. There are 30 of these photos in the collection. All information has a broad statistical feature set because it was not developed from a unicellular organism or obtained from a specific organ.
4.2 Pre-processing
4.2.1 Extraction of tissue
Since the majority of WSI areas have the background and hence are useless in our research, the areas having liver tissue must be removed ahead of time. The colored image is first transformed into the HSV color system (HSV). The ideal threshold for every channel is then calculated using the threshold operator. Even though the weighted K-means approach is frequently used as a clustering algorithm, Thresholding is more computationally efficient. Lastly, the tissue regions were created by combining the filters of channels H and S.
4.2.2 Normalizing color
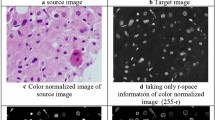
In HIA, color normalization is a critical step. Several parameters, including tissue section fixing, embedding, trimming, and dyeing techniques, affect the appearances of liver tissue slides colored with H&E. The two H&E coloring tissue samples differed significantly as a result of these circumstances. Even though these disparities only limit pathologists’ assessment of the image in portion, they have a major impact on automatic feature processing algorithms’ effectiveness. For relocating the colored picture channels to the left, a histogram equalization technique (HEA) is performed and afterward the pixels to [0-255]. The StainGAN approach is an advanced image-normalizing technique. Table 1; Fig. 3 contain the results. It is discovered that using advanced color normalizing algorithms is beneficial but not especially substantial in this case. As a result, HEA is used as the color-normalizing approach in the research to identify and concentrate on the fusing of patch-level information as well as the selecting of feature representation.
4.2.3 Extracting patch
However, a CNN is competent in picture segmentation on its own, utilizing a neural network to classify histopathology images with more than 100,000 pixels is unfeasible. Nevertheless, reducing the histopathological images to a suitable size for a CNN would result in a loss of detail [21]. The histological liver cancer images are tiled into patches with some of these concerns in mind and then adjusted to (114*114) pixels. A background that has patches in areas of more than 40% is deleted as well.
4.3 Feature extraction
4.3.1 Edge detection with threshold operator
Gradients, or the finite luminance differences among the two pixels, were calculated in edge detection algorithms. That is why such approaches calculate a certain amount of gradients including modest intensity changes (due to noise). As a result, every gradient-based edge detection method is thought to be prone to noise. Edge identification in the suggested method is done by determining the local standard deviation value in a’s*s’ window over every pixel. Because it relies on the idea of standard deviation rather than the calculation of gradients, the method that was proposed is less susceptible to being affected by noise. gradient-based edge detector that can detect edges at varying length scales and is less susceptible to noise in the input image than detectors that have been employed in the past.
Algorithm for edge detection
-
Step 1- The first stage in edge detection is to analyze simply this space info of a color input image because it largely includes foreground data (nuclei), that may be found across the database. Furthermore, as described in the preceding sub-section, the stains are immediately divided in this space.
-
Step 2- A non-linear filtering is employed for the space picture, which seems to be the inverse logarithmic (1 log2) of a digital Gaussian filter in a 3*3 frame. The filter decreases distortion in liver cancer histopathological images to a considerable extent.
-
Step 3- The user needs to enter a value (r) that corresponds to the microscopic zoom (10, 20, or 40) of both the associated image. ‘r’ is then multiplied by 0.15 and then placed in the variable ‘p’. Thus p = 6 in the case of 40 images as well as 3 in the instance of 20 images.
-
Step 4- The filtering grey scale image is then convolved with such a 3*3 mask, the coefficients of which would be dynamic instead of static. First, the standard deviation of ‘p*p’ pixels is calculated around the first pixel of an image and then the similar variable through every 9 locations of a 3*3 mask. Following that, the window coefficient is increased by the appropriate pixel intensities (in which the 3*3 mask is inserted), and those values are summarized in a variable ‘sum,’ substituting the first-pixel intensity with ‘sum,’ as indicated in Algorithm 1. Afterward, when, this 3*3 window (mask) is moved from one pixel to the other side of the image, a similar technique for allocating intensity distribution to every pixel in the image is repeated. The entire frame is convolutional with that 3*3 mask in this fashion, using the sliding window approach; the Python code is supplied in Algorithm 1.
4.3.2 Thresholding operator
An automated and non-parametric thresholding method is utilized which might also ideally pick the specified threshold in an image, maximizing inter-class variation in a bi-modal distribution. Equation 1 represents the weighted intra-class variation.
\({E}_{1}\left(H\right)\) and \({E}_{2}\left(H\right)\) are the frequency of groups \({E}_{1}\) and \({E}_{2}\), correspondingly, whilst \(H\) is the threshold value. The threshold was chosen as the best threshold value \(H\) to minimize \({\alpha }^{2}\left(H\right)\) in Eq. (1). It is explained that the entire logical derivation is in detail. Numerous investigations have found that the thresholding method is efficient as well as commonly utilized for nuclei segmentation. In required to the conversion of edge-detected (greyscale) images to binary images, Otsu’s thresholding method is used.
4.3.3 Segmentation using three blocks of CNN
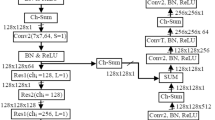
The difficulties encountered while automated segmentation of the nucleus of H&E stained images was mostly related to the uneven appearances of various types of cells and tissues, and the nucleus overlapped in immediate contact. Because a simple algorithm might not tackle this problem with effective results, deep neural networks are recommended. Along with its better-segmented images as well as fewer parameters; the suggested CNN for the segmentation as well as identification of nuclear pictures was principally influenced through the encoder-decoder paradigm of UNet [22]. A resilient residual block with a grouping of convolution layers in the encoder is used. Standard convolution layers, including depth-wise as well as point-wise, separated convolutions, have been employed in the grouped convolutions to improve confidence in high-level conceptual aspects of histopathological images as well as decrease the process parameters. Batch normalization as well as a rectified linear unit (ReLU) activation function were utilized in all of these convolution operations. After that, every residual block was down-sampled using a 2 × 2 max pooling procedure using phase 2. Because each following stage’s characteristics are half the size of the preceding stages, the procedure aids in rapidly extending the receptive field. A four-step robust residual block for feature extraction is employed, and after every down-sampling phase, the quantity of feature channels is multiplied. The bottleneck block of three 3 × 3 convolutions preceded the last down-sampled step. The block verified that the feature representations were compacted by the networks to obtain the greatest fitting in the available area and the least losses throughout learning [23]. The attention decoder block, which included attention gating, consolidated the retrieved characteristics, following a transposition convolution to rebuild the images in their initial dimensions. The fundamental goal of the concentration gating would have been to inhibit feature actions in all irrelevant background areas. The varieties of different channels were reduced in half after the fourth stage of feature fusion. A 1 × 1 convolution has been utilized for mapping features of vectors to the required amount of categories in the final layer.
4.3.4 Block of residual
The residual block retrieved the high-level latent semantic mappings from histopathological images. The block, shown in Fig. 4, consists of a collection of convolution layers also the depth-wise as well as point-wise separated convolution layers. I denote the feature mappings. 3 × 3 filter kernels having step 1 were utilized throughout all convolutional as well as depth-wise convolution layers. 1 × 1 filter kernels having step 1 were utilized in the pointwise convolution layer. As shown in Fig. 3, the depth-wise and point-wise convolutions have been conducted in the separated blocks. Batch normalization, as well as a rectified linear unit, is being used in every convolution (ReLU). To improve the channel’s capability as well as segmentation outcomes, the residual connections combined the features that are inputted with the features produced by the convolution layers [24]. The residual framework retrieved images’ tiny, good features that are essential in histopathological imaging, since losing just a minor feature might result in nuclei loss, resulting in bad identification. To improve processing performance and reduce the complexity of the algorithm while compromising the quality of the extracted features, different convolution versions had to be chosen. The feature channel capacity was gradually expanded by a factor of two using four such blocks and is evaluated as follows Eq. 2:
Here \({A}_{1}\) signifies the input features, \({A}_{mn}\) indicates the features learned following the convolution layer at the \(Xth\) stage and \(Yth\) layer, \({R}_{k*k}{S}_{k*k }{T}_{k*k}\) indicates standard convolution and also depth-wise and point-wise differentiable convolution layers, including both, with \(k\) depicting the number of filter kernels, and signifies feature concatenation.
