Abstract
Apple Scab and Apple Rust are the major classes of apple leaf diseases that gravely affect the apple yield. Seeking an automatic, less expensive, fast yet precise method to detect plant diseases is crucial. Traditional approaches to detect plant diseases using computer vision involve complex and labor-intensive methodologies that rely on image enhancement methods and hand-engineered features. The deep convolutional neural network models are highly favourable in performing image classification with many target classes without involving the arduous phase of feature engineering. In this paper, we utilized the Capsule Neural Network (CapsNet) architecture and modified the network structure by adding additional convolution layers to enhance the model’s learning capacity to classify the apple diseases into apple rust, apple scab, healthy, and multiple diseases on the same leaf. Model training was performed on a dataset of images that reflected complex growing conditions observed in the real world. The ability of the model to learn was improved by enhancing the images. Experimentation was conducted on the Kaggle Plant Pathology 2020 - FGVC7 dataset. Experimental study demonstrated a recognition accuracy of 91.37% on the test set, with an overall improvement of 3.67% in accuracy when compared to the research work utilizing the same dataset in literature. Therefore, the proposed method effectively achieves Apple foliar leaf disease detection and surpasses existing state-of-the-art techniques applied to the same dataset. “(Dataset Link: https://www.kaggle.com/c/plant-pathology-2020-fgvc7/data)"
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Apples are highly sought-after fruits worldwide due to their exceptional medicinal and nutritional value [17]. Apples support a healthy immune system, promote gut health and are known as diabetes-friendly fruit. The apple production worldwide is estimated to be around 63.9 million tonnes in 2020. However, foliar fungal diseases may pose severe threats to the apple production rate. Apple Scab and Apple Rust are the main categories of diseases that severely affect apple yield. Sometimes, the apple leaves may also be affected by multiple diseases. In apple rust, large yellow spots appear on the leaves’ upper surface turning yellow-orange to gray-brown as the spores mature. Small velvety brown to olive-green spots appear on the leaves that enlarge and darken to become more or less circular in the case of apple scab. When infections are numerous, young leaves become curled and distorted. These diseases can have catastrophic effect on the quantity and quality of apple production. In the worst case, it can even result in a complete loss of harvest. Therefore, it is crucial to employ various techniques that can swiftly identify leaf diseases in their early stages to avoid any adverse impact on agricultural production.
Traditional methods of disease detection are labor-intensive and lack accuracy. The manual diagnosis becomes even more challenging when multiple diseases are present on the same leaf. To address these challenges, artificial intelligence, particularly deep learning, has emerged as a viable method for automating plant disease detection and classification. Deep learning algorithms aim to create automated systems capable of accurately and efficiently detecting and categorizing different types of apple leaf diseases. Deep learning models are trained using extensive datasets of annotated images that include both healthy and diseased apple leaves. Through this training process, the models acquire the ability to extract essential features and patterns from the images, enabling them to effectively distinguish between healthy and infected leaves. Once trained, these models can be used to analyze new images and provide rapid and reliable detection of apple leaf diseases. Deep learning offers numerous advantages when applied to apple leaf disease detection. It allows for early and accurate disease identification, facilitating timely interventions to prevent further spread and damage. Additionally, it reduces the reliance on manual labor and expertise, making disease detection more accessible and cost-effective for farmers.
In this paper, an enhanced Capsule Network model is introduced for the identification of apple leaf diseases. Most of the existing works in literature use leaf images with homogeneous background under identical lighting conditions which makes the disease detection slightly easier. However, only a few works have discussed techniques to handle leaf images under complex conditions. This provided the impetus for the present study. The Kaggle Plant Pathology 2020 - FGVC7 dataset [36] is utilized in this paper, which consists of images taken under actual field conditions. The findings from the experiments confirm the effectiveness of the enhanced Capsule Network model in promptly detecting apple leaf diseases. Early identification of leaf diseases helps minimize the unnecessary or excessive application of chemicals, leading to reduced production costs. Moreover, the proposed model exhibits superior real-time detection capabilities for apple leaf diseases, surpassing existing methods in terms of recognition accuracy. The paper is structures as follows: Section 2 presents a comprehensive review of previous studies focusing on detecting different plant diseases and highlights the limitations of existing methods. In Section 3, the dataset and model employed for apple leaf disease detection are described. The findings obtained from the experiments are reported in Section 4. Lastly, Section 5 summarizes the key findings and conclusions derived from this study.
