Abstract
Today, skin cancer is considered one of the most dangerous and common cancers in the world, demanding special attention. Skin cancer can be developed in different types, including melanoma, actinic keratosis, basal cell carcinoma, squamous cell carcinoma, and Merkel cell carcinoma. Among them, melanoma is considered to be more unpredictable. However, melanoma cancer can be diagnosed at early stages, which increases the possibility of successful treatment. Automatic classification of skin lesions is a challenging task due to diverse forms and grades of the disease, which demands the implementation of novel methods. Deep convolutional neural networks (CNNs) have shown an excellent potential for data and image classification. In this article, we examine the problem of skin lesion classification using CNN techniques. Remarkably, we present that prominent classification accuracy of lesion detection can be achieved through proper design and application of transfer learning framework on pre-trained neural networks. This can be accomplished without the need for data augmentation techniques; specifically, we merged the core architectures of VGG16 and VGG19, which were pretrained on a generic dataset, into a modified AlexNet network. We then fine-tuned this combined architecture using a subject-specific dataset consisting of dermatology images. The convolutional neural network was trained using 2541 images. In particular, dropout was employed to mitigate overfitting. Finally, we assessed the model’s performance by applying the K-fold cross validation method. The proposed model improved classification accuracy with an increase of 3% (from 94.2% to 98.18%) compared to other methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer encompasses various types of malignant tumors, commonly referred to as neoplasms in medicine. Skin cancer originates from the skin cells, which comprise the primary components of the skin. These skin cells undergo growth and division to generate new cells. Subsequently, the cells undergo aging and eventual death, with new cells emerging to replace them. Deviations can occur in the cell’s lifecycle; new cells appear when they are not required, and old cells stay alive beyond their lifespan. This accumulation of excess cells takes the form of an abnormal tissue known as a tumor. This might happen when one of the body’s cells undergoes abnormal growth due to various possible factors, primarily, continuous exposure to the sunlight, that eventually leading to the development of cancerous tumor. The tumor then invades and damages the affected area of the body before potentially spreading to other parts [13]. Interest in skin cancer diagnosis and therapy has significantly increased in recent years due to the irreparable damage caused by this type of cancer and its widespread prevalence. Skin cancer lesions can be classified into two main categories: malignant lesions and benign moles.
Among the malignant lesions, melanoma is considered one of the most deadly forms of cancers. Approximately 70% of worldwide deaths caused by skin cancer are attributed to melanoma. Skin cancer primarily manifests as extensive damage to the epidermal layer of the skin.
In this regard, early diagnosis plays a crucial role in increasing the chances of patient recovery. Therefore, significant efforts have been dedicated to develop effective methods for diagnosing the disease at early stages. Traditional image feature classification techniques have been employed to undertake this crucial task. However, given that human lives are at stake, the utmost accuracy in detection is imperative. For this purpose, deep learning algorithms have been exploited recently to ensure the highest possible accuracy of the results. In their research, Jayalakshmi et al achieved an accuracy of 89.3% by customizing and tuning the CNN model while using the PH2 dataset in a two-class classification scenario [14]. In general, the excellency of convolutional neural network in image classification have been widely approved across various applications. For instance, CNNs have been successfully utilized for tasks such as car license plates recognition, and aerial target tracking, resulting in high performance and accuracy [16, 23].
Brindha et al. unrevailed the superiority of the CNN algorithm over the SVM algorithm in the classification of ISIC image dataset, resulting in a significant increase in accuracy from 61% to 83%. [4].
Pham and his colleagues achieved an accuracy of 79.5% and 87% in classifying the ISIC dataset by utilizing Transfer Learning methods, specially, Reznet50, and InceptionV3, respectively [19].
Mijwil exploited and compared three different architectures; namely, VGG19, ResNet, and Inception V3, to detect skin cancer using ISIC2019 and ISIC2020 archives. The dataset consisted of a significant number of more than 24,000 images. They found an accuracy of 73.11%, and the best 86.9% for the mentioned architectures [17].