4.3.5 Block of bottleneck
As shown in Fig. 5, the last down-sampled phases were preceded by a bottleneck block consisting of three 3 × 3 convolution operations. The block enabled the attention decoder block to quickly eliminate unwanted parts from the backgrounds by recording huge global information on the position including histopathological images. Because it lowered the number of features, the blocks were accountable for the least amount of training loss. Computational costs declined significantly since the number of operations required dropped due to the input tensor’s decreased characteristics. The procedure could be expressed mathematically as Eq. 3:
\({Q}_{k*k}\) means conventional convolution, batch normalization, and ReLU, in which \(k\) describes the amount of filter kernels. \({A}_{XY}\) indicates the outcome features from the residual block, \({AX}_{n}\) indicates the features only after convolution operation.
4.3.6 Block of attention decoder
Coarse extracted features, as expected, acquired contextual information as well as the positioning of foreground items. However, because of the wide range of shapes and appearances of cells and nuclei in histopathological images, false-positive assumptions were made. Attention gates (AGs) are employed to increase accuracy since they have several parameters as well as produce better outcomes than other strategies. To exclude task-irrelevant characteristics, the semantic attributes of the residual block as well as the coarse feature mappings out from the bottleneck block have been merged. The goal of the attention technique was to give weight to the input pattern, emphasizing important features and minimizing background noise. The skip connections were supplied as 1st Feature, and the coarse attribute obtained from the bottleneck block was provided as 2nd Feature (gating signal). The skipped connections were an essential component in the process of transferring feature information to the bottom layer. This was done to prevent the loss of gradient information that would have been caused by bypassing certain middle levels. The link provided representations of less significant features, some of which found their way into the attention mechanism over time. Through examination of the kernel weight distribution, we can identify it. There is a gradient that is diminishing if the weights are consistently falling closer and closer to zero. The accuracy of the procedure was increased by reducing the amount of vanishing gradient. Between the gating signal and skipping connections, further, 3 × 3 padded convolutional layers are employed to create finer coarse maps capable of identifying key input images as depicted in Fig. 6. Rather than downsampling the skip connections, it is chosen to increase limited feature maps by upsampling the gating signal, which comprises significant features transmitted through the bottleneck block. The sigmoid’s skip characteristics, as well as vector coefficients, are subsequently combined with a multiplier to eliminate unnecessary background patches and then sent across a 1 × 1 convolution layer to change the filtering dimensions as well as add depth without massively increasing variables and calculations. Such steps could be expressed mathematically as Eq. 4:
Here, \({I}_{1}\) represents feature inputs 1 and \({I}_{2}\) indicates feature inputs 2, \({\alpha }_{1}\) represents an element-wise nonlinear function, \({\alpha }_{2}\)a2 denotes the sigmoid function used for normalizing, and \({Q}_{k*k}\) stands for standard convolution, whereby\(k\) is the number of filter kernels, \({A}_{Y}\) stands for sigmoid operation attributes having pixels in the range [0,1], \({A}_{N}\) stands for convolution operation features, and is for multiplication.
From every pixel-wise feature space, wherein 0,1, the attention coefficients in the attention gate have been calculated. Such coefficients in lowered features output from task-irrelevant feature information, allowing for the identification of important image regions. The outcome of the attention gate was features adjusted by the coefficients, which reduced the unnecessary areas.
Figure 7 depicts the overall process of nuclei segmentation. In comparison to certain other reference nuclei segmentation algorithms for histopathological images, the suggested CNN model produces better outcomes with fewer variables [25].