2 Literature review
In the field of plant disease detection, extensive application of machine learning algorithms has been observed to analyze features extracted from the region of interest, which is obtained through image processing methods. Machine Learning algorithms have been widely employed in this context, especially when the available dataset is limited in size. Liu et al. [22] extracted the diseased part of the unhealthy leaves by using the HSI color space. The images were processed to extract color and texture features and the classification task was performed using the Support Vector Machine (SVM) algorithm. The experimentation was done on cucumber, walnut, and grape leaves and average accuracy obtained was 90%. Dubey et al. [8] employed K-means clustering and Multi-class Support Vector Machine (SVM) to classify apple leaves into healthy and infected categories in a binary classification task. Li et al. [20] employed Artificial Neural Networks to detect diseases in orchid leaves to obtain an accuracy of 92%. N. Petrellis [28] created a mobile application that utilizes shape and color analysis of spots, along with historical weather data, for plant disease detection. Wozniak et al. [39] detected defects in fruit peels by employing an Adaptive Artificial Neural Network achieving an accuracy of about 80%. The traditional machine learning algorithms exhibit satisfactory performance.Nevertheless, these approaches necessitate manual feature engineering, which is a time-consuming task and demands domain expertise. As a result, the emergence of deep learning algorithms has opened new avenues for plant disease detection.
Deep learning has gained significant traction in the agricultural sector due to its superior accuracy and its ability to surpass conventional image processing techniques. It is being employed to assist farmers in predicting harvest yields, evaluating crop quality, detecting crop diseases or weed infestations, and identifying plant species. Several works have been proposed to detect and classify the disease in plants by employing Deep Learning Algorithms. Park et al. [25] automated the identification of diseased strawberry crops by using a CNN model which yielded an accuracy of 89.7%. In their study, Sardogan et al. [31] achieved an overall accuracy of 86% in the automatic detection and classification of tomato leaf diseases by utilizing a Convolutional Neural Network (CNN) combined with the Learning Vector Quantization algorithm. By employing the AlexNet and GoogleNet architectures, Mohanty et al. [23] achieved an overall accuracy of 85.53% and 99.34% respectively in their smartphone-based plant disease diagnosis study. Hu et al. [12] employed Improved GoogleNet architecture to classify the corn leaf diseases. With this approach, an overall accuracy of 97.6% and a per-class accuracy of 95% were achieved. Jearanaiwongkul et al. [16] presented a two-layer ontology-based model for modeling plant diseases. Jakjoud et al. [15] developed four different models using various Neural Network optimizers to achieve plant disease detection. Li et al. conducted a study [20] where they investigated the utilization of Deep Learning Artificial Neural Networks to detect diseases in orchid leaves by analyzing the patterns present on the leaves. This model yielded a test accuracy of 90%. Wan et al. [38] used various pretrained models to diagnose agricultural diseases. Among the various models evaluated, the Inception-V3 based model exhibited comparatively superior performance and achieved an accuracy rate of 87.9%. A novel approach was presented by Ays et al. [2] utilizing the MobileNetV2 model for the classification of Cassava leaf disease, distinguishing it into five distinct classes. Patil et al. [27] employed CNN in conjunction with an IoT-based platform to capture images of cotton leaf diseases and collect relevant external factors such as weather conditions. The proposed model achieved an impressive accuracy of 98%. Jaiswal et al. [14] focused on the detection of diseases in fruit and vegetable leaves using the Plant Village Dataset. They employed GoogLeNet and Sequential Model for this task, achieving remarkable recognition accuracies of 98.48% and 97.47%, respectively. Trivedi et al. [37] presented a CNN model for classifying leaf diseases. The model achieved an impressive accuracy of 95.81%. Bhatia et al. [4] proposed a hybrid solution combining deep learning and cloud-based technology for plant disease detection. The ResNet50 model yielded the best results, achieving a test accuracy of 92.52%. Patidar et al. [26] developed a solution for rice crop disease classification by implementing the Residual Neural Network. The approach achieved an accuracy of 95.83%. Sladojevic et al. [33] introduced a model based on the CaffeNet architecture to identify 13 distinct categories of plant diseases. The model achieved an average precision of 96.3%. Brahimi et al. [6] employed the GoogLeNet and AlexNet architectures to detect 9 types of diseases in tomato leaves. The GoogleNet model outperformed AlexNet, achieving an impressive accuracy of 99.185%. Oyewala et al. [24] developed a deep CNN Model for Cassava Mosaic Disease detection with an overall accuracy of 96.75%.
Many researchers have worked on apple leaf disease detection using various datasets available online [3, 24, 29] or using self-made datasets [5, 17, 33, 39]. In their research, Baranwal et al. [3] employed a LeNet-5 inspired architecture for apple leaf disease recognition. The model achieved a commendable test accuracy of 98.54%. Bi et al. [5] presented a system that could identify apple leaf diseases using MobileNet architecture. The system performed disease identification with an accuracy of 73.65%. Jiang et al. [17] proposed an improvised model using the characteristics of VGG and Inception models to detect diseases in apple leaves. This model performed well, yielding an accuracy of 97.14%. Yu et al. [41] introduced a deep learning model designed to identify apple leaf disease using region of interest analysis. The proposed architecture demonstrated an accuracy of 84.3%. Liu et al. [21] employed pathological images for the identification of Apple Leaf diseases utilizing a Convolutional Neural Network Architecture based on AlexNet. The proposed architecture achieved a test accuracy of 97.62%. Rehman et al. [29] employed a regional Convolutional Neural Network (CNN) with a Mask-based approach, utilizing a parallel processing framework for the detection of apple leaf diseases. The suggested method achieved an impressive accuracy of 99.1%. Subetha et al. [34] trained ResNet50 and VGG19 model to achieve the apple plant disease classification. Both models exhibited improved performance in predicting apple leaf diseases, achieving an accuracy of 87.7%.
However, the approaches suggested in literature have a few downsides associated with them. [3, 17, 21, 28, 29, 39] used images with a homogeneous background that did not depict actual field situations. Specific to the apple leaf disease detection, [17, 33, 41] did not take into consideration the images of leaves infected with multiple diseases. Identifying leaves that have multiple diseases allows for a more comprehensive and accurate diagnosis. By detecting and identifying multiple diseases present on a single leaf, it provides valuable information to farmers or plant pathologists to take appropriate actions in managing the diseases effectively. Additionally, monitoring and managing multiple diseases on leaves can significantly impact crop health and overall yield. Detecting multiple diseases allows for early intervention and proactive management practices, which can help prevent severe damage and optimize crop production. This served as the motivation for the proposed work. This study focuses on a dataset consisting of images with intricate backgrounds, encompassing four classes: Healthy, Apple Scab, Apple Rust, and a combination of Rust and Scab. The original Capsule Network architecture, when applied directly to apple leaf disease detection, exhibits suboptimal performance. This research focuses on enhancing the model architecture by incorporating additional convolutional layers into the base Capsule Network architecture, thereby enhancing its recognition accuracy. Numerous experiments were conducted to identify the optimal combination of hyperparameters for developing a model capable of effectively distinguishing between the four disease categories. This novel contribution significantly contributes to the field of apple leaf disease detection.
2.1 Contributions of the proposed work
-
This work deals with the images taken under real-life variable conditions regardless of light, angle, shade and physiological age of the leaf.
-
The dataset is augmented by applying operations such as image rotation, image shift, and image zoom to address the limited number of images in the training set and improve the model’s resilience. Additionally, to handle the class imbalance, images belonging to the ’Multiple’ class are subjected to additional augmentation techniques separately. The unsharp masking method and contrast adjustment is employed to sharpen the images and enhance the images respectively.
