Abstract
Medical Visual Question answering (MedVQA) systems provide answers to questions based on radiology images. Medical images are more complex than general images. They have low contrast and are very similar to one another. The difference between medical images can only be understood by medical practitioners. While general images have very high quality and their differences can easily be spotted by anyone. Therefore, methods used for general-domain Visual Question Answering (VQA) Systems can not be used directly. The performance of MedVQA systems depends mainly on the method used to combine the features of the two input modalities: medical image and question. In this work, we propose an architecturally simple fusion strategy that uses multi-head self-attention to combine medical images and questions of the VQA-Med dataset of the ImageCLEF 2019 challenge. The model captures long-range dependencies between input modalities using the attention mechanism of the Transformer. We have experimentally shown that the representational power of the model is improved by increasing the length of the embeddings, used in the transformer. We have achieved an overall accuracy of 60.0% which improves by 1.35% from the existing model. We have also performed the ablation study to elucidate the importance of each model component.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Medical imaging is the process of obtaining images of the body’s internal organs with the help of technologies like CT, MRI, XRay, Nuclear medicine, etc. Radiologists interpret these medical images and present their analyses to doctors. Based on the radiologists’ diagnosis, doctors determine the course of treatment for the patients. Nowadays, doctors rely immensely on the report of radiologists for diagnosis, staging, and treatment. Doctors’ dependence on radiologists’ reports has increased the radiologists’ workload drastically. An average radiologist has a few seconds to examine a single image [1]. Behavioral studies have shown that as the decision speed of humans increases, decision accuracy decreases. A study [2] examining the factors responsible for diagnostic errors found that increased workload and fatigue caused by long workdays contribute to diagnostic errors. At times diagnostic inconsistency is seen among radiologists. They vary in their interpretations of medical images. Therefore, accurate interpretation of medical images requires experience, expertise, and ample time from radiologists. Efforts are made to enhance diagnostic accuracy and reduce interobserver variability. An automated system can solve the abovementioned problems by efficiently analyzing medical images and answering questions a medical practitioner may have regarding radiology images.
Visual Question Answering (VQA) systems analyze input images and answer textual questions based on them. The input question and the output answer are in a human-readable form. The success of VQA for general images has attracted the research community’s attention in developing Medical Visual Question Answering (MedVQA) systems. MedVQA takes as input a medical image and a question based on the input medical image. It provides as output an answer to the question. It can reduce the burden on doctors by assisting them in understanding complex medical images rapidly. A complete understanding of medical images requires time and specialization. Medical practitioners can ask specific questions to MedVQA systems to better understand the medical image and use the answers provided by the MedVQA system as a second opinion while making diagnoses, thereby saving time and reducing the rate of diagnostic errors. These systems can enhance telemedicine and remote patient care as doctors can use them to probe patient reports remotely. These systems can act as a source of reliable information for patients who are keen to gather more information regarding their condition. It will increase the involvement of patients in their treatment process and bring transparency to it. Medical students can use MedVQA systems for learning and improving their diagnostic skills. Therefore, these systems can revolutionize the healthcare sector by rapidly assessing medical images and streamlining the triage process. However, this is possible only by an efficient and complete MedVQA model. Unlike VQA for general images, MedVQA is still in its nascent stages of development. Therefore, further research is needed to improve MedVQA.
VQA systems, in general, consist of three main modules. The first module is the input feature encoder which extracts information from the input images and questions. The second module is feature fusion, where extracted image and text features are combined. The third module is an answer generator that takes the fused representation of input modalities and outputs the most appropriate answer. Figure 1. describes the overall architecture. VQA systems require deeper comprehension of the image and question features and the semantic relationship between them to answer the questions correctly. Therefore, we can improve VQA systems through advancements in two directions of research: feature encoder and feature fusion. Ample research has been done in the area of image classification, object detection, and text encoding, but feature fusion for VQA systems needs to be explored further.
Developing a MedVQA model is comparatively more difficult than developing a VQA for general images. The first challenge is the size of the dataset. Unlike VQA for general domain, obtaining a high-quality large dataset for MedVQA is tough, as annotation requires professional medical experts. If we synthetically generate question-answer pairs from the image, it also needs to be assessed by medical experts. The second challenge is handling different modalities (i.e., Xray, CT, Ultrasound, MRI) of medical images related to different organs. Also, Medical images are of low contrast, and the region of interest can be very small. Therefore, the model must be able to focus on a fine-grained scale. In contrast, general-domain VQA has very good-quality images, and the region of interest is not microscopic. Finally, questions can contain medical terminology. Therefore, the model must be able to understand both medical and non-medical terminologies. These differences highlight the need to develop a MedVQA model that can learn to efficiently align and fuse/combine text and image features using a small dataset.
Motivation
Recent medical VQA models use various techniques of Natural Language Processing such as the attention mechanism of Transformer [3] and the BERT architecture [4]. The basic block of these schemes projects the input into query, key, and value vectors. These vectors play a significant role in capturing the relationship between regions of input. Many MedVQA [5, 6] modules have shown the efficiency of self-attention in fusion. However, they have yet to explore the impact of varying the query, key, and value dimensions on the model performance and size. The recent works in MedVQA directly use the same length of the mentioned vectors from BERT [5, 6]. The length of the vectors is comparatively small to capture the complex information of the medical images. This motivates us to increase the representational power of the model by increasing the length of the vectors. As we have seen from the previous work of WideResNet, widening the intermediate feature maps helps improve the representational power of the model. This in turn reduces the depth of the model [7]. It motivates us to widen the query, key, and value vectors in the model. Thus, widening helps us to reduce the length of the network. Thereby, reducing the total number of parameters by half from the existing model [5].
Contribution
The main contributions of this paper are as follows:
-
We propose a Multi-modal multi-head self-attention model for Medical VQA, with improved representational power in a significantly less number of parameters.
-
We have evaluated the results on VQA-Med-2019 of ImageCLEF [8] and compared our fusion strategy’s results with previous work on this dataset, and shown improvements in terms of accuracy, and computational efficiency.
