Abstract
Breast cancer (BC) is one of the most common malignant tumors in women and still poses a significant threat to women worldwide. Recurrence of BC in situ, metastasis to distant organs, and resistance to chemotherapy are all attached to high mortality in patients with BC. Non-coding RNA (ncRNA) of the type known as “circRNA” links together from one end to another to create a covalently closed, single-stranded circular molecule. With characteristics including plurality, evolutionary conservation, stability, and particularity, they are extensively prevalent in various species and a range of human cells. CircRNAs are new and significant contributors to several kinds of disorders, including cardiovascular disease, multiple organ inflammatory responses and malignancies. Recent studies have shown that circRNAs play crucial roles in the occurrence of breast cancer by interacting with miRNAs to regulate gene expression at the transcriptional or post-transcriptional levels. CircRNAs offer the potential to be therapeutic targets for breast cancer treatment as well as prospective biomarkers for early diagnosis and prognosis of BC. Here, we are about to present an overview of the functions of circRNAs in the proliferation, invasion, migration, and resistance to medicines of breast cancer cells and serve as a promising resource for future investigations on the pathogenesis and therapeutic strategies.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common cancer in the world and the leading cause of cancer deaths in women. Despite the progress achieved in surgery, chemotherapy, radiation, endocrine therapy, targeted therapy, immunotherapy and other treatments, the incidence of BC has been on the rise globally over the past decade. The most prevalent cancer among women in the world today is breast cancer, overtaking lung cancer [1]. Based on the expression of estrogen receptors (ER), progestin receptors (PR), KI67, and the human epidermal growth factor receptors (EGFR2/HER2), BC is categorized into four types: luminal A, luminal B, ErbB2 overexpression, and triple-negative breast cancer (TNBC) [2]. Increased mortality in BC patients is associated with tumor recurrence and metastasis to distant organs such as the bone, lungs, brain, and liver [3, 4].
The occurrence of BC is influenced by external elements, familial genetic predisposition, estrogen replacement therapy and long-term high-dose radiation exposure. Mutations in genetic disorders such as P53, BRCA1 and BRCA2 mutations, as well as people with a history of ovarian or breast cancer, are more likely to develop BC [5]. Today, it is believed that epigenetic modifications and genetic alterations play significant roles in the pathogenesis of BC. DNA methylation and histone modification are examples of epigenetic modifications [6]. Additionally, a number of ncRNAs, including long non-coding RNA (lncRNA), microRNA (miRNA) [7], and circular RNA (circRNA) [8] were found to influence the onset and progression of BC.
CircRNA is a kind of covalently closed RNA that was first discovered by Sanger et al. in a plant-infected virus in 1976 [9]. However, due to the inadequate technical conditions at that time, this circRNA was once dismissed as a product of exon splicing errors. CircRNA is an endogenous short ncRNA with a closed-loop shape that originates primarily from gene exons [10]. Compared with linear mRNA, circRNA lacks an uncovered 5′ end cap and 3′-polyadenylate terminal structure, resulting in its less susceptible to nucleic acid exonuclease. