Abstract
The present study introduces an efficient algorithm for automatic segmentation and detection of mass present in the mammograms. The problem of over and under-segmentation of low-contrast mammographic images has been solved by applying preprocessing on original mammograms. Subtraction operation performed between enhanced and enhanced inverted mammogram significantly highlights the suspicious mass region in mammograms. The segmentation accuracy of suspicious region has been improved by combining wavelet transform and fast fuzzy c-means clustering algorithm. The accuracy of mass segmentation has been quantified by means of Jaccard coefficients. Better sensitivity, specificity, accuracy, and area under the curve (AUC) are observed with support vector machine using radial basis kernel function. The proposed algorithm is validated on Mini-Mammographic Image Analysis Society (MIAS) and Digital Database for Screening Mammography (DDSM) datasets. Highest 91.76% sensitivity, 96.26% specificity, 95.46% accuracy, and 96.29% AUC on DDSM dataset and 94.63% sensitivity, 92.74% specificity, 92.02% accuracy, and 95.33% AUC on MIAS dataset are observed. Also, shape analysis of mass is performed by using moment invariant and Radon transform based features. The best results are obtained with Radon based features and achieved accuracies for round, oval, lobulated, and irregular shape of mass are 100%, 70%, 64%, and 96%, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer is a problem of extreme significance with social and financial implications to the public health. Different kinds of cancers have been already reported in literature and can be classified by the type of cells that are initially affected. Breast cancer is the major health issue among women. Early detection of breast cancer can be achieved by mammography techniques which allow the visualization of tissue structure of the breast. Various types of lesions present in mammograms can be classified as micro-calcification, mass, architectural distortion, and bilateral asymmetry. Detection of micro-calcification has achieved a high degree of accuracy [27, 35]. Automatic mass segmentation is the crucial task due to (a) the presence of background information in the mammograms; (b) variability in the mass size, mass shape, and margin; and (c) breast tissue density. Hence, the accuracy obtained through automatic mass detection needs further improvement. Interestingly enough, local segmentation of mass has been done by manually cropping the mass regions [8, 13, 46].
Several techniques have already been applied for automatic mass detection by several authors [2, 5, 22]. The range of sensitivity in these works varies between 80% to 95% with 0.31 to 4.8 false positive per image (FP/I). Various approaches for mass segmentation is categorized as unsupervised and supervised [33]. Unsupervised segmentation includes region based, contour based, and clustering based techniques. Region based methods employ region growing, spilt, and merges techniques. Edge information is utilized in contour based approach to identify the suspicious region [19, 23]. Mass segmentation using density weighted contrast enhancement and Laplacian-of-Gaussian edge detector is carried out by Petrick et al. [36, 37]. Pattern matching approach is used in model-based mass segmentation. Ng and Bischof, Che et al., and Constantinidis et al. applied cross-correlation method for mass detection [6, 9, 28]. Information clustering based algorithm for mass detection is proposed by Cao et al. [4]. Segmentation of mass region with growing neural gas network algorithm has been proposed by Oliveira et al. [32]. A sensitivity of 89.3% has been achieved by applying the support vector machine (SVM) with Riply’s k-function on 997 mammograms of Digital Database for Screening Mammography (DDSM) dataset. Nunes et al. applied k-means clustering and template matching techniques for mass segmentation [30]. The classification accuracy of 83.94% along with 83.24% sensitivity and 84.14% specificity is observed with geometry and texture based Simpson’s diversity index features. Sampaio et al. applied the cellular neural network based mass segmentation with shape and geostatic based texture features [39]. Sensitivity of 80% with 0.84 FP/I is obtained by using SVM classifier to classify as mass and non-mass. Combination of wavelet and genetic algorithm based mass detection is employed by Pereira et al. [35]. Sensitivity of 95% is reported on testing with 640 images of DDSM dataset. Kurt et al. applied Otsu as well as Havrda & Charvat thresholding techniques for mass detection [24]. The proposed technique obtained 93% sensitivity with 96 mammograms of Mini-Mammographic Image Analysis Society (MIAS) dataset. Jen et al. proposed automatic mass detection by using morphological image processing techniques. Sensitivity values of 88% and 86% are reported for sample images of DDSM and MIAS dataset respectively, by using intensity based features and principle component analysis (PCA) [20]. Classification between malignant and non-malignant mass using Zernike based features and SVM classifier is carried out by Sharma and Khanna [41]. Fractal dimension is applied by Nguyen and Rangayyan for shape analysis of masses to determine its malignancy [29]. Surendiran and Vadivel employed region based features for shape analysis of mass to differentiate it as benign and malignant [43]. Moment invariant and Radon based features for shape analysis of micro-calcification are employed by Bocchi and Nori [3].