-
We have performed an extensive ablation study to validate the significance of individual modules.
-
We have integrated our model with GradCam [9] explainability technique to identify the image regions the model emphasizes while answering.
2 Related work
2.1 General VQA
VQA challenge in the general domain started in 2016 and has been held yearly since then. The earliest VQA models used simple methods like concatenation, elementwise sum, or product for feature fusion. However, such methods failed to capture the hidden semantic relationship between the two modalities efficiently. This motivated the use of the outer product for fusion as they involve interaction between every element of one modality with every other element of another modality. However, the major drawback of using the outer product is that they are computationally expensive. Therefore, ways to reduce the computational overhead of outer product while preserving their discriminating capacity has become an active area of research. In Multimodal Compact Bilinear Pooling (MCB), [10] Fukui et al. used Count Sketch [11] to represent image and question features. They chose count sketch because of their exciting property, which Pham et al. proves in [12]. The major drawback of this method is that count sketch can introduce errors due to hash collisions. Therefore, for efficient results, the dimension of the count sketch vector needs to be significant. In the original paper, the dimension of this count sketch was set to 16000. Though the computational cost of MCB [10] is less than the original outer product, but it is still very high. In Multi-modal Factorized Bilinear Pooling (MFB) [13], another strategy is proposed to compute the outer product. It is based on the efficient matrix factorization technique. Multimodal Tucker Fusion (MUTAN) [14] uses Tucker decomposition to reduce the size of the parameter tensor. It rewrites the very large bilinear interaction as a smaller bilinear interaction between input projections.
The attention mechanism is another technique for feature fusion in VQA, as one modality is used as a context to generate the weights for another modality. The attention mechanism enables VQA models to focus on desired features while answering the question. Stacked Attention Networks (SAN) [15] achieved significant improvement on the VQA benchmark by using a multilevel attention mechanism. SAN identifies important image regions with the help of question features. Hierarchical Image-Question Co-Attention (HieCoAtt) [16] further emphasizes the importance of identifying key question words in addition to finding important image regions. In HieCoAtt [16], the co-attention network uses image features to determine attention over question words, and similarly, question features determine attention on image features. With the success of Transformers [3] in NLP tasks, several VQA models have started using self-attention for fusion and achieved higher accuracy value.
2.2 Medical VQA
The Medical VQA challenge began in 2018. For the first edition of the Medical VQA challenge, five out of 28 registered teams successfully submitted their models. This considerable participation indicated the immense interest in the Medical VQA task. The following subsections briefly describe the work proposed in VQA-Med 2018 and VQA-Med 2019 challenges.
2.2.1 VQA-med 2018
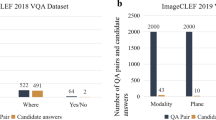
The dataset proposed for the inaugural version of the Medical VQA challenge, called VQA-Med 2018, consists of 2866 medical images and 6413 question-answer pairs, divided into the train, validation, and test set. The train set has 2278 medical images and 5413 question-answer pairs. The validation set has 324 medical images and 500 question-answer pairs. The test set has 264 medical images and 500 questions. UMMS [17] secured first rank in the 2018 challenge. They used ResNet-152 [18] for image features and LSTM [19] for Question features. They combined both features using MFH [13]. TU [20] extracted image features using the Inception-Resnet-v2 [21] network and question features using BiLSTM [22]. They added an attention layer to combine image and text features. The attended features and question features are concatenated and passed through the fully connected layer, followed by the softmax layer. NLM [23] used VGG16 [24], pre-trained on Imagenet, for image features and LSTM [19] for question features. They used a 2-layer SAN [15] to identify important image regions for answering. JUST [25] proposed an encoder-decoder model. Encoder has a VGG network and an LSTM for encoding image and question features. The two features are concatenated and passed to the decoder, which is an LSTM. FSTT [26] treated the VQA task as a multi-label classification problem. They used pre-trained VGG16 [24] and Bi-LSTM [22] for image and question feature extraction, respectively. Image and question features are combined using two fully connected layers. Combined image and question features pass through a multi-label Decision Tree classifier.
2.2.2 VQA-med 2019
Medical VQA models, like general VQA models, contain four main components. The first component is the Image/Visual Encoder. Medical VQA models use CNNs like VGGNet [24], ResNet [18] predominantly for visual features. The second component is the Text/Question Encoder which extracts question features. LSTM [19], BiLSTM [22], GRU [27], and the Transformers e.g. BERT [4] are the widely used Text Encoders. The third component of Med VQA models, the Fusion module, combines the extracted question and image features. The output of the Fusion module passes through the Classification/Generation module, the fourth component, for the final answer. This section briefly describes the fusion techniques prevalent in Medical VQA.
In general, strategies proposed for general VQA like SAN [15], BAN [28], HieCoAtt [16], MFB [13] are used in MedVQA. The winning model of ImageCLEF 2019 competition Hanlin [29] used MFB with coattention [13] to combine image and question features. Recently, new strategies have been developed specifically for the task of MedVQA. Some of them are MedFuseNet [30] and Question-Centric Multimodal Low-rank Bilinear (QC-MLB) [31]. MedFuseNet [30] uses two types of attention mechanism. It first performs question attention on the input question to identify important question words. This attended question representation is passed to the image attention module. The image attention module first captures the robust interaction between the image and attended question features by fusing them using MFB [13] and generates a combined feature vector. This feature vector is used for computing multiple attention maps on image features based on attended question features. These attention glimpses are added to form attended image features. Inspired by the findings reported in [32] that image-only models perform poorly compared to question-only models, a novel fusion strategy that lays more emphasis on question features is proposed in QC-MLB [31]. In this method, first, the multi-glimpse attention mechanism proposed by Xu et al. [33] is used to compute image regions that are highly correlated to question features. The final global image features are obtained by concatenating these multiple attention maps. Question features are transformed and concatenated (or tiled) to create global question features. Both this global image and question features are combined to form a fused representation that is passed to the answer generation module.
Multimodal Medical BERT(MMBERT) [5] is a Transformer-based model. It passes image features obtained from ResNet152 [18] and text token embeddings to a BERT-like model. It has four BERT layers. Ye et al. [34] proposed a fusion strategy called CMSA that uses self-attention for Referring Image Segmentation task and is later adopted for the Medical VQA task [35] on VQARad dataset [36]. They used two layers of CMSA for the Medical VQA task [35] to achieve better accuracy. We adopt a fusion technique similar to [35] for the VQA-Med 2019 ImageCLEF dataset. We observe that by varying the hidden dimension of query, key, and value, we are able to achieve better performance than MMBERT [5] for the VQA-Med 2019 ImageCLEF dataset with a single layer multi-head self-attention. Therefore, we used multi-head self-attention to capture the interaction between the two input modalities in this work.
Table 1 gives an overview of the work done in Medical Visual Question Answering. Along with the models proposed for VQA-Med-2019 dataset, we have included works proposed for other datasets. We have added models proposed for VQA-RAD Dataset [36], Med-VQA-2020 [37], Med-VQA-2021 [38], SLAKE [39]
3 Proposed methodology
We have formulated the problem of Medical VQA as a classification task. We aim to estimate our model’s most likely response \(\hat{r}\) of our model from a fixed set of responses R, given a radiology image I and a natural language textual question T. The task is summarised in (1):
where \(\theta \) represents model parameters.
Our model consists of four modules: visual encoder for extracting visual features from the radiology image, textual encoder for extracting text features from the input question, cross-modal fusion network for combining the two input modalities, and answer predictor for generating the most likely answer. Figure 2. depicts the architecture of our MedVQA system
3.1 Visual encoder
We use Wide ResNet-50-2 [7] pre-trained on ImageNet. Wide ResNet-50-2 has the same architecture as ResNet50 [18] except that it has wider \(3\times 3\) layers than ResNet50 by a factor of 2. We have shown a detailed architecture of ResNet50 and Wide ResNet-50-2 in Table 2. The first convolution layer is the same; both have 64 Kernels of size \(7\times 7\) with stride 2. Each residual block of WideResNet-50-2 has same kernel size but twice the number of Kernels than ResNet50. By widening the convolutional layer, WideResNet50-2 performs better and faster than ResNet152, which has three times more layers. The widening of convolutional layers increases the representational power of residual blocks, which is beneficial for medical tasks as it can capture more features. We process the raw input medical images before sending them through the Visual encoder. We have explained the preprocessing technique in Section 4.3 Implementation datails. We are using the output of the last convolution layer before the global average pooling layer of pre-trained Wide ResNet-50-2 for high-level feature representation of the medical image. Our visual features I is a tensor of dimensions \(R^{2048\times 8\times 8}\).
3.2 Textual encoder
We use DistilBERT [48] for encoding questions. DistilBERT, which stands for Distilled BERT, is a compressed version of a large BERT-base-uncased model. It has 40% fewer parameters and is 60% faster than the original BERT model. Despite being smaller, it preserves 97% of the original model’s language understanding. It is pre-trained using the knowledge Distillation process where the small student model(DistilBERT) is trained to mimic the output probability distribution of the large teacher model(large BERT-base-uncased). We first tokenize each question word using DistilBERT Tokenizer. Each tokenized question i is encoded into the word embedding \(T_{i} = \{t_{1}, t_{2}, t_{3}, ..., t_{L}\}\) where \(t_{j} \in R^D\), D is the word embedding dimension since we are using DistilBERT, its dimension is 768. L is the length of each question. We have set it to 12 as the mode of the distribution of question length is 12. We obtained T by averaging the output of the last and the penultimate layers of DistilBERT. Our question is a tensor of dimension \(R^{12\times 768}\).
3.3 Multi-modal multi-head self-attention
The Visual I and Question T features obtained from the Visual and Textual encoders, respectively are concatenated. Visual features \(I\in \,R^{C\times H\times W}\) and Question features \(T\in \,R^{L\times D}\) are concatenated such that each question word is concatenated at each spatial location of the image features resulting in a concatenated multi-modal representation \(P= [I \vert \vert T] \in \,R^{L\times H\times W\times (C+D)}\) . This multi-modal representation is linearly projected to generate Query(Q), Key(K), and Value(V) vectors as \(Q=PW_{QP}\), \(K=PW_{KP}\) and \(V=PW_{VP}\) where \(W_{QP}, W_{KP}, W_{VP} \in R^{(C+D)\times d}\). The Q, K, and V are reshaped to the dimension \(R^{S\times d}\) where \(S=L\times H\times W\). We compute the similarity between the Query and Key vectors using a scaled dot product followed by a row-wise softmax function to obtain a score matrix. The final output \(\hat{V} \in R^{S\times d}\) is generated by computing the weighted sum of the Value vector weighted by the similarity score as shown in (2):
Similar to [3], for richer multi-modal representation, we perform multi-head attention wherein Query, Key, and Value are linearly transformed h times to obtain h (Q, K, V) heads, and the scaled dot-product is performed independently by these attention heads. In multi-head attention, the dimension of query, key, and value is \(d_{q}, d_{k}, d_{v}\). The output from these h independent heads is concatenated and linearly transformed by \(W^{O}\) to produce the final multi-modal attended features as shown in (3):
where, \(head_i = Multi\_Attn(Q,K,V) = Attn(QW_{i}^{Q}, KW_{i}^{K}, VW_{i}^{V})\), \(W_{i}^{Q} \in R^{(d\times d_{q})}\), \(W_{i}^{K} \in R^{(d\times d_{k})}\), \(W_{i}^{V} \in R^{(d\times d_{v})}\) and \(W^{O} \in R^{(h*d_q\times d)}\) are the learnable matrices. To limit the size of the multi-head attention layer like [3], we set \(d_{v}=d_{k}=d_{q}=d/h\). The multi-modal attended features \(V'\) is projected and reshaped to the same dimension as P to obtain linearly transformed feature \(\hat{P} = V'W_P\) where \(W_{\hat{P}} \in R^{d\times (C+D)}\). We add a residual connection between transformed multi-modal attended features and concatenated multi-modal representation P followed by layer normalization [49] to obtain \(P'\). Thus, \(P'= LayerNorm(\hat{P}+P)\). It contains rich multimodal information and we pass it to next module.