4.3.7 Evaluation metrics
The image quality variables F- score (F1), Precision (Pre), Accuracy (A), Recall, and Jaccard indexes (JI) have been used to examine the simulation performance of benchmark deep-learning algorithms for segmenting nucleus of different histopathological image datasets. The proportions of pixels were overlapped between segmented results and the ground certainties were measured for both quality metrics. Such image variables were expressed in mathematical Eqs. 5–8:
Here TP, TN, and FN are True Positive, True Negative, and False Negative.
5 Result and discussions
A distinctive non-linear filter is being used in the study, which would be inverted logarithms (1 log2) of a 3*3 digital Gaussian filter. Because it’s a non-linear filter that provides higher weight to the center regions which is the only filtering that allows us to properly localize including two of the spatial and frequency domains concurrently, the Gaussian filter has become an obvious option for reducing distortion for above a decade. Furthermore, by introducing more nonlinearity (that is, adopting the inverse of logarithmic base 2), the nonlinear filter significantly reduces noise in histopathological images. Moreover, the level of noise in the image processed by the suggested edge detection approach is substantially lower than that of other traditional gradient-based methods. Every state-of-the-art CNN design employed very similar hyper-parameters during the training stage. All of the algorithms were extensively examined, and the measurement results shown in Tables 2 and 3 are the arithmetic averages following the execution of every model three times for both databases. For the dataset, all of the existing test images are used to achieve proper assessment. The test photos have been used to analyze the metrics findings for both databases. Figure 8 shows the comparison chart of existing and proposed methods of the liver dataset.
Tables 2 and 3 show the performance metrics for every model. These performance criteria were evaluated by inferring algorithms after training. Having a higher F score and a higher IOU (JI) score for both databases, the tables demonstrate that the proposed CNN architecture surpasses current models. In comparison to the current model, the suggested model produced more than a 1% increase in Table 3, while it provided roughly a 3% increase in Table 2, the liver dataset.
Figure 9 presents the evaluation metrics for nuclei segmentation on the liver dataset, comparing different methods. Precision values range from 73.42% (UNet++) to 88.74% (Proposed CNN), indicating the proportion of true positive predictions among all positive predictions. Recall varies from 68.52% (UNet) to 89.53% (Proposed CNN), representing the proportion of true positives correctly identified. Accuracy spans from 85.43% (FCN) to 94.85% (Proposed CNN), reflecting the overall correctness of the segmentation method. Among the methods, Proposed CNN demonstrates superior performance across all metrics, with precision, recall, and accuracy values of 88.74%, 89.53%, and 94.85%, respectively. SegNet follows closely with precision, recall, and accuracy values of 79.55%, 83.43%, and 91.35%, respectively. FCDenseNet also performs well with precision, recall, and accuracy values of 76.41%, 73.63%, and 90.21%, respectively. UNet and UNet + + show lower performance compared to the other methods, indicating the effectiveness of the proposed CNN architecture in nuclei segmentation on the liver dataset.
Figure 10 shows the comparison chart of existing and proposed methods of multi-organ datasets. Nuclei segmentation in histopathological images is an important step in accurately identifying then treating cancer. DNN plays a significant part in such activity, and they perform admirably. Because nuclei are the building components of tissues, their correct detection predicts future medical consequences. Convolution neural networking is the most ideal for segmenting the nucleus from histopathological images, according to recent advancements in medical imaging, because it produces better performance in terms of accuracy and also is quicker than several other methods. The issue of segmenting nuclei is addressed in the study by employing extensive attentiveness processes that help emphasize the salient sections of features.