The purpose of this study is to develop an efficient algorithm with reduced computational time, by applying key image processing techniques in a way which improves the overall segmentation and classification accuracy. Segmentation of suspicious mass region is performed by fast fuzzy c-means (FCM) clustering in which histogram of pixel intensity values of images are used instead of actual pixel intensity values for clustering. Texture based features, namely first and second order statistical features using gray level co-occurrences matrix (GLCM), gray level run length matrix (GLRLM), and local binary patterns (LBP) have been used for classification of suspicious region as mass or non-mass. After detection of mass regions, shape analysis of mass has been carried out to classify it as benign or malignant.
The present study is structured into the following sections. The proposed methodology is introduced in Section 2. Classification of mass shape is described in Section 3. Validation of algorithm and experimental results are discussed Section 4. Conclusion and future work are presented in Section 5.
2 Proposed methodology
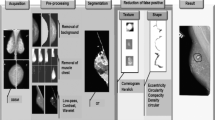
Block diagram of the proposed methodology is presented in the Fig. 1. It consists of various steps, i.e., preprocessing, mass segmentation, feature extraction of segmented mass, feature selection and classification. Each step is briefly described in the following subsections.
2.1 Preprocessing
Mammograms consist of patient’s information, labels and artifacts. Preprocessing operation is executed for breast region identification and to remove undesirable information from the original mammogram. At first, binarization is achieved by using global thresholding method to identify the breast area. Background information and labels are removed by employing the morphological opening operation [15]. A “Square” structuring element of 12 pixel width is applied for morphological opening operation in the present work. Further, object labeling is performed by applying connected component labeling technique. Breast area is the largest connected object and it is superimposed on the original mammogram to obtain the artifacts free mammogram. Artifacts free mammogram is inverted. Further, unsharp masking is applied to enhance low contrast mammograms [15]. The enhanced inverted and artifacts free mammogram is subtracted from enhanced artifacts free mammogram to obtain the subtracted mammogram. Overall noise present in the subtracted image is removed by applying median filtering of 3 × 3 neighborhood.
2.2 Mass segmentation
This step consists of wavelet decomposition and segmentation of suspicious mass region. Frequency and temporal information can be obtained by wavelet transformation. The filtered image I F is decomposed by using wavelet transform which produces four coefficients, i.e., approximation, vertical detailed, horizontal detailed, and diagonal detailed coefficients. Mother wavelet functions are used as a basis function which produces set of other functions by applying dilation and translation operations. The scaled and translated basis functions φ j , l , p (a, b) and \( {\phi}_{j, l, p}^i\left( a, b\right) \) are expressed as [15]:
here, h , v , d represents the detailed coefficients in horizontal, vertical, and diagonal directions, respectively. Integer variables j , l , p scale and dilate the mother wavelet. Index j is scaling parameter while l and p are translation parameters. 2-D wavelet transform of an image I F (a, b) of size L × P can be represented as:
here, j 0 is an arbitrary starting scale. Daubechies-4 wavelet decomposition is used in the present work. Approximation coefficients of level-1 are used for segmentation of suspicious mass.
A mammogram is divided into homogeneous and non-homogeneous regions to locate the suspicious region-of-interest (ROI). Mass detection accuracy depends on the proper segmentation of ROI. FCM gives optimal number of partitioning [34]. It returns the value between 0 and 1 which represents the degree of membership between each data and cluster center. Image pixels are represented as one-dimensional feature vector for clustering.