Algorithm 1 and 2 describe the algorithm of the Multi-Modal Multi-Head self-attention while, Fig. 3 represents the detailed architecture.
3.4 Answer predictor
Kafle et. in [32] reported that the question-only model performs better than image-only models. Therefore, we average pool \(P'\) at all image regions to obtain an image attended question embedding \(\hat{T}\) as in (4). This \(\hat{T}\) is again projected to the same dimension as the question embedding T and summed with T to obtain \(T'\) as shown in (5).
where \(W_T \in R^{(C+D)\times D}\) is a projection matrix. \(T'\) summed over all the words to obtain a single attended context vector \(\hat{c}\) as shown in (6)
For the final answer, we pass \(\hat{c}\) through MLP, followed by a softmax layer to obtain probabilities over the answer set:
4 Experiment
4.1 Dataset description
We use the ImageCLEF VQA-Med-2019 dataset [8] in our work. The dataset has 2-D medical images and question-answer pairs. Medical images in this dataset are collected from MedPix databaseFootnote 1. The training set contains 3200 2-D images of different sizes. The smallest image in the dataset is of dimension \(106\times 109\), and the largest is of dimension \(2268\times 2040\). Except for eight images, every dataset image has questions related to four categories. Therefore, there are 12792 question-answer pairs. There are 500 images in the Validation set. Similar to the trainset, the validation set also has four types of questions based on them. Therefore, the validation set contains 2000 question-answer pairs. The dataset contains both open-ended and closed-ended questions. These questions in the dataset belong to four different categories. These four categories of questions are:
-
Modality:The main aim of questions from this category is to determine the modality of the radiology image. It contains images from eight major modalities: CT, XR, MR, US, MA, GI, AG, and PT. These modalities are further sub-categorized.
-
Plane: questions in this category are posed to identify the direction or plane of the given medical image. This dataset has questions related to identifying the 16 planes from eight major modalities.
-
Organ system: deals with questions about identifying organs/anatomy displayed in the image. There are ten different organ systems.
-
Abnormality: determines the abnormality depicted in the image. There are around 1600 different types of diseases in the dataset. Several diseases in this dataset have only one or two images.
The test set contains 125 image and question-answer pairs for each question category. Therefore the size of the testset is 500 question-image pairs. Figure 4. shows an image and its related questions. We use an additional VQA-Med-2020 test and validation datasets [37]. It contains 500 image and question-answer pairs; each question is related to the abnormality category.
4.2 Evaluation metric
Since our model is a classification model, we have used the accuracy metric to check the quantitative performance of our method. Accuracy is the total number of samples correctly classified per the total number of samples in the dataset. Besides accuracy, we have also used Recall, Precision, and F1 score to evaluate the performance of our classification model.
In binary classification, there are only two classes, positive and negative. For binary classification:
-
1.
True Positive(TP): when the predicted and actual labels are the same and equal to the positive class.
-
2.
False Positive(FP): when the predicted label is positive but the actual label is negative.
-
3.
False Negative(FN): when the predicted label is negative but the actual label is positive.
-
4.
True Negative(TN): when the predicted and actual labels are the same and equal to the negative class.
Multiclass classification problem has n number of classes where n is more than two. However, if we consider each class positive and all others negative, we will get n binary classification problems. Hence, a multiclass classification problem can be assumed as a collection of binary classification problems, one for each class. Therefore, we can compute TP, FP, FN, and FP in the same way we calculate for binary classification problems. The class we are considering is positive and others negative.
Recall measures the performance of a classification model in predicting all examples from a given class accurately. Mathematically it is defined as:
Precision informs us about the accuracy of a classification model in predicting a positive example i.e. total positive examples correctly predicted per total number of examples predicted as positive by the model. It is given as follows:
Here, TP is when a predicted class is the same as the actual class I.
F1 score is an essential metric for assessing the classification model’s performance, especially in an imbalanced dataset. It is a single value between 0 and 1 obtained by combining recall and precision by computing their harmonic mean. Mathematically it is defined as:
Diagnostic Odds Ratio (DOR) is a single indicator for analyzing the discriminative capability of the classification model. The odds ratio computes the relation between two events. Here we refer to the events as outcome and property. For example, in our case, the outcome is the prediction of our classification models, and the property is the actual label. DOR is the odds of a positive outcome where the property holds (positive actual label) divided by the odds where the property does not hold. Mathematically, DOR is:
The value of DOR lies between zero and infinity. A DOR value less than 1 indicates that the classification model cannot discriminate and is worse than chance. A DOR value equal to one shows that the model cannot discriminate between positive and negative cases. A higher value of DOR, greater than one, indicates a higher discriminatory power of the model.
4.3 Implementation details
We use a single NVIDIA Tesla v100 GPU for training and testing purposes. As described in Section 4.1, VQA-Med-2019 dataset contains questions related to four categories. Therefore, we train four models. Each model specializes in answering questions from a single category. We use the method given in [50] for classifying the questions into four categories during testing. Based on the question category, we call the corresponding trained model.