In their study, Nawaz et al. combined a region-based CNN technique with the support vector Machine (SVM) classifier and utilized the ISIC2016 dataset for melanoma classification. To increase the dataset size, they employed data augmentation techniques, resulting in more than 7,000 images. Their approach achieved an accuracy of 89.1% [18].
In their investigation, Alzubaidi and his colleagues achieved a classification accuracy of 97.5% for skin lesion images using a deep learning method. They employed a multi-phase training scenario and a multistage CNN model with the aim of surpassing the limitations posed by limited number of labeled data for medical applications [2].
In their paper, Ashraf et al. conducted an examination of skin lesion images with the help of deep learning method. They employed region of interest segmentation preprocessing and image augmentation. The initial result without region of interest segmentation and augmentation was approximately 81.3%. However, by implementing the segmentation and augmentation, they acquired an increase to 97.2% in the classification accuracy [3]. Rafi and coworkers achieved an accuracy of 98.7% by applying transfer learning architectures based on Efficient NET-B7. Their approach involved extensive image pre-processing, including resizing, conversion, augmentation, and in particular a post scaling step [20].
Lafraxo and coworkers proposed a CNN architecture for recognizing malignancy in dermoscopic images. In their approach, they employed regularization, as well as geometric and color augmentations to enlarge the datasets. Specifically, they augmented the ISBI dataset to 18,000 images, the PH2 dataset to 2,880 images, and the MED-NODE dataset to 1,800 images. The achieved accuracies were 98.44%, 97.39%, and 87.77% respectively [15].
Rasel and his colleagues implemented a deep CNN model based on transfer learning, with the main ideas borrowed from LeNet. Their model consists of a total of 31 layers and utilized nonlinear variable Leaky ReLU activation function. The training was conducted over 250 epochs. They achieved accuracies of 75.50%, 97.50%, and 98.33% for PH2, augmented (rotated) PH2, and a smaller subset of images from ISIC archives, respectively [21].
Hassan et al. conducted a comprehensive literature survey to assess the performance of different optimization algorithms. Additionally, they demonstrated accuracies of 97.3% (92% up to 98%) and 99.07% for their deep learning model applied to the ISIC dataset (with 6000 iterations) and the COVIDx dataset (with 300 iterations), respectively. These impressive results were obtained by utilizing the Adam optimizer [10].
Furthermore, Hassan et al. achieved a superior accuracy of 97.47%, employing ResNet50 and Adam optimizer for the classification of retinal optical coherence tomography images with 84495 total number of images [9].
Alahmadi and coworkers presented a CNN/transformer coupled network, that incorporated both supervised and unsupervised training techniques. Their approach yielded accuracy rates of 95.51% and 97.11% for ISIC and PH2 datasets, respectively [1].
Wu et al. proposed and developed a novel two-stream network, that efficiently capture both local features and global long-range dependencies by combining a CNN with an additional transformer branch. They achieved accuracies of 95.78% (ISIC2018), 93.26% (ISIC2017), 96.04% (ISIC2016), and 97.03% (PH2) for the respective datasets. For a better model initialization, they used deit-tiny-distilled-patch16-224 and ResNet34. They also utilized dynamic polynomial learning rate decay [26].
In this manuscript, our aim is to achieve superior performance and precision through the utilization of a transfer learning model. Our approach involves an innovative adaptation and fusion of network architecture and weights, with the primary objective of attaining better detection accuracy while reducing computational burden. Notably, our methodology yields remarkable results in prime detection accuracy without resorting to any data augmentation techniques.
The rest of the paper is organized as follows: Section 2 discusses the methods, model architecture, and the dataset used. Then, in Section 3 experiments and results are presented along with a discussion on the outputs. Finally, Section 4 concludes the paper.
2 Methods
In this research, we undertook the task of redesigning and training deep neural networks using images of skin lesions. A deep convolutional neural network (CNN) is trained using a dataset consisting of skin lesions images. To update the network weights, we employed the Adam optimizer and implemented early stopping. The experiments are performed on Google Colaboratory [12]. Eventually, the output layer was dedicated to performing the final binary classification. In order to prevent any increase in the loss value a random removal method has been employed.