Figure 11 displays the evaluation metrics for nuclei segmentation on the multi-organ dataset, showcasing various methods’ performance. Precision values range from 62.75% (UNet) to 91.5% (Proposed CNN), representing the proportion of true positive predictions among all positive predictions. Recall varies from 61.43% (UNet) to 91.1% (Proposed CNN), indicating the proportion of true positives correctly identified. Accuracy spans from 69.42% (FCN) to 95.42% (Proposed CNN), reflecting the overall correctness of the segmentation method. Among the methods, Proposed CNN demonstrates superior performance across all metrics, with precision, recall, and accuracy values of 91.5%, 91.1%, and 95.42%, respectively. SegNet follows closely with precision, recall, and accuracy values of 89.1%, 89.2%, and 91.49%, respectively. FCDenseNet also performs well with precision, recall, and accuracy values of 88.78%, 88.93%, and 90.23%, respectively. UNet and UNet + + show comparatively lower performance compared to the other methods, emphasizing the effectiveness of the proposed CNN architecture in nuclei segmentation on the multi-organ dataset.
A nuclei segmentation approach that is based on determining local standard deviation as suggested in this study paper for stained H&E liver cancer histopathological images. The suggested edge detection approach, like other current edge detection methods, wasn’t susceptible to distortion since it used the concept of standard deviation instead of calculating gradients. Furthermore, it was demonstrated that the suggested segmentation approaches overcome all of the drawbacks of prior unsupervised techniques.
6 Conclusion
In conclusion, our proposed system addresses the critical task of nuclei segmentation in histopathological images, particularly focusing on stained H&E liver disease images. By integrating a novel edge detection technique based on local standard deviation and leveraging a robust CNN architecture, we achieved significant advancements in automated nuclei segmentation. The proposed method demonstrated promising results on multi-organ with impressive metrics such as Jaccard index (89.99%), Precision (91.5%), Recall (91.1%), F1-measure (91.27%), and Accuracy (95.42%). and liver datasets attain the Jaccard index (65.72%), Precision (88.74%), Recall (89.53%), F1-measure (90.12%), and Accuracy (94.85%). However, despite the success of our approach, there are limitations to consider. One limitation is the reliance on stained H&E images, which may not cover the full spectrum of histopathological variations. Additionally, the proposed method may require further refinement to handle more complex scenarios such as overlapping nuclei or rare cell types. Future research could explore the integration of additional imaging modalities or advanced deep-learning techniques to enhance segmentation accuracy and robustness. Moreover, extending the evaluation to include diverse datasets from various organs and pathologies would provide a more comprehensive understanding of the proposed method’s generalizability. Overall, while our study presents a significant advancement in nuclei segmentation, there remains ample opportunity for further refinement and exploration in this important area of computer-aided diagnostics for cancer classification and prognosis.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Mirmozaffari M (2019) Developing an expert system for diagnosing liver diseases. Eur J Eng Technol Res 4(3):1–5
McGlynn KA, London WT (2011) The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis 15(2):223–243
Iwatsuki SHUNZABURO, Gordon RD, Shaw BW Jr, Starzl TE (1985) Role of liver transplantation in cancer therapy. Ann Surg 202(4):401
Sun C, Xu A, Liu D, Xiong Z, Zhao F, Ding W (2019) Deep learning-based classification of liver cancer histopathology images using only global labels. IEEE J Biomed Health Inf 24(6):1643–1651
Esgiar AN, Naguib RN, Sharif BS, Bennett MK, Murray A (1998) Microscopic image analysis for quantitative measurement and feature identification of normal and cancerous colonic mucosa. IEEE Trans Inf Technol Biomed 2(3):197–203
Lu C, Mandal M (2015) Automated analysis and diagnosis of skin melanoma on whole slide histopathological images. Pattern Recogn 48(8):2738–2750
Al-Kofahi Y, Lassoued W, Lee W, Roysam B (2009) Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng 57(4):841–852