Before passing the images to the model, we resize them to \(256\times 256\). Therefore, the dimensions of image features I obtained from WideResNet-50-2 is \((2048\times 8\times 8)\). To make our model robust, we apply Data augmentation techniques. We use random rotation, horizontal flip, and contrast enhancement. The length of all the input questions is set to 12 words. Any question over 12 words are trimmed, and smaller than 12 words are padded to make all the questions of the same length. For the MMHSA module, we set the head h as eight and experiment with three different values of \(d_q = d_k = d_v\) for our four models to determine the best value. Table 3 shows the result of our experiment. Based on the accuracy metric, we chose the value of \(d_q = d_k = d_v\) for our four models. We have three fully connected layers in the Answer predictor module of Plane, Organ, and Modality models, each with 512, 2048, and 1024 neurons. The Answer predictor module of the Abnormality model gave good results with a single layer having 2048 neurons. We used LeakyReLU [51] activation function between our fully connected layers of the Answer classifier. We trained all the models with Adam optimiser [52] with default values of \(\beta _{1}\) & \(\beta _{2}\). We start the training process with a learning rate of 0.001, which is reduced by 0.1 after every ten epochs. Figure 5 shows the train loss as learning rate decreases after every ten epochs. We train the models for 150 epochs with an early stopping criterion to prevent overfitting. During training, we save the model having a minimum loss on validation dataset for testing. The training accuracy and loss over different epochs are shown in Figs. 6 and 7 respectively. From the plots it can be observed that for all the question categories the accuracy of the model on the validation set increases with the decreasing loss. This shows that as the training progresses our model is learning the generalized patterns from the training data. Table 4 shows the performance of our trained model on both training and test data sets.
4.4 Ablation
To evaluate the effectiveness of each component of our model, we perform an ablation study on our data. A detailed explanation of the model architecture in each experiment is given below.
-
Different image encoder and text encoder: In this experiment we try different combinations for image and text encoder to identify the best image encoder and text encoder pair for our task. For image encoder, we use Wide ResNet-50-2, ResNet50 [18], and VGG16 [24]. For text encoder, we use DistilBERT and Clinical BERT [53].
-
Without MMHSA: In this experiment, we remove the proposed Multi-Modal Multi-Head Self Attention(MMHSA) module from the model architecture. We concatenate visual and textual features such that each word is concatenated at each image region resulting in a concatenated representation of dimension\((L\times H\times W\times (C+D))\) where L is the length of the question, D is the question word embedding dimension, C is image channel size, H and W are the height and the width of the visual feature. This concatenated representation is summed along all spatial positions of the image and along all the words to obtain a single vector. This single vector is passed through MLP for answer generation.
4.5 VQA benchmarking
We choose the attention model proposed by Kazemi et al. [54] as the baseline model because this model outperformed previous models in VQA 1.0 [55] and VQA 2.0 [56] datasets and became a baseline. It uses an attention module similar to SAN [15]. The model concatenates the image and question features and passes through two convolution layers, generating two image attention glimpses. Image attention glimpses and question features are concatenated and passed through the classifier.
5 Results
5.1 Ablation study result
In our first ablation study, we try different image and text encoder combinations to find the most appropriate image encoder and text encoder combination. Table 5 shows the results with different Image and Text Encoders. From Table 5 we find that combining WideResNet-50-2 with DistilBERT is a better choice for our network architecture. It outperforms VGG16+DistilBERT by 12.78%, ResNet50+DistilBERT by 4.53%, VGG16+ClinicalBERT by 15.67%, ResNet50+ClinicalBERT by 13.63%, and WideResNet-50-2 with ClinicalBERT by 3.8%. Therefore, for all our experiments we use WideResNet-50-2 with DistilBERT. In our second ablation study, we see the contribution of our proposed attention MMHSA module in the performance of the model. From Table 6 we see that the MMHSA module plays a significant role in the performance of our model. With MMHSA, the accuracy of the model increases from 26.9% to 60.0%. Therefore, the proposed MMHSA module is able to effectively combine the features of the two input modalities.
5.2 Comparision with VQA benchmark
VQA Benchmark achieved an accuracy of 64.6% in VQA 1.0 [55] and 59.7% in VQA 2.0 [56] datasets. It surpassed the previous best model accuracies by 0.4% in VQA 1.0 and by 0.5% in VQA 2.0 datasets. From the results shown in Table 6, we see that our model achieves higher accuracy in all question categories than VQA Benchmark [54]. Our attention model achieves an accuracy of 60.0% while the VQA Benchmark achieves 40.6% on ImageCLEF 2019 dataset. Therefore, we can say that our attention mechanism is able to capture the relationship between text and image features.
5.3 Comparison with state-of-the-art
We compare the overall accuracy achieved by our model with the overall accuracy achieved by some of the top-performing models in the ImageCLEF 2019 challenge. Since we have treated MedVQA task as a classification problem, we compare our model with classification models only. We have not compared our model with answer generation models. Hanlin [29] secured the first position in ImageCLEF 2019 challenge. It uses MFB with co-attention [13] for fusion. UMMS [40] also uses MFB with co-attention for multi-modal fusion. IBM Research AI [41] and Team_PwC_-Med [42] use attention mechanism for fusion. Table 7 shows the overall accuracy comparison of these models. In Table 8, we compare our results with the Transformer-based model MMBERT [5]. Since the MMBERT model uses additional medical data for pretraining, for fair comparision we use the image encoder pretrained on ImageNet only for MMBERT. We observe that our model performes better than MMBERT [5] by 1.35% with fewer parameters. Our model uses 46.78% less parameters than MMBERT as shown in Table 8.
5.4 Qualitative analysis
For qualitative analysis, we examine the output of GradCam [9] to determine the regions in the image where the model is actually looking while providing answers. Figure 8. shows the output of GradCam. We have shown the output of the image-question pair, which the model correctly classified. These image-question answer pairs are from the organ category. We have included the output of the organ category because identifying organs from radiology images is a slightly simpler task for a non-medical person than identifying other categories. In the GradCam output, we see that the model focuses on the regions where the organ is present while making predictions.