The purpose of FCM is to minimize the group sum of squared error objective function J FCM
where I = x 1 , x 2 , x 3 . … … x n ⊆ R m represents the set of pixel values in vector space of m dimension, n denotes the number of pixel values, and c represents the number of clusters and its values lies between 2 ≤ c < n. Degree of fuzzy membership of pixel x j in the r th cluster is represented by u rj , k denotes the weighting exponent to control the fuzziness of member function, v r denotes the prototype of cluster center r, and the distance between j th pixel x j and r th cluster center v r is given by d 2(x j , v r ). The similarity between pixel value and cluster center is measure by Euclidean distance [34]. Objective function is minimized by updating the cluster center v r and membership value u rj
Here suspicious region is segmented from the approximated image by employing fast fuzzy c-means (FFCM) clustering algorithm. In FFCM algorithm, histogram of the pixel value is created and subsequent iterations of FCM clustering are updated by using the histogram instead of actual pixel values. The histogram is defined as small number of bins which are fixed. The runtime of iteration can be reduced by using histogram bins in clustering step. We can easily identify the cluster assigned to pixel value, because a unique correspondence exists between pixel value and mapped histogram bin. The value of cluster center and weighting exponent are taken as 3 and 2 respectively in the present work. Cluster 3 map is further used for ROI extraction.
2.3 Feature extraction of segmented mass and feature selection
Feature extraction step is used to characterize the segmented suspicious mass region. Textural features play a significant role to extract the meaningful information from mammograms [12, 14, 44]. Texture features contain spatial relationship among pixels and gray value variations in the suspicious region. Following textural features are extracted from suspicious regions.
2.3.1 First order statistical features
Six first order statistical features, i.e., mean, average contrast, skewness, kurtosis, energy, and entropy are extracted from the segmented suspicious regions [26].
2.3.2 Gray level co-occurrences matrix features (GLCM)
Second-order features are generated by GLCM [16]. GLCM represents intensity distribution and the relative positions of neighboring pixels. Fourteen GLCM features, i.e., energy, entropy, contrast, correlation, auto-correlation, maximum probability, homogeneity, variance, sum variance, sum entropy, sum average, difference variance, information measure of correlation1, and information measure of correlation2 are extracted for four orientations (00,450,900,1350) from the segmented regions.
2.3.3 Gray level run length matrix features (GLRLM)
GLRLM calculates the number of gray level runs of various length [45]. Seven GLRLM features, i.e., short run emphasis, long run emphasis, gray level distribution, run percentage, short run low gray level emphasis, long run high level emphasis are also extracted for four directions (00,450,900,1350).
2.3.4 Local binary pattern (LBP)
LBP performs rotation invariant textural analysis [25, 31] to describe the local texture property of ROI. In basic LBP, image pixels are labeled by taking the thresholding on 3 × 3 neighborhood of each pixel with the center pixel value and get the binary number. LBP operator can also be used for circular neighborhood [31, 45]. Considering M sampling points and Q radius of neighborhood, the sampling points around pixel (a, b) lie at coordinates
Pixel value is bilinearly interpolated when sampling point does not belong to integer coordinates. LBP code for the center pixel of an image I(a, b) is computed by summation of the binary string which is weighted by power of two as:
r(y) represents the thresholding function with value 1 if y ≥ 0, otherwise 0.
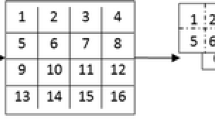
The labeling of pixel values depends on the size of neighborhood M. Different 2m combinations of binary strings can be generated in each neighborhood. Total 36 rotation invariants pattern can be obtained from combination of 28 binary strings by considering M = 8. The whole procedure of LBP generation for circular neighborhood of radius q = 1 and neighboring pixels M = 8 is presented in Fig. 2. Rotation invariant LBP is achieved by specifying the minimum value of the binary string which gives unique bit sequences. Basic LBP pattern is extended into uniform pattern in case of circular bit pattern which consists of at most 2 bitwise transitions from 1 to 0 or 0 to 1. Histogram labeling is computed after labeling of each pixel with LBP codes and used as a texture descriptor. Basic LBP produces 256 texture descriptor based on 3 × 3 neighborhood. Uniform, rotation invariant, and rotation invariant uniform LBP generate 59, 36, and 10 descriptors, respectively. Rotation invariant uniform LBP is extracted from segmented ROI in the present work.