2.1 Convolutional neural network (CNN)
Deep learning methods have broadened the borders of machine learning technology for practical applications. In this class of methods, intermediate layers are employed for data mapping and feature learning, which allows the elimination of non-automatic feature engineering, as the most advantageous distinction of the method. In this regard, for instance, convolution layers operate as the kernel in one of the most promising deep learning algorithm, known as CNN. Various architectures can be used for processing and classifying the input image as well as the intermediate feature maps. Subsequently, a pooling layer is used to reduce the size of the feature maps and network parameters. In our model, we incorporate the max-pooling strategy. After the final pooling layer, the fully connected layer is positioned. This layer is primarily responsible for converting the output of the neural network into a one-dimensional representation. The softmax function is placed as the last layer responsible for performing binary indexing (0 and 1) to represent the two classes of the images under investigation i.e. normal versus cancerous [5]. The described model is sketched in Fig. 1.
2.2 Model architecture
The basis of our proposed model lies in the integration of transfer learning principles with the renowned AlexNet architecture, thereby enhancing its performance within the context of our specific dataset. To accomplish this, we embark on a layered approach, supplementing the pre-trained architecture with additional layers through the application of transfer learning techniques. In essence, we amalgamate the weights garnered from the training of the ImageNet dataset using VGG16 and VGG19 architectures with those associated with both the initial three layers and the concluding two layers of our tailored AlexNet variant.
This intricate fusion of weights and architectural components not only imparts a sophisticated depth to our network but also endows it with a broader capacity to discern intricate patterns within the data. Moreover, the amalgamation of these diverse sources of knowledge mitigates overfitting tendencies, a feat that can be attributed to our strategic implementation of the dropout method. This approach introduces a deliberate element of randomness during training, thereby curbing the network’s inclination to excessively fit the training data. Through these meticulous steps, our model emerges as a robust solution that not only harnesses the strengths of transfer learning and architectural customization but also effectively manages the delicate balance between model complexity and overfitting prevention [25].
The proposed model has been implemented on Google Colab along with the other reference architecture. For the training of each network, we conducted 100 epochs while incorporating. This technique enables us to halt the training process once the highest attainable performance is reached, ensuring optimal results in the shortest time possible [11]. Finally, to address the task of classifying the image set into two classes, we implemented the last layer of the neural network with two neurons.
For the central component of our proposed model, we leveraged the frozen ImageNet weights from the VGG16 and VGG19 architectures. Adhering to the established protocol of transfer learning implementation, we fine-tuned and trained the last two layers of our model to facilitate custom classification based on our specific image dataset. It’s worth noting that our dataset comprised medical dermatological images, a category not explicitly represented in the 1000 classes of the ImageNet dataset.
In order to better tackle this challenge, we made the strategic decision to retrain the first three layers of the pre-trained AlexNet network. Notably, this choice had a substantial positive impact on the model’s ability to accurately delineate the boundaries of the lesions, as evidenced in our results. Throughout the training process, we retrained the weights of these three layers in addition to the last two layers. In this context, our approach can be described as a dual transfer learning methodology. [7, 27].
Figure 2 depicts the schematic of our CNN model, which utilizes a customized transfer learning scheme. This customization enhances the algorithm’s capability to accurately detect lesions, improves the speed of convergence, and also ensures high model accuracy.
2.3 Dataset
Medical datasets often suffer from unbalanced data, with a much larger number of benign samples than malignant ones. There are several methods to tackle this problem, one of which is random undersampling, as discussed in [24]. This method randomly reduces the number of benign samples to balance the dataset. To increase the number of malignant samples, we combined the image samples from the Complete-MedNode-Dataset.
In the current study, a total of 2541 input images were utilized, comprising 1200 melanoma lesions and 1341 benign mole images. To ensure dataset balance, a reduced number of benign samples were randomly selected [6]. For model evaluation, 762 images (30%) were set aside, while the remaining 1779 images were allocated for model training.