6 Discussion
According to the results of the ablation study and comparison with the VQA Benchmark model shown in Table 6, we see that the MMHSA module effectively fuses image and question features. Our model achieves accuracy similar to MMBERT in organ, modality, and plane categories and outperforms MMBERT in the abnormality category with 46.78% less parameters as shown in Table 8. Besides accuracy, we have also computed Recall, Precision, and F1 score using scikit-learn [57] to analyze the performance of our proposed model. Table 9 shows the Recall, Precision and F1 score. A high F1 score for classification models indicate a high recall and precision value. A classification model with high recall and precision value is considered suitable for the classification task. Therefore, the higher the F1 score, the better the model. Our model achieves an f1 score of 0.83, 0.84 for modality, plane categories, which is higher than MMBERT and VQA Benchmark. MMBERT achieves 0.81 and 0.83 f1 score in modality and plane categories, while VQA Benchmark scores 0.53 and 0.45 in the same categories respectively. In the organ category, our model and MMBERT achieves the same f1 score of 0.75, while VQA Benchmark achieves 0.25. For the abnormality category, our model achieves a high recall value in comparison to MMBERT and VQA Benchmark. However, the overall performance in the abnormality category is low for all three models.
To analyze the discriminative ability of our models, we compute the DOR values for each class in four categories. There are 13 classes in the test set of plane category. Among all predicted classes, the plane classifier perfectly classifies all the instances of “mammo-cc" class. Therefore, for the " mammo-cc" class, the classifier achieves 100% accuracy and infinite DOR value. The “ap" class has the lowest DOR of 61.1 among all the predicted classes. The plane classifier incorrectly predicts all the instances of “longitudinal", “frontal", and “oblique" classes. The DOR value for these three classes is zero, signifying that the classifier cannot learn the discriminating features of these classes. The train set has the least proportion of these classes. Therefore, due to fewer images from these classes, the classifier cannot learn the discriminating features and, hence, misclassifies them. However, for all the predicted classes the plane classifier achieves a DOR value much higher than one.
The modality category has 25 different classes in the testset. For classes “cta - ct angiography", “ct w/contrast (iv)", “mr - flair", “mr - adc map (app diff coeff)", the classifier classifies perfectly with no false positve and false negative cases. Hence, the classifier obtains infinite DOR and 100% accuracy in these classes. The “yes" class has a minimum DOR value of 245.3 among all the predicted classes. The classifier cannot predict “us-d-doppler ultrasound", “gi and iv", “ct with gi and iv", “non contrast(mri)", and “sbft - small bowel" classes. Therefore, the classifier achieves a zero DOR for these classes. The“sbft" is a submodality of modality X-ray, since it is a particular type of X-ray. Due to less number of images of this class in the training set, the classifier cannot learn the discriminating features and classify “sbft - small bowel" as “xr - plain film". Similarly, due to less number of images in the trainset and overlapping concepts, all the instances of “us-d-doppler ultrasound", “gi and iv", and “non contrast(mri)" are mapped to “us - ultrasound", “iv", and “noncontrast" classes.
The organ category has ten classes. For the class “skull & contents", the classifier achieves the highest DOR value of 586.5 among all the predicted classes. The “face, sinuses, and neck" class has the lowest DOR value of 28.25. Organ classifier performs worst in classifying “heart and great vessels" and “vascular and lymphatic" classes. The classifier misclassifies all the instances of these classes in the test set. The train set has less images from these two classes. Due to the lowest proportion in the train set, the classifier cannot capture their discriminating features and, hence, misclassifies them. Overall, organ classifier achieves DOR value greater than one for all the predicted classes.
Abnormality category has 111 different classes in its testset. The abnormality classifier perfectly classifies all the examples of “yes", “acute appendicitis", “enchondroma", “hypertrophic pyloric stenosis" and “appendicitis" from the test set. For these classes, the classifier achieves 100% accuracy and infinite DOR value. However, the model misclassifies all the instances from other classes. Therefore, for all other classes, the DOR value is zero, and the overall accuracy of the abnormality classifier reduces to 0.08%.
According to the DOR values of each class, our model efficiently captures the discriminative features for maximum classes of plane, modality and organ categories. However, our model does not perform well in the abnormality category. This is because of the high data imbalance in the abnormality category. Also, the testset of this category has classes that are absent in the trainset. In the present form, the MedVQA model can only answer basic questions related to plane, organ, and modality identification from medical images. Doctors cannot use it for disease diagnosis. However, we can overcome this problem by training the model with more medical data and adding clinical knowledge. In Table 10 we discuss the strengths, weaknesses, opportunities, and threats.
7 Conclusion
This paper proposes a Multi-Modal Multi-head Self-Attention mechanism for Medical VQA. We use ImageCLEF 2019 VQA-Med data. It contains questions related to four categories. Therefore, we train four models, each specializing in answering questions from a single category. We apply data augmentation techniques to increase the size of the data. We resize all images to 256X256. We apply random rotation, contrast enhancement, and horizontal and vertical flips on the images. For question tokenization, we use the DistilBERT tokenizer. We use WideResNet50-2 for extracting image features. We pass question tokens through DistilBERT to obtain question word embedding. We feed the image features and question word embeddings into the MMHSA module. In the MMHSA module, question word embeddings are concatenated with the image features. We concatenate image and question words such that each word is concatenated at each spatial location. These concatenated image and question word embeddings, called as concatenated multimodal features, pass through a multi-head self-attention module. The multi-head self-attention module transforms the concatenated input into multiple query, key, and value vectors. It computes the scaled dot product between the query and key vectors and applies softmax to obtain weights on the value vector. The value vector is projected and resized to the same dimension as concatenated multimodal features. The output of the multi-head self-attention module is added with concatenated multimodal features. This representation contains relevant question and image features. We average it spatially along the image dimension. The resulting vector is linearly transformed to the same dimension as question word embedding. We add the transformed vector with the question word embeddings and sum along question words to obtain a single context vector that contains relevant image and text information. This context vector passes through Multi-Layer Perceptron followed by softmax to obtain the answer distribution. In the multi-head self-attention module, we widened the dimension of query, key, and value and observe the model’s performance for the MedVQA classification task on different question categories. We find that different question categories perform well at different query, key, and value dimensions. We obtain an accuracy of 60.0% with a single multi-head self-attention module. It is comparable to several models with multiple multi-head self-attention layers. Since our model uses a single attention module, we achieve this accuracy with almost half the parameters used by models using multiple self-attention layers.