-
The CapsNet model is enhanced by incorporating supplementary convolutional layers to effectively detect various types of apple diseases. The model demonstrates favorable training accuracy when applied to a diverse dataset comprising images captured with varying focus settings, at different time intervals, and depicting multiple diseases in plants at different maturity stages. The model performs well, with test accuracy greater than the accuracy achieved by the related works that utilized the same dataset.
3 Materials and methods
3.1 Dataset
The dataset utilized in this research is derived from the Plant Pathology 2020 challenge dataset, which comprises a total of 3,651 RGB images. Among these images, there are 1,200 samples depicting apple scab, 1,399 samples illustrating cedar apple rust, 187 samples representing complex disease symptoms, and 865 samples portraying healthy leaves. Seven duplicate images found were taken out from the image dataset to obtain final set of images consisting of 3,642 high-quality RGB images with annotations. The images were captured under various angles, illumination, surface, and noise conditions.
3.2 Data pre-processing and augmentation
Initially, the images are sharpened using the unsharp masking method [9]. The contrast of the images is then adjusted (enhanced) using the imadjust function in Matlab [10]. The contrast range for the input and output image are set as [0.3,0.7] and [0,1] respectively. Fig. 1 shows the images from the training set after being subjected to pre-processing. Since the size of the dataset, which comprises 3,642 images distributed among four classes, is insufficient for training a deep learning model effectively, data augmentation techniques are employed. Random rotations are applied within the range of \(0^{\circ }\) to \(360^{\circ }\), random horizontal and vertical shifts within the range of [-0.2, +0.2), and random zoom within the range of [0.8, 1.2]. These augmentations help to expand the dataset and enhance the model’s generalization capability. Furthermore, to address the issue of class imbalance, additional data augmentation techniques were employed specifically to the ’multiple’ class images. Random brightness adjustments, horizontal flips, and vertical flips were performed on these samples. The augmented training set, which includes the augmented ’multiple’ class images, is visually represented in Fig. 2.
3.3 Deep neural network architecture
3.3.1 Capsule neural network
CapsNet is an equivariant network containing detailed information on every pixel in the image contrary to the Convolutional Neural Networks. Pooling operations in CNNs can introduce a limitation by potentially causing information loss in the images. This loss of information necessitates huge amount of training data to compensate for it. In contrast, CapsNet overcomes this limitation by preserving spatial information and retaining important features, thus minimizing information loss [40].
In a capsule network, each capsule comprises multiple neurons, and each neuron’s output represents a distinct aspect of the same feature. This allows for recognition of the entire entity through a process of recognizing its individual parts. The input to a capsule consists of features extracted from a CNN, and the output of the capsule is always in the form of a vector. A Capsule Network is constructed with Convolutional Layers, which are then followed by a Primary Capsule Layer and a Class Capsule Layer. The Convolutional Layer is responsible for extracting fundamental features from the input images. The Primary Capsule Layer takes the basic features extracted by the Convolutional Layer and identifies complex patterns among them. The number of capsules utilized in both the Primary Capsule Layer and Class Capsule Layer varies depending on the dataset employed. The dynamic routing algorithm determines the mapping between capsules in the Primary Capsule Layer and the Class Capsule Layer by considering the weights of the lower-level capsules (Primary Capsules) and the higher-level capsule (Class Capsule). When there is a high agreement between the weights of higher and lower-level capsules, the coupling coefficient is increased, and vice versa. The weighted sum of all the lower-level capsules mapping to higher-level capsules is computed. Since the output vector’s length represents probabilities, the squashing function is employed to compress the value within the range of 0 and 1. Equations (1), (2), and (3) provide the mathematical expressions for capsule transformation, weighted sum, and squashing function, respectively. Figure 3 illustrates the functionality of a capsule in the Capsule Network.
where, \(\mathrm {u_{i}}\) represents the activity vector of capsule i in the Primary Capsule Layer, \(\mathrm {W_{ij}}\) denotes the weight matrix of capsule i in the Primary Capsule Layer corresponding to capsule j in the Class Capsule Layer, and \({\hat{\text {u}}_{j|i}}\) refers to the capsule i’s vote for capsule j.
where \(\mathrm {c_{ij}}\) represents the coupling coefficient, while \(\mathrm {s_{j}}\) denotes the weighted sum of the predictions for capsule j.
where \(\mathrm {v_{j}}\) refers to the output of capsule j in the Class Capsule Layer.
3.3.2 Improved CapsNet architecture
In this work, the Capsule Network Architecture proposed for MNIST Digit Classification is improved by adding additional convolutional layers. The architecture contains 3 convolutional Layers in addition to the basic CapsNet architecture.The mentioned layers have been incorporated to enhance the feature extraction process, leading to the generation of high-level characteristics that highly contribute to the model’s performance. To preserve the spatial information contained in the input images and avoid potential information loss, pooling layers have been excluded from the architecture. The enhanced CapsNet structure is depicted in Fig. 4. The proposed architecture comprises an Input Layer, four convolutional Layers, a Primary Capsule Layer (PrimaryCaps), and a Class Capsule Layer. Various combinations of convolutional layers and parameter values were experimented with to ensure that the model does not overfit and achieves optimal performance. The input Layer is fed with the images of size 130 x 130. Initially, the images are of variable sizes. The images are resized to 130 x 130 to reduce the computational cost. Convolutional layers with a varying number of filters and stride are used to reduce the features’ dimensionality.
Feature map obtained from different convolution layers. Row 1: Healthy leaf, Row 2: Apple leaf with multiple diseases, Row 3: Apple Rust, Row 4: Apple Scab. a. Apple leaves. b. Feature Map for first Convolution Layer. c. Feature Map from second Convolution Layer. d. Feature Map from third Convolution Layer. e. Feature Map from forth Convolution Layer
The parameters for the convolutional layers are outlined in Table 1. These filters play a crucial role in determining which pixels the model focuses on. After passing through the pixel values of an input image, the filters generate feature maps. These feature maps depict what the convolutional layer detects or retains from the input. The objective of visualization is to comprehend the detected or preserved features in the feature maps. Fig. 5 displays the feature maps corresponding to images from different classes. In the case of apple rust, the feature map obtained from the final convolutional layer reveals that large yellow spots on the upper surface of the leaves are a prominent characteristic.
The features obtained from the final convolutional layer are fed as input to the Primary Capsule Layer, and the model is trained iteratively utilizing the dynamic routing algorithm. The Adam optimizer was employed with various learning rates (0.001, 0.01, and 0.1), and the most favorable outcome was achieved with 0.1 as learning rate. The training process consisted of 200 epochs, with a batch size of 10. The loss for each class was computed using the margin loss function, as defined by (4).
where the first term \(\mathrm {T_{i} \ max(0, x^+-||v_{i}||)^2}\) stands for class present and the second term \(\mathrm {\lambda (1 - T_{i}) \ max(0, ||v_{i}|| - x^-)^2}\) stands for class not present.
The parameters \(\mathrm {x^+}\) and \(\mathrm {x^-}\) are used with the values 0.9 and 0.1 respectively and \(\mathrm {\lambda }\) is set to a value of 0.5.
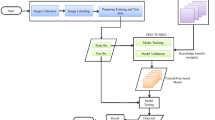
The proposed model’s overall structure is illustrated in Fig. 6. The initial step involves preprocessing the images in the dataset, which includes applying the unsharp masking method and contrast enhancement to enhance their suitability for model generation. Next, the dataset is randomly split into training and test sets, with an 80:20 ratio. The training dataset undergoes image augmentation to increase its size and address any class imbalance concerns. These augmented training images are then used as input for the architecture depicted in Fig. 4, resulting in a model trained to classify different classes. Finally, the generated model is evaluated on the test image set to assess its performance in detecting apple leaf diseases.