The image set is from the International Skin Imaging Collaboration dataset (ISIC) [22], which comprises images labeled by various institutions including the Hospital Clinics de Barcelona, Medical University of Vienna, Memorial Sloan Kettering Cancer Center, Melanoma Institute Australia, the University of Queensland, and the University of Athens Medical School.
In addition, other images of benign and malignant have been taken from the Complete-Mednode-Dataset, published by the Department of Dermatology of the University Medical Center Groningen [8]. To conduct the experiments in this study, we combined and balanced the image datasets from these two sources.
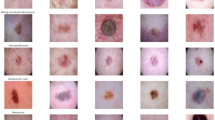
In general, these lesions are typically categorized into two groups: melanoma lesions and moles Benign (nevus). These categories are used to identify and detect suspected malignant melanoma lesions. Figure 3 shows some examples of both cases. In general, the size of the images are \(224\times 224\) pixels. It is noteworthy that, especially, since each network architecture implementation requires a particular specifications for the input images, we employed a pre-processing function for each case. Some researches conducted in the field has attempted to augment their dataset by cropping or rotating the images or applying data weighting techniques. However, in the present study, we integrated multiple datasets to prevent the use of duplicate image.
3 Results and discussion
To demonstrate the effectiveness of the proposed model, Fig. 4 illustrates the accuracy and performance of our customized transfer learning network, which is based on, VGG-16 in comparison to the performance of the reference transfer learning network [24]. As can be observed, the detection accuracy shows an increase from 96.5% to 97.51%. Furthermore, Fig. 5 illustrates the distinction between the utilization of a simple transfer learning network based on VGG-19 and the model that we have developed. In particular, it is evident that higher accuracy can be achieved by reducing the number of epochs required (from 97% to 98.4%).
3.1 Ablation study
In this experiment, we conducted three separate runs, systematically excluding each of the newly introduced layers, and assessed the resulting impact on the network’s performance. The outcomes clearly underscored the remarkable efficacy of the added layers, as the omission of any single layer invariably led to a noticeable decline in accuracy. This compelling evidence highlights the indispensable contribution of each layer to the overall functionality and effectiveness of the network, reaffirming their role in enhancing the model’s performance and robustness.
3.2 Optimizer selection
In this experiment, we looked at different optimizers. We focused on two specific ones: SGD and Adam. We compared how well they worked and put the results into a graph shown in Fig. 6. From the graph, it’s pretty clear that the Adam optimizer performed better than the SGD optimizer. This finding is important because it helps us understand which optimizer is more effective for our specific experiment.
3.3 K-fold cross validation
We employed K-fold cross-validation algorithm in order to evaluate and obtain a reliable perdiction of the true performance of the proposed model in accurately detecting skin lesion in unseen data. The incorporation of the K-fold cross-validation technique enabled the effective determination of optimal hyperparameter values for the implemented neural network. In this experiment, we employed a K value of 10. By utilizing the K-fold method, the modified VGG-16 and VGG-19 architectures achieved an average accuracy exceeding 97.5%. A summary of the details is presented in Table 1.
3.4 Early stopping
Two methods have been utilized to mitigate overfitting: dropout and early stopping. The graphs illustrate that early stopping not only helps prevent overfitting but also contributes to a relative reduction in the processing time required for data analysis.
Figures 7 and 8 compare the results obtained from the proposed method that employs early stopping with the reference cases for the both VGG-16 and VGG-19 based architectures networks. As evident from the figures, we avoided extra unnecessary data processing (shortened green plots).
In order to evaluate the performance of the proposed model in comparison with other models, Table 2 summarizes and compares the results of the present study and researches reported in the literature. It can be observed that the proposed (dual) transfer learning method achieves a significant level of accuracy while requiring relatively less workflow compared to other methods.
As the wrap-up, the analysis of skin lesion images is a challenging task due to high degree of similarity between these images. However, with the modification that was introduced in the transfer learning method, a significant increase in accuracy for lesion detection could be achieved. Table 2 summarizes the superiority of the proposed model in present paper compared to the reference studies. The table present the average values.