Our work’s primary focus is to enhance our model’s overall performance using the minimum number of parameters. We have yet to consider the problem of data imbalance. In our future work, we aim to explore image-pretraining techniques to use the vast amount of publicly available unlabelled medical data. We also want to examine the impact of adding a medical knowledge base to the model’s efficiency, especially in the abnormality category. In future work, we will examine the performance of the model on other MedVQA datasets.
Data Availability
We have used VQA-Med-2019 and VQA-Med-2020 datasets for this task. The complete VQA-Med-2019 dataset is publicly available at https://github.com/abachaa/VQA-Med-2019. VQA-Med-2020 is available at https://github.com/abachaa/VQA-Med-2020. Only the Validation set and Test set of VQA-Med-2020 is available publicly so we have used only that for training purpose.
Notes
https://medpix.nlm.nih.gov/
References
McDonald RJ, Schwartz KM, Eckel LJ, Diehn FE, Hunt CH, Bartholmai BJ, Erickson BJ, Kallmes DF (2015) The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Academic radiology 22(9):1191–1198
Itri JN, Tappouni RR, McEachern RO, Pesch AJ, Patel SH (2018) Fundamentals of diagnostic error in imaging. Radiographics 38(6):1845–1865
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst 30
Kenton JDM-WC, Toutanova LK (2019) Bert: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of naacL-HLT, vol 1, p 2
Khare Y, Bagal V, Mathew M, Devi A, Priyakumar UD, Jawahar C (2021) Mmbert: Multimodal bert pretraining for improved medical vqa. In: 2021 IEEE 18th international symposium on biomedical imaging (ISBI), pp 1033–1036. IEEE
Ren F, Zhou Y (2020) Cgmvqa: A new classification and generative model for medical visual question answering. IEEE Access. 8:50626–50636
Zagoruyko S, Komodakis N (2016) Wide residual networks. In: Wilson RC, Hancock ER, Smith WAP (eds) Proceedings of the British Machine Vision Conference 2016, BMVC 2016, York, UK, September 19–22, 2016. http://www.bmva.org/bmvc/2016/papers/paper087/index.html
Abacha AB, Hasan SA, Datla VV, Liu J, Demner-Fushman D, Müller H (2019) Vqa–med: Overview of the medical visual question answering task at imageclef 2019. CLEF (working notes) 2
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626
Fukui A, Park DH, Yang D, Rohrbach A, Darrell T, Rohrbach M (2016) Multimodal compact bilinear pooling for visual question answering and visual grounding. In: Su J, Carreras X, Duh K (eds) Proceedings of the 2016 conference on empirical methods in natural language processing, EMNLP 2016, Austin, Texas, USA, November 1–4, 2016, pp 457–468 . https://doi.org/10.18653/v1/d16-1044
Charikar M, Chen K, Farach-Colton M (2004) Finding frequent items in data streams. Theoretical Comput Sci 312(1):3–15
Pham N, Pagh R (2013) Fast and scalable polynomial kernels via explicit feature maps. In: Proceedings of the 19th ACM SIGKDD international conference on knowledge discovery and data mining, pp 239–247
Yu Z, Yu J, Fan J, Tao D (2017) Multi-modal factorized bilinear pooling with co-attention learning for visual question answering. In: Proceedings of the IEEE international conference on computer vision, pp 1821–1830
Ben-Younes H, Cadene R, Cord M, Thome N (2017) Mutan: Multimodal tucker fusion for visual question answering. In: Proceedings of the IEEE international conference on computer vision, pp 2612–2620
Yang Z, He X, Gao J, Deng L, Smola A (2016) Stacked attention networks for image question answering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 21–29
Lu J, Yang J, Batra D, Parikh D (2016) Hierarchical question-image co-attention for visual question answering. Adv Neural Inf Process Syst 29
Peng Y, Liu F, Rosen MP (2018) Umass at imageclef medical visual question answering (med-vqa) 2018 task. In: CLEF (working notes)
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural computation. 9(8):1735–1780
Zhou Y, Kang X, Ren F (2018) Employing inception-resnet-v2 and bi-lstm for medical domain visual question answering. In: CLEF (working notes)
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence, vol 31
Schuster M, Paliwal KK (1997) Bidirectional recurrent neural networks. IEEE Trans Signal Proces 45(11):2673–2681
Abacha AB, Gayen S, Lau JJ, Rajaraman S, Demner-Fushman D (2018) Nlm at imageclef 2018 visual question answering in the medical domain. In: CLEF (working notes)
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: Bengio Y, LeCun Y (eds) 3rd International conference on learning representations, ICLR 2015, San Diego, CA, USA, May 7–9, 2015, conference track proceedings
Talafha B, Al-Ayyoub M (2018) Just at vqa-med: A vgg-seq2seq model. In: CLEF (working notes)
Allaouzi I, Ahmed MB (2018) Deep neural networks and decision tree classifier for visual question answering in the medical domain. In: CLEF (working notes)
Cho K, Merrienboer B, Gülçehre Ç, Bahdanau D, Bougares F, Schwenk H, Bengio Y (2014) Learning phrase representations using RNN encoder-decoder for statistical machine translation. In: Moschitti A, Pang B, Daelemans W (eds) Proceedings of the 2014 conference on empirical methods in natural language processing, EMNLP 2014, October 25–29, 2014, Doha, Qatar, A Meeting of SIGDAT, a special interest group of The ACL, pp 1724–1734 . https://doi.org/10.3115/v1/d14-1179
Kim J-H, Jun J, Zhang B-T (2018) Bilinear attention networks. Adv Neural Inf Process Syst 31
Yan X, Li L, Xie C, Xiao J, Gu L (2019) Zhejiang university at imageclef 2019 visual question answering in the medical domain. CLEF (working notes) 85