A feature vector of 102 dimensions is created, which includes 7 first order statistical features, 56 GLCM, 28 GLRLM, and 10 LBP features. Features are rescaled with zero mean (μ) and unit variance (σ) with the Z-score normalization as:
Further, dimensionality reduction of feature vector is performed by applying PCA to get accurate result in less computation time [21]. PCA mathematically transforms the correlated features into small number of uncorrelated features which are called principle components (PC). First principle component shows the high variability and succeeding components show remaining variability.
2.4 Classification
Segmented ROIs are classified as mass or non-mass regions by employing SVM, which is a supervised machine learning classifier [10]. Reduced feature vector from feature selection step is given as input data to the SVM classifier. It creates support vectors to identify the boundaries between mass and non-mass. Support vector determines the position of the hyperplane. SVM classifier is tested with various kernel functions, i.e., RBF, linear, polynomial, and MLP. The parameters used for different SVM kernel function are shown in Table 1.
3 Shape analysis
Breast imaging reporting and data system (BI-RADS) introduced by Radiology department of American college reported various categories of masses according to shape, size, and density [1]. According to this classification, shape of the masses is round, oval, lobulated, microlobulated, ill-defined, and architectural distortion. Benign masses are characterized with round or oval shape with circumscribed margin. Malignant masses are characterized with irregular shape and ill-defined margin as shown in Table 2.
Segmented masses are further processed for analysis of various shapes of mass. Analysis of mass shape is a significant step to evaluate the malignancy of the lesion. Classification of masses in four categories namely round, oval, lobulated, and irregular masses is evaluated by using two different sets of features. First set uses rotation and translation invariant features computed from second and third order moment of inertia. Second set of features uses radon descriptor, which is rotation and scale invariant of the image. Multi-class SVM is used to assign each mass into one of the four classes. Same classifier is used for both set of features and the performance of both feature sets has been compared.
3.1 Moment invariant features
Moment invariants provide the shape characteristics of an object. Rotation invariant Hu moments are used for translation, scale, and rotation invariant pattern identification [17]. Seven moment invariant features extracted from segmented masses are given below, where μ represents central moment of the objectI(a, b), and (p + q)denote order of moments(p, q = 0, 1, 2…).
Where
3.2 Radon transform
Rotation and scale invariant properties of image can be obtain by Radon descriptor which is useful for shape analysis of mass [11]. Radon transform is used in various applications such as image reconstruction, face recognition, and object identification. Radon transform of an image I(a, b)can be expressed as:
It gives the projection of an image I(a, b)along different value of φ and projection line. Radon transform maps the Cartesian co-ordinate (a, b)into a polar coordinate(ρ, φ). Complete representation of the image can be obtained by computing the radon transform for each value of φ and ρ.Radon transform of the segmented mass is computed for 180 values of φ, i.e.,φ= 0, 1, 2….179. Energy of each projection is computed as:
Rotation and scale invariant feature vector of energy is obtained by normalizing it with respect to the largest component of the energy. Elements of the features are rotated until the largest element becomes the first element. Due to rotation, the first element of feature is 1 and other elements are in the range between 0 and 1.
4 Experiments and results
Privacy issue is the major constraint in acquiring medical images for experimentation. Publicly available MIAS and DDSM data sets are used to validate the proposed methodology [38, 42]. Total 322 mediolateral oblique (MLO) views of mammograms are available in MIAS dataset. All mammograms are digitized with 1024 × 1024 pixels resolution and 256 Gy levels. The information about the location of abnormalities like the center of a circle surrounding the tumor, its radius, breast position (left or right), type of breast tissues (fatty, fatty-glandular, or dense), and tumor type if exists (benign or malign) are present in the dataset. 2620 cases with contrast resolution of 12-bits and 16-bits are available in the DDSM dataset. It also contains patient age, screening exam date, and date on which mammograms have been digitized. Pixel level ground truth of abnormalities is also given. Experiments have been performed using 60 mass containing mammogram sample from MIAS dataset and randomly chosen 700 mammogram samples from the DDSM dataset. Complete description of selected sample mammograms from DDSM and MIAS dataset has been given in Table 3. Scaling of all sample images of DDSM dataset to 1024 × 1024 pixels is done by applying nearest neighbor interpolation method. Proposed algorithm has been implemented on MATLAB® 2012a, 3.2GHz processor with 4GB RAM.