4 Results and discussion
Image pre-processing was performed using Matlab 2018a for the experiments. The data augmentation and implementation of the CapsNet architecture were performed using Anaconda3 (Python 3.6), along with the OpenCV-python3 and Keras-GPU libraries. Training of deep CNN was accelerated by GPU with Intel(R) CoreTM i7-2600 CPU at 3.40 GHz and NVIDIA GTX470 Card as the experimental hardware environment.
The ultimate dataset was divided into an 80% training set and a 20% test set. The model described in Section 3 was trained using the training set. Subsequently, the trained model was utilized to classify the type of disease based on the leaf images in the test set. Two experiments were conducted. One was including the leaves with multiple diseases, and the other was by excluding the leaves with multiple diseases. Fig. 7 represents the sample images in the test set. Figures 8 and 9 illustrate the accuracy and loss of the model during the training process, considering both the inclusion and exclusion of leaves with multiple diseases. The classification procedure was repeated five times to account for the variability in the selection of training, validation, and test sets. This approach helps prevent overfitting and ensures robustness in model performance. The model’s evaluation was based on Precision, Specificity, Recall, and F1-Score, which are represented by (5), (7), (6), and (8) respectively. Dalianis [7] where FP - False Positive, TP - True Positive, FN - False Negative and TN - True Negative. Figures 10 and 11 depicts the confusion matrices of the identification results and ROC curves for the model with and without the ’multiple’ class respectively.
The confusion matrix reveals that the model demonstrates strong performance when the category of leaves with multiple diseases is excluded from consideration. Table 2 represents the performance parameter values recorded over 5-folds cross-validation. The recorded values in the table correspond to two instances: one where the class of leaves with multiple diseases is considered, and the other where this class is excluded. The zero values in the table indicate the latter instance. The performance of the model is compared to diverse CNN models such as VGGNet [32], DenseNet [13], ResNet [11], Inception V3 [35], LeNet [19], MobileNet-V2 [30] and AlexNet [18]. Transfer learning is utilized with the aforementioned models, which are pre-trained using the ImageNet dataset. A fully-connected Softmax layer is then added to the models to classify the target classes for apple disease identification.
The training accuracy and test accuracy of various models are presented in Table 3. The models were trained for 200 epochs, considering all categories of leaves. The results in Table 3 clearly demonstrate that the proposed model outperforms other pre-trained models on the same dataset. The comparative analysis between training accuracy, validation accuracy, and test accuracy achieved by different models is depicted in Fig. 12 demonstrating the superior performance of the enhanced CapsNet model. Table 4 presents a comparative analysis between the enhanced Capsule Network and the original Capsule Network to showcase the superior performance of the enhanced model. The experimental results underscore the importance of enhancing the CapsNet architecture by incorporating convolutional layers. From Table 4 it can be seen that training CapsNet model without enhancement is around 6 times slower than the CapsNet model with enhancement. Also, accuracy has seen a whopping 40% increase for the latter compared to the former. It can be concluded that the modification in CapsNet architecture not only decreases the training time but also results in a substantial improvement in the model’s accuracy. The experimental results underscore the importance of enhancing the CapsNet architecture by incorporating convolutional layers. The proposed model is contrasted with state-of-the-art methods from the literature in terms of plant disease classification, as presented in Table 5. This comparative study aims to highlight the improved performance of the proposed model and provide insights into the datasets used in previous studies. This work proposes a model for performing apple leaf disease detection under real-life conditions considering, complex backgrounds with uneven lighting conditions and outperforms the work discussed in [34] with an increase in the recognition accuracy by 3.67%, hence demonstrating the efficiency of the proposed model.
5 Conclusion & future work
Accurate identification and classification of plant leaf diseases using images depicting the real life conditions is of great importance in ensuring the health and enhancing the plant production quality. It is of great significance as it enables proactive disease management, optimizes crop yield, reduces costs, and promotes sustainable agricultural practices. It empowers farmers with the knowledge and tools to protect their apple orchards and ensure the long-term health and productivity of their crops.
This work introduced an enhanced CapsNet architecture that incorporates multiple convolutional layers for the purpose of identifying apple foliar leaf disease. The input images were pre-processed using enhancement techniques before feeding it to the model, to improve the model generalization capabilities. Adam Optimizer with different learning rates was used to train the model, and a learning rate of 0.1 was chosen as it generated the best results. Experimental findings demonstrated that the proposed model outperformed the other pre-trained models with a recognition rate of 91.37% and 96.67% with inclusion and exclusion of multiple class from the dataset respectively. The decreased accuracy observed on considering ’Multiple’ class is majorly due to fewer samples in the ’Multiple’ class and the dominance of Apple Rust or Apple Scab characteristics in the leaf images belonging to ‘Multiple’ class. The experimental results highlight the significance of enhancing the CapsNet architecture through the inclusion of convolutional layers. This modification not only reduces the training time but also leads to a noteworthy improvement in the model’s accuracy.
As part of future work, to tackle the dataset imbalance caused by samples from the multiple class, we can leverage Generative Adversarial Networks (GANs) to generate synthetic images that depict leaves with multiple diseases. This technique helps in addressing the scarcity of data in the multiple class and enhances the model’s ability to handle such cases. Additionally, we anticipate that incorporating an attention mechanism into the proposed model would enhance its ability to diagnose and differentiate leaves with multiple diseases. Furthermore, developing a normalization-free network can enhance the performance of the model with larger batch sizes and improved learning rates.
Availability of data and material
The data that support the findings of this study are available from the first author upon reasonable request.
Code Availability
Programming codes implemented in this study are available from the first author upon reasonable request.
References
Amara J, Bouaziz B, Algergawy A (2017) A deep learning-based approach for banana leaf diseases classification. Datenbanksysteme für business, technologie und web (BTW 2017)-workshopband
Ayu H, Surtono A, Apriyanto D (2021) Deep learning for detection cassava leaf disease. In: Journal of physics: conference series, vol. 1751, p 012072. IOP Publishing
Baranwal S, Khandelwal S, Arora A (2019) Deep learning convolutional neural network for apple leaves disease detection. In: Proceedings of international conference on sustainable computing in science, technology and management (SUSCOM), Amity University Rajasthan, Jaipur-India
Bhatia GS, Ahuja P, Chaudhari D, Paratkar S, Patil A (2020) Plant disease detection using deep learning. In: Smys S, Senjyu T, Lafata P (eds) Second International Conference on Computer Networks and Communication Technologies. Springer International Publishing, Cham, pp 408–415
Bi C, Wang J, Duan Y, Fu B, Kang JR, Shi Y (2020) Mobilenet based apple leaf diseases identification. Mob Netw Appl 1–9
Brahimi M, Boukhalfa K, Moussaoui A (2017) Deep learning for tomato diseases: classification and symptoms visualization. Appl Artif Intell 31(4):299–315
Dalianis H (2018) Evaluation metrics and evaluation. In: Clinical text mining, pp 45–53. Springer
Dubey SR, Jalal AS (2016) Apple disease classification using color, texture and shape features from images. Signal, Image Video Process 10(5):819–826
Gabbouj M, Alaya Cheikh F, Khriji L (1998) Unsharp masking based approach for color image processing. In: Proceedings IX european signal processing conference EUSIPCO 1998, Island of Rhodes, Greece
Gupta S, Porwal R (2016) Appropriate contrast enhancement measures for brain and breast cancer images. Int J Biomed Imaging 2016
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hu R, Zhang S, Wang P, Xu G, Wang D, Qian Y (2020) The identification of corn leaf diseases based on transfer learning and data augmentation. In: Proceedings of the 2020 3rd international conference on computer science and software engineering, pp 58–65
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Jaiswal A, Pathak S, Rathore YK, Janghel RR (2020) Detection of disease from leaf of vegetables and fruits using deep learning technique. In: Advances in biomedical engineering and technology, pp 199–206. Springer