Sharma D, Purushotham S, Reddy CK (2021) Medfusenet: An attention-based multimodal deep learning model for visual question answering in the medical domain. Scientific Reports 11(1):1–18
Vu MH, Löfstedt T, Nyholm T, Sznitman R (2020) A question-centric model for visual question answering in medical imaging. IEEE Trans Medical Imaging 39(9):2856–2868
Kafle K, Kanan C (2017) Visual question answering: Datasets, algorithms, and future challenges. Comput Vis Image Underst 163:3–20
Xu K, Ba J, Kiros R, Cho K, Courville A, Salakhudinov R, Zemel R, Bengio Y (2015) Show, attend and tell: Neural image caption generation with visual attention. In: International conference on machine learning, pp 2048–2057. PMLR
Ye L, Rochan M, Liu Z, Wang Y (2019) Cross-modal self-attention network for referring image segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 10502–10511
Gong H, Chen G, Liu S, Yu Y, Li G (2021) Cross-modal self-attention with multi-task pre-training for medical visual question answering. In: Proceedings of the 2021 international conference on multimedia retrieval, pp 456–460
Lau JJ, Gayen S, Ben Abacha A, Demner-Fushman D (2018) A dataset of clinically generated visual questions and answers about radiology images. Scientific data. 5(1):1–10
Abacha AB, Datla VV, Hasan SA, Demner-Fushman D, Müller H (2020) Overview of the vqa-med task at imageclef 2020: Visual question answering and generation in the medical domain. In: CLEF (working notes)
Ben Abacha A, Sarrouti M, Demner-Fushman D, Hasan SA, Müller H (2021) Overview of the vqa-med task at imageclef 2021: Visual question answering and generation in the medical domain. In: Proceedings of the CLEF 2021 conference and labs of the evaluation forum-working notes. 21–24 Sept 2021
Liu B, Zhan L-M, Xu L, Ma L, Yang Y, Wu X-M (2021) Slake: A semantically-labeled knowledge-enhanced dataset for medical visual question answering. In: 2021 IEEE 18th international symposium on biomedical imaging (ISBI), pp 1650–1654. IEEE
Shi L, Liu F, Rosen MP (2019) Deep multimodal learning for medical visual question answering. In: CLEF (working notes)
Kornuta T, Rajan D, Shivade C, Asseman A, Ozcan AS (2019) Leveraging medical visual question answering with supporting facts. arXiv preprint arXiv:1905.12008
Bansal M, Gadgil T, Shah R, Verma P (2019) Medical visual question answering at image clef 2019-vqa med. In: CLEF (working notes)
Verma H, Ramachandran S (2020) Harendrakv at vqa-med 2020: Sequential vqa with attention for medical visual question answering. In: CLEF (working notes)
Liu S, Ding H, Zhou X (2020) Shengyan at vqa-med 2020: An encoder-decoder model for medical domain visual question answering task. In: CLEF (working notes)
Sitara NMS, Srinivasan K (2021) Ssn mlrg at vqa-med 2021: An approach for vqa to solve abnormality related queries using improved datasets. In: CLEF (working Notes), pp 1329–1335
Manmadhan S, Kovoor BC (2023) Parallel multi-head attention and term-weighted question embedding for medical visual question answering. Multimedia Tools and Applications 1–22
Liu B, Zhan L-M, Wu X-M (2021) Contrastive pre-training and representation distillation for medical visual question answering based on radiology images. In: Medical image computing and computer assisted intervention–MICCAI 2021: 24th International conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part II 24, pp 210–220 . Springer
Sanh V, Debut L, Chaumond J, Wolf T (2019) Distilbert, a distilled version of bert: smaller, faster, cheaper and lighter. arXiv preprint arXiv:1910.01108
Xu J, Sun X, Zhang Z, Zhao G, Lin J (2019) Understanding and improving layer normalization. Adv Neural Inf Process Syst 32
Al-Sadi A, Al-Ayyoub M, Jararweh Y, Costen F (2021) Visual question answering in the medical domain based on deep learning approaches: A comprehensive study. Pattern Recogn Lett 150:57–75
Xu J, Li Z, Du B, Zhang M, Liu J (2020) Reluplex made more practical: Leaky relu. In: 2020 IEEE symposium on computers and communications (ISCC), pp 1–7 . IEEE
Kingma DP, Ba J (2015) Adam: A method for stochastic optimization. In: Bengio Y, LeCun Y (eds) 3rd International conference on learning representations, ICLR 2015, San Diego, CA, USA, May 7–9, 2015, conference track proceedings . arXiv:1412.6980
Alsentzer E, Murphy J, Boag W, Weng W-H, Jin D, Naumann T, McDermott M (2019) Publicly available clinical BERT embeddings. In: Proceedings of the 2nd clinical natural language processing workshop, pp 72–78. Association for computational linguistics, Minneapolis, Minnesota, USA. https://doi.org/10.18653/v1/W19-1909, https://www.aclweb.org/anthology/W19-1909
Kazemi V, Elqursh A (2017) Show ask attend and answer: A strong baseline for visual question answering. CoRR. arXiv:1704.03162
Antol S, Agrawal A, Lu J, Mitchell M, Batra D, Zitnick CL, Parikh D (2015) Vqa: Visual question answering. In: Proceedings of the IEEE international conference on computer vision, pp 2425–2433
Goyal Y, Khot T, Summers-Stay D, Batra D, Parikh D (2017) Making the v in vqa matter: elevating the role of image understanding in visual question answering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 6904–6913
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V et al (2011) Scikit-learn: Machine learning in python. J Mach Learn Res 12:2825–2830
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Joshi, V., Mitra, P. & Bose, S. Multi-modal multi-head self-attention for medical VQA. Multimed Tools Appl 83, 42585–42608 (2024). https://doi.org/10.1007/s11042-023-17162-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17162-3