4.1 Results of mass detection
Intermediate results of various operation performed in breast region extraction step are shown in Fig. 3. The original mammogram, binarized image, opened image after performing morphological opening, and extracted breast region are shown in Fig. 3(a-d), respectively. Fig 3 clearly shows that breast region extraction and removal of background information has been done successfully. Fig 4(a-d) show the enhanced extracted breast region mammogram after applying unsharp masking operation, inverted mammogram, enhanced inverted mammogram, and subtracted mammogram, respectively. The suspicious region is visually improved in the subtracted images as compared to the original image. Subtraction operation is a significant step to highlight the suspicious region and suppress breast tissues. It helps to segment the suspicious region accurately.
Intermediate results for mammogram enhancement on the output from Fig. 3 (a) Enhanced, (b) Inverted, (c) Enhanced inverted, and (d) subtracted mammogram
Filtered image, segmented mammograms using FFCM with cluster 1, cluster 2 and cluster 3, and class membership and class assignment of pixel intensity values in FFCM are presented in Fig. 5(a-e), respectively. Segmentation results show that better segmentation of suspicious region is found in cluster 3.
Segmentation results with FFCM algorithm on the sample mammogram from MIAS dataset in Fig. 3 (a) Filtered mammogram, (b) Cluster1 map, (c) Cluster 2 map, (d) Cluster 3 map, and (e) Class membership and class assignment
Description of some randomly chosen sample mammograms of MIAS and DDSM dataset are given in Table 4. Mass segmentation results for both benign and malignant sample mammogram images of MIAS and DDSM dataset are given in Fig. 6 and Fig. 7, respectively. Mass regions are encircled and shown in the original mammograms. It can be seen from the Fig. 6 and Fig. 7 that proposed algorithm can segment various types of benign and malignant masses from mammograms with different breast tissue density.
Segmentation accuracy is quantified in terms of Jaccard similarity co-efficient [7] which is expressed as:
Here, P is ground truth of mass region and Q is segmented mass region obtained by the proposed algorithm. Jaccard value 0 indicates no common area between segmented region and ground truth region and value 1 indicates maximum overlap. Jaccard values of randomly chosen sample images of MIAS and DDSM datasets are presented in Fig. 8(a) and 8(b), respectively. Average value of Jaccard coefficients on selected set of sample images of DDSM and MIAS is obtained as 0.90 and 0.88, which is considerably good. Also, CPU time taken by FFCM and conventional FCM for randomly chosen sample mammograms of MIAS and DDSM is presented in Table 5. It can be seen that the FFCM is computationally efficient as compared to FCM algorithm.
Performance of the classifier is measured in terms of sensitivity, specificity, accuracy, and area under curve (AUC). Sensitivity measures the proportion of actual mass which are correctly identified as mass. Specificity measures the proportion of actual non-mass which is correctly identified as non-mass. These are expressed as follows:
here, T P , T N , F P and F N represent the true positive, true negative, false positive and false negative, respectively. T P represents the number of mass samples that are correctly classified as mass. T N denotes the number of non-mass samples that are correctly classified as non-mass. F P represents the number of non-mass samples that are incorrectly classified as mass. F N denotes the number of mass samples that are classified as non-mass. ROC curve is a plot in 2-dimensional space between true-positive rate (TPR) and false-positive rate (FPR). AUC is measured for the better understanding of ROC. Performance of SVM classifier with MLP, linear, polynomial and RBF kernel function on MIAS and DDSM dataset for 10-fold cross-validation is presented in Table 6. Highest sensitivity, specificity, accuracy, and AUC are observed with RBF kernel function. Also, ROC curves obtained with different kernel functions are shown in Fig. 9.
4.2 Comparison with the existing techniques
Performance comparison of the proposed method with other existing methods is summarized in Table 7.