4 Conclusion
In recent years, the adoption of the transfer learning method has gained considerable attention among researchers, owing to its advantages in enhancing model performance. However, it remains imperative to tailor the network’s training to suit the specifics of each dataset. This paper has delved into this intricate landscape, striving to enhance the capabilities of deep networks by meticulously adjusting the layer configuration and weight distribution to align with the demands of detecting lesion-affected regions within images. As a testament to our endeavors, achievements have been realized, with accuracy levels reaching 92.5% for the VGG-16 architecture and an even more impressive 94.2% for the VGG-19 architecture. We also used k-fold cross-validation methodology, which ensures a robust and unbiased assessment of our proposed model’s performance. Employing k-fold the accuracy of 97.51% for the VGG-16 architecture and 98.1% for the VGG-19 architecture have been achieved.
Looking ahead, our work opens paths for future exploration. It would be worthwhile to consider the impact of different pre-trained architectures, as well as to explore how varying degrees of fine-tuning could further enhance the model’s efficacy. Additionally, while our study showcases promising outcomes, it’s essential to acknowledge its limitations. As with any methodology, there are constraints to consider, such as the potential for overfitting in more complex datasets or the challenges associated with domain shifts. Addressing these shortcomings and expanding upon the strengths of our approach will undoubtedly pave the way for the continued evolution of accurate and efficient lesion detection methods.
References
Alahmadi MD, Alghamdi W (2022) Semi-supervised skin lesion segmentation with coupling CNN and transformer features. IEEE Access 10:122560–122569. https://doi.org/10.1109/ACCESS.2022.3224005
Alzubaidi L, Al-Amidie M, Al-Asadi A et al (2021) Novel transfer learning approach for medical imaging with limited labeled data. Cancers 13(7):1590. https://doi.org/10.3390/cancers13071590
Ashraf R, Afzal S, Rehman AU et al (2020) Region-of-interest based transfer learning assisted framework for skin cancer detection. IEEE Access 8:147858–147871. https://doi.org/10.1109/ACCESS.2020.3014701
Brindha PG, Rajalaxmi R, Kabhilan S et al (2020) Comparative study of svm and cnn in identifying the types of skin cancer. J Crit Rev 7(11):640–643. https://doi.org/10.31838/jcr.07.11.117
Coşkun M, YILDIRIM Ö, Ayşegül U et al (2017) An overview of popular deep learning methods. Eur J Techn (EJT) 7(2):165–176
Fahad NM, Sakib S, Khan Raiaan MA et al (2023) Skinnet-8: an efficient CNN architecture for classifying skin cancer on an imbalanced dataset. In: 2023 International conference on electrical, computer and communication engineering (ECCE), pp 1–6. https://doi.org/10.1109/ECCE57851.2023.10101527
Gao Y, Mosalam KM (2018) Deep transfer learning for image-based structural damage recognition. Comput Aided Civ Infrastruct Eng 33(9):748–768. https://doi.org/10.1111/mice.12363
Giotis I, Molders N, Land S et al (2015) MED-NODE: a computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst Appl 42:6578–6585. https://doi.org/10.1016/j.eswa.2015.04.034
Hassan E, Elmougy S, Ibraheem MR et al (2023) Enhanced deep learning model for classification of retinal optical coherence tomography images. Sensors 23(12):5393. https://doi.org/10.3390/s23125393
Hassan E, Shams MY, Hikal NA et al (2023) The effect of choosing optimizer algorithms to improve computer vision tasks: a comparative study. Multimedia Tools Appl 82(11):16591–16633. https://doi.org/10.1007/s11042-022-13820-0
Hinton GE, Srivastava N, Krizhevsky A et al (2012) Improving neural networks by preventing co-adaptation of feature detectors. arXiv:1207.0580
Hoefler T, Alistarh D, Ben-Nun T et al (2021) Sparsity in deep learning: pruning and growth for efficient inference and training in neural networks. J Mach Learn Res 22(241):1–124. https://doi.org/10.1145/3578356.3592583