Jakjoud F, Hatim A, Bouaaddi A (2019) Deep learning application for plant diseases detection. In: Proceedings of the 4th international conference on big data and internet of things, pp 1–6
Jearanaiwongkul W, Anutariya C, Andres F (2018) An ontology-based approach to plant disease identification system. In: Proceedings of the 10th international conference on advances in information technology, pp 1–8
Jiang P, Chen Y, Liu B, He D, Liang C (2019) Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access 7:59069–59080
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD (1989) Backpropagation applied to handwritten zip code recognition. Neural Comput 1(4):541–551
Li LH, Chu YS, Chu JY, Guo SH (2019) A machine learning approach for detection plant disease: Taking orchid as example. In: Proceedings of the 3rd international conference on vision, image and signal processing, pp 1–6
Liu B, Zhang Y, He D, Li Y (2018) Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 10(1):11
Liu L, Li J, Sun Y (2019) Research on the plant leaf disease region extraction. In: Proceedings of the 2019 international conference on video, signal and image processing, pp 49–54
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Fronti Plant Sci 7:1419
Oyewola DO, Dada EG, Misra S, Damaševičius R (2021) Detecting cassava mosaic disease using a deep residual convolutional neural network with distinct block processing. PeerJ Comput Sci 7:e352
Park H, Eun JS, Kim SH (2017) Image-based disease diagnosing and predicting of the crops through the deep learning mechanism. In: 2017 international conference on information and communication technology convergence (ICTC), pp 129–131. IEEE
Patidar S, Pandey A, Shirish BA, Sriram A (2020) Rice plant disease detection and classification using deep residual learning. In: International conference on machine learning, image processing, network security and data sciences, pp 278–293. Springer
Patil BV, Patil PS (2021) Computational method for cotton plant disease detection of crop management using deep learning and internet of things platforms. In: Evolutionary computing and mobile sustainable networks, pp. 875–885. Springer
Petrellis N (2017) Mobile application for plant disease classification based on symptom signatures. In: Proceedings of the 21st pan-hellenic conference on informatics, pp 1–6
Rehman Zu, Khan MA, Ahmed F, Damaševičius R, Naqvi SR, Nisar W, Javed K (2021) Recognizing apple leaf diseases using a novel parallel real-time processing framework based on mask rcnn and transfer learning: An application for smart agriculture. IET Image Process
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4510–4520
Sardogan M, Tuncer A, Ozen Y (2018) Plant leaf disease detection and classification based on cnn with lvq algorithm. In: 2018 3rd international conference on computer science and engineering (UBMK), pp 382–385. IEEE
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci 2016
Subetha T, Khilar R, Christo MS (2021) A comparative analysis on plant pathology classification using deep learning architecture-resnet and vgg19. Materials today: Proceedings
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2818–2826
Thapa R, Zhang K, Snavely N, Belongie S, Khan A (2020) The plant pathology challenge 2020 data set to classify foliar disease of apples. Appl Plant Sci 8(9):e11390
Trivedi J, Shamnani Y, Gajjar R (2020) Plant leaf disease detection using machine learning. In: International conference on emerging technology trends in electronics communication and networking, pp 267–276. Springer
Wan H, Lu Z, Qi W, Chen Y (2020) Plant disease classification using deep learning methods. In: Proceedings of the 4th international conference on machine learning and soft computing, pp 5–9
Woźniak M, Połap D (2018) Adaptive neuro-heuristic hybrid model for fruit peel defects detection. Neural Netw 98:16–33
Xu C, Wang X, Zhang S (2023) Dilated convolution capsule network for apple leaf disease identification. Convolutional Neural Networks and Deep Learning For Crop Improvement and Production 16648714:182
Yu HJ, Son CH, Lee DH (2020) Apple leaf disease identification through region-ofinterest-aware deep convolutional neural network. J Imaging Sci Technol 64(2):20507–1
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
S, S., S, S., Acharya, V. et al. Apple foliar leaf disease detection through improved capsule neural network architecture. Multimed Tools Appl 83, 48585–48605 (2024). https://doi.org/10.1007/s11042-023-17463-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17463-7