The algorithm proposed by Cao et al. achieved 90.7% sensitivity with 2.57 FPI. The method applied a combination of robust information clustering and adaptive thresholding for extraction of both breast region and mass region on 60 sample mammograms of MIAS. Dominguez et al. obtained 80% sensitivity with 2.3 FPI by applying statistically based enhancement of mammograms and multilevel thresholding based mass segmentation on 57 sample images of MIAS dataset. Sampaio et al. reported 80% sensitivity using SVM classifier and 10-fold cross validation. The method utilized cellular neural network and geostatic function for mass detection on 623 mammograms of DDSM dataset. Pereira et al. reported 95% sensitivity on 640 mammogram images of DDSM dataset. They applied multi-thresholding, wavelet, and genetic based mass segmentation algorithm on both CC and MLO view of mammograms. Jen and Yu reported sensitivity of 88% and 86% on 200 sample images of DDSM and 322 sample images of MIAS, respectively. Sampaio et al. reported 92.99% sensitivity in non-dense and 83.70% sensitivity in dense breast of 1727 mammogram of DDSM dataset. They utilized genetic algorithm for mass detection system along with density estimation. Phylogenetic trees and LBP is used as input feature vector for training and testing of SVM classifier. Sensitivity, specificity, and accuracy of 94.63%, 92.74%, 92.02% have been observed on MIAS dataset, whereas 91.76% sensitivity, 96.26% specificity, and 95.46% accuracy have been achieved on DDSM data set by proposed algorithm. Promising results has been achieved by the proposed algorithm as compared to the existing mass detection techniques.
4.3 Results of shape analysis of mass
Shape analysis of mass is performed using the moment invariant and Radon based features. Total 270 sample mammograms which contain different shape of mass have been taken from both MIAS and DDSM datasets for experiment. Sample mammograms include 60 round, 48 ovals, 60 lobulated, and 102 irregular shape masses. Extracted masses of different shapes and their Radon transforms are shown in Fig. 10(a-h), respectively.
Performance of the proposed feature vectors is evaluated using multi-class SVM with RBF kernel. Moment invariant and radon based features are used separately for training and testing of SVM classifier. 180 samples are used for training and 80 samples are used for testing. Total 80 testing mass images contain 18 round shape, 13 oval, 25 irregular, and 24 lobulated type masses. Performance comparison of classification is also done by combined set of both the features. Confusion matrix is shown in Table 8. Shape discrimination power of Radon based features is more effective than moment based features. Classification accuracy decreases by using combined features as compared to Radon based features; however, it increases as compared to moment based features.
5 Conclusions
This work presents an algorithm for fully automatic segmentation and classification of suspicious regions in the mammograms. Image processing operations have been applied on mammograms to clearly highlight the suspicious mass region. Multi-resolution based wavelet decomposition is used to reduce the noise and computational time. Segmentation of suspicious mass region has been performed by fast FCM clustering algorithm. Less computational time has been achieved by using the fast FCM as compared to conventional FCM algorithm. Classification between mass and normal tissues has been done using linear, RBF, polynomial and MLP kernel function. Highest sensitivity, specificity, and accuracy have been achieved by SVM with RBF kernel function. The classification of various mass shapes has also been performed using moment invariant and Radon based features. The validation results show that Radon based features give more accurate results as compared to moment-based features for discrimination of the shape of mass. In future work, classification accuracy will be enhanced by extracting new features and classification of different types of benign and malignant masses.
References
American College Radiology (1998) BI-RADS Committee. Breast imaging reporting and data system. American College of Radiology, Reston