Belsare AD, Mushrif MM (2012) Histopathological image analysis using image processing techniques: an overview. Signal Image Process 3(4):23
Chow KH, Factor RE, Ullman KS (2012) The nuclear envelope environment and its cancer connections. Nat Rev Cancer 12(3):196–209
Veta M, Pluim JP, Van Diest PJ, Viergever MA (2014) Breast cancer histopathology image analysis: a review. IEEE Trans Biomed Eng 61(5):1400–1411
Hassan L, Abdel-Nasser M, Saleh A, Omer A, Puig D (2021) Efficient stain-aware nuclei segmentation deep learning framework for multi-center histopathological images. Electronics 10(8):954
Baidoshvili A, Khacheishvili M, van der Laak JA, van Diest PJ (2023) A whole-slide imaging based workflow reduces the reading time of pathologists. Pathol Int 73(3):127–134
Liao H, Xiong T, Peng J, Xu L, Liao M, Zhang Z, Zeng Y (2020) Classification and prognosis prediction from histopathological images of hepatocellular carcinoma by a fully automated pipeline based on machine learning. Ann Surg Oncol 27:2359–2369
Feng S, Yu X, Liang W, Li X, Zhong W, Hu W, ..., Zhang X (2021) Development of a deep learning model to assist with diagnosis of hepatocellular carcinoma. Front Oncol 11:762733
Wang X, Fang Y, Yang S, Zhu D, Wang M, Zhang J, ..., Han X (2021) A hybrid network for automatic hepatocellular carcinoma segmentation in H&E-stained whole slide images. Med Image Anal 68:101914
Aatresh AA, Alabhya K, Lal S, Kini J, Saxena PP (2021) LiverNet: efficient and robust deep learning model for automatic diagnosis of sub-types of liver hepatocellular carcinoma cancer from H&E stained liver histopathology images. Int J Comput Assist Radiol Surg 16:1549–1563
Mahmood T, Owais M, Noh KJ, Yoon HS, Koo JH, Haider A, ... Park KR (2021) Accurate segmentation of nuclear regions with multi-organ histopathology images using artificial intelligence for cancer diagnosis in personalized medicine. J Personalized Med 11(6):515
Babu PA, Rai AK, Ramesh JVN, Nithyasri A, Sangeetha S, Kshirsagar PR, Rajendran A, Rajaram A, Dilipkumar S (2024) An explainable deep learning approach for oral cancer detection. J Electr Eng Technol 19(3):1837–1848
Kalaivani K, Kshirsagarr PR, Sirisha Devi J, Bandela SR, Colak I, Nageswara Rao J, Rajaram A (2023) Prediction of biomedical signals using deep learning techniques. J Intell Fuzzy Syst (Preprint) 1–14
Ahmad I, Xia Y, Cui H, Islam ZU (2023) DAN-NucNet: a dual attention based framework for nuclei segmentation in cancer histology images under wild clinical conditions. Expert Syst Appl 213:118945
Zaki G, Gudla PR, Lee K, Kim J, Ozbun L, Shachar S, ..., Pegoraro G (2020) A deep learning pipeline for nucleus segmentation. Cytometry Part A 97(12):1248–1264
Singh LK, Khanna M, Thawkar S, Singh R (2024) Deep-learning based system for effective and automatic blood vessel segmentation from retinal fundus images. Multimed Tools Appl 83(2):6005–6049
Singh LK, Pooja, Garg H, Khanna M (2022) Deep learning system applicability for rapid glaucoma prediction from fundus images across various data sets. Evol Syst 13(6):807–836
Khanna M, Singh LK, Thawkar S, Goyal M (2023) Deep learning based computer-aided automatic prediction and grading system for diabetic retinopathy. Multimed Tools Appl 82(25):39255–39302
Khanna M, Singh LK, Thawkar S, Goyal M (2024) PlaNet: a robust deep convolutional neural network model for plant leaves disease recognition. Multimed Tools Appl 83(2):4465–4517
Acknowledgements
This project was supported by Researchers Supporting Project number (RSP2024R283) King Saud University, Riyadh, Saudi Arabia.
Funding
No funding is involved in this work.
Author information
Authors and Affiliations
Contributions
All authors are contributed equally to this work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No participation of humans takes place in this implementation process.
Human and animal rights
No violation of Human and Animal Rights is involved.
Conflict of interest
Conflict of Interest is not applicable in this work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sunesh, Tripathi, J., Saini, A. et al. Nucleus segmentation from the histopathological images of liver cancer through an efficient deep learning framework. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18705-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18705-y