Jain S, Pise N et al (2015) Computer aided melanoma skin cancer detection using image processing. Procedia Comput Sci 48:735–740. https://doi.org/10.1016/j.procs.2015.04.209
Jayalakshmi G, Kumar VS (2019) Performance analysis of convolutional neural network (CNN) based cancerous skin lesion detection system. In: 2019 International conference on computational intelligence in data science (ICCIDS), IEEE, pp 1–6. https://doi.org/10.1109/ICCIDS.2019.8862143
Lafraxo S, Ansari ME, Charfi S (2022) MelaNet: an effective deep learning framework for melanoma detection using dermoscopic images. Multimedia Tools Appl 81(11):16021–16045. https://doi.org/10.1007/s11042-022-12521-y
Mahdavi F, Rajabi R (2020) Drone detection using convolutional neural networks. In: 2020 6th Iranian conference on signal processing and intelligent systems (ICSPIS), IEEE, pp 1–5. https://doi.org/10.1109/ICSPIS51611.2020.9349620
Mijwil MM (2021) Skin cancer disease images classification using deep learning solutions. Multimedia Tools Appl 80(17):26255–26271. https://doi.org/10.1007/s11042-021-10952-7
Nawaz M, Masood M, Javed A et al (2021) Melanoma localization and classification through faster region-based convolutional neural network and SVM. Multimedia Tools Appl 80(19):28953–28974. https://doi.org/10.1007/s11042-021-11120-7
Pham TC, Tran CT, Luu MSK et al (2020) Improving binary skin cancer classification based on best model selection method combined with optimizing full connected layers of deep CNN. In: 2020 International conference on multimedia analysis and pattern recognition (MAPR), IEEE, pp 1–6. https://doi.org/10.1109/MAPR49794.2020.9237778
Rafi TH, Shubair RM (2021) A scaled-2D CNN for skin cancer diagnosis. In: 2021 IEEE conference on computational intelligence in bioinformatics and computational biology (CIBCB), IEEE, pp 1–6. https://doi.org/10.1109/CIBCB49929.2021.9562888
Rasel M, Obaidellah UH, Kareem SA (2022) convolutional neural network-based skin lesion classification with variable nonlinear activation functions. IEEE Access 10:83398–83414. https://doi.org/10.1109/ACCESS.2022.3196911
Rotemberg V, Kurtansky N, Betz-Stablein B et al (2021) A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci Data 8(34):1–8. https://doi.org/10.34970/2020-ds01
Shahidi Zandi M, Rajabi R (2022) Deep learning based framework for Iranian license plate detection and recognition. Multimedia Tools Appl 81(11):15841–15858. https://doi.org/10.1007/s11042-022-12023-x
Sonsare PM, Gunavathi C (2021) Cascading 1D-convnet bidirectional long short term memory network with modified COCOB optimizer: a novel approach for protein secondary structure prediction. Chaos Solitons Fractals 153:111446. https://doi.org/10.1016/j.chaos.2021.111446
Wu H, Gu X (2015) Towards dropout training for convolutional neural networks. Neural Netw 71:1–10. https://doi.org/10.1016/j.neunet.2015.07.007
Wu H, Chen S, Chen G et al (2022) FAT-Net: feature adaptive transformers for automated skin lesion segmentation. Med Image Anal 76:102327. https://doi.org/10.1016/j.media.2021.102327
Zhang T, Zhang X (2021) Squeeze-and-excitation Laplacian pyramid network with dual-polarization feature fusion for ship classification in SAR images. IEEE Geosci Remote Sens Lett 19:1–5. https://doi.org/10.1109/LGRS.2021.3119875
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Availability of Data and Materials
The datasets analysed during the current study are available in the ISIC 2020 repository https://challenge2020.isic-archive.com/, and MED-NODE repository https://www.cs.rug.nl/ imaging/databases/melanoma_naevi/
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Faghihi, A., Fathollahi, M. & Rajabi, R. Diagnosis of skin cancer using VGG16 and VGG19 based transfer learning models. Multimed Tools Appl 83, 57495–57510 (2024). https://doi.org/10.1007/s11042-023-17735-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17735-2