Bellotti R, De Carlo F, Tangaro S, Gargano G, Maggipinto G, Castellano M, Massafra R, Cascio D, Fauci F, Magro R, Raso G (2006) A completely automated CAD system for mass detection in a large mammographic database. Med phy 33:3066–3075. doi:10.1118/1.2214177
Bocchi L, Nori J (2007) Shape analysis of micro-calcifications using radon transform. Med Eng Phy 29:691–698. doi:10.1016/j.medengphy.2006.07.012
Cao A, Song Q, Yang X, Wang L (2004) Breast mass segmentation based on information theory. In: Pattern Recognition, 2004. ICPR 2004. Proceedings of the 17th IEEE International Conference vol. 3, pp 758–761. Doi:10.1016/j.cviu.2007.07.004
Cascio D, Fauci F, Magro R, Raso G, Bellotti R, De Carlo F, Tangaro S, De Nunzio G, Quarta M, Forni G et al (2006) Mammogram segmentation by contour searching and mass lesions classification with neural network. IEEE Trans Nucl Sci 53:2827–2833. doi:10.1109/TNS.2006.878003
Che FN, Fairhust MC, Wells CP, Hanson M (1996) Evaluation of a two-stage model for detection of abnormalities in digital mammograms. In: IEEE Colloquium Digest Mammography, pp 13–1. Doi: 10.1049/ic:19960496
Cheetham AH, Hazel JE (1969) Binary (presence-absence) similarity coefficients. J Paleontol 43(5):1130–1136
Christoyianni I, Dermatas E, Kokkinakis G (2000) Fast detection of masses in computer-aided mammography. Sig Proc Mag 17:54–64. doi:10.1109/79.814646
Constantinidis AS, Fairhust MC, Deravi F, Hanson M, Wells CP, Chapman-Jones C (1999) Evaluating classification strategies for detection of circumscribed masses in digital mammograms. In: International Conference on Image Processing and its Application. pp 435–439. Doi:10.1049/cp:19990359
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297
Deans SR (2007) The Radon transform and some of its applications. Courier Corporation, North Chelmsford
Do Nascimento MZ, Martins AS, Neves LA, Ramos RP, Flores EL, Carrijo GA (2013) Classification of masses in mammographic image using wavelet domain features and polynomial classifier. Expert Sys Appl 40:6213–6221. doi:10.1016/j.eswa.2013.04.036
Dominguez AR, Nandi AK (2009) Toward breast cancer diagnosis based on automated segmentation of masses in mammograms. Pattern Recog 42:1138–1148. doi:10.1016/j.patcog.2008.08.006
Ganesan K, Acharya UR, Chua CK, Min LC, Abraham KT, Ng KH (2013) Computer-aided breast cancer detection using mammograms: a review. IEEE Rev Biomed Eng 6:77–98. doi:10.1109/ICoCS.2014.7060995
Gonzalez RC, Woods RE (2008) Digital Image Processing, 3rd edn. Pearson Prentice Hall, Upper Saddle River
Haralick RM, Shanmugam K, Dinstein IH (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 6:610–621. doi:10.1109/TSMC.1973.4309314
Hu MK (1962) Visual pattern recognition by moment invariants. IRE Trans Inf Theory 8:179–187
Hu K, Gao X, Li F (2011) Detection of suspicious lesions by adaptive thresholding based on multi-resolution analysis in mammograms. IEEE Trans Instrum Meas 60:462–472. doi:10.1109/TIM.2010.2051060
Huo Z, Giger ML, Vyborny CJ, Bick U, Lu P, Wolverton DE, Schmidt RA (1995) Analysis of spiculation in the computerized classification of mammographic masses. Med Phys 22:1569–1579. doi:10.1118/1.597626
Jen CC, Yu SS (2015) Automatic detection of abnormal mammograms in mammographic images. Expert Syst Appl 42:3048–3055. doi:10.1016/j.eswa.2014.11.061
Jolliffe I (2002) Principal component analysis. John Wiley & Sons, Ltd., New York
Kom G, Tiedeu A, Kom M (2007) Automated detection of masses in mammograms by local adaptive thresholding. Comput Biol Med 37:37–48. doi:10.1016/j.compbiomed.2005.12.004
Kupinski MA, Giger ML (1998) Automated seeded lesion segmentation on digital mammograms. IEEE Trans Med Imag 17:510–517. doi:10.1109/42.730396
Kurt B, Nabiyev VV, Turhan K (2014) A novel automatic suspicious mass regions identification using Havrda & Charvat entropy and Otsu's N thresholding. Comput Methods Prog Biomed 114:349–360. doi:10.1016/j.cmpb.2014.02.014
Llado X, Oliver A, Freixenet J, Mart R, Mart J (2009) A textural approach for mass false positive reduction in mammography. Comput Med Imaging Graph 33:415–422. doi:10.1016/j.compmedimag.2009.03.007
Materka A, Strzelecki M et al (1998) Texture analysis methods–a review. Technical University of Lodz, Institute of Electronics, COST B11 Report, Brussels, pp 9–11
Muramatsu C, Hara T, Endo T, Fujita H (2016) Breast mass classification on mammograms using radial local ternary patterns. Comput Biol Med 72:43–53. doi:10.1016/j.compbiomed.2016.03.007
Ng SL, Bischof WF (1992) Automated detection and classification of breast tumors. Comput Biomed Res 25:218–237. doi:10.1016/0010-4809(92)90040-H
Nguyen TM, Rangayyan RM (2006) Shape analysis of breast masses in mammograms via the fractal dimension. In: Engineering in Medicine and Biology Society, 2005. IEEE-EMBS 2005. 27th Annual International Conference pp 3210–3213. doi:10.1109/IEMBS.2005.1617159
Nunes AP, Silva AC, de Paiva AC (2010) Detection of masses in mammographic images using geometry, Simpson’s diversity index and SVM. Int J Signal Imaging Syst Eng 3:43–51. doi:10.1504/IJSISE.2010.034631
Ojala T, Pietikainen M, Maenpaa T (2002) Multi-resolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24:971–987. doi:10.1109/TPAMI.2002.1017623
Oliveira Martins L, Silva AC, De Paiva AC, Gattass M (2009) Detection of breast masses in mammogram images using growing neural gas algorithm and Ripley’s K function. J Signal Process Syst 55:77–90. doi:10.1007/s11265-008-0209-3
Oliver A, Freixenet J, Martí J, Pérez E, Pont J, Denton ERE, Zwiggelaar R (2010) A review of automatic mass detection and segmentation in mammographic images. Med Image Anal 14:87–110. doi:10.1016/j.media.2009.12.005
Pal NR, Bezdek JC (1995) On cluster validity for the fuzzy c-means model. IEEE Trans Fuzzy Syst 3:370–379. doi:10.1109/91.413225
Pereira DC, Ramos RP, Do Nascimento MZ (2014) Segmentation and detection of breast cancer in mammograms combining wavelet analysis and genetic algorithm. Comput Methods Prog Biomed 114:88–10. doi:10.1016/j.cmpb.2014.01.014
Petrick N, Chan HP, Sahiner B, Wei D (1996a) An adaptive density-weighted contrast enhancement filter for mammographic breast mass detection. IEEE Trans Med Imaging 15:59–67. doi:10.1109/42.481441
Petrick N, Chan HP, Wei D, Sahiner B, Helvie MA, Adler DD (1996b) Automated detection of breast masses on mammograms using adaptive contrast enhancement and texture classification. Med Phys 23:1685–1696. doi:10.1118/1.597756
Rose C, Turi D, Williams A, Wolstencroft K, Taylor C (2006) Web services for the ddsm and digital mammography research. In: Digital Mammography pp 376–383
Sampaio WB et al (2011) Detection of masses in mammogram images using CNN, geostatistic functions and SVM. Comput Biol Med 41:653–664. doi:10.1016/j.compbiomed.2005.12.004
de Sampaio WB, Silva AC, de Paiva AC, Gattass M (2015) Detection of masses in mammograms with adaption to breast density using genetic algorithm, phylogenetic trees, LBP and SVM. Expert Syst Appl 42:8911–8928. doi:10.1016/j.eswa.2015.07.046
Sharma S, Khanna P (2015) Computer-aided diagnosis of malignant mammograms using Zernike moments and SVM. J Digit Imaging 28:77–90. doi:10.1007/s10278-014-9719-7
Suckling J, Parker J, Dance D, Astley S, Hutt I, Boggis C, Ricketts I, Stamatakis E, Cerneaz N, Kok S, Taylor P (1994) The mammographic image analysis society digital mammogram database. Excerpta Med Int Congr Ser 1069:375–378
Surendiran B, Vadivel A (2012) Mammogram mass classification using various geometric shape and margin features for early detection of breast cancer. Int J Med Eng Inf 4:36–54. doi:10.1504/IJMEI.2012.045302
Tai SC, Chen ZS, Tsai WT (2014) An automatic mass detection system in mammograms based on complex texture features. IEEE J Biomed Health Inform 18:618–627. doi:10.1109/JBHI.2013.2279097
Tang X (1998) Texture information in run length matrices. IEEE Trans Image Process 7:1602–1609. doi:10.1109/83.725367
Wang D, Shi L, Heng PA (2009) Automatic detection of breast cancers in mammograms using structured support vector machines. Neurocomputing 72:3296–3302. doi:10.1016/j.neucom.2009.02.015
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kashyap, K.L., Bajpai, M.K. & Khanna, P. An efficient algorithm for mass detection and shape analysis of different masses present in digital mammograms. Multimed Tools Appl 77, 9249–9269 (2018). https://doi.org/10.1007/s11042-017-4751-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-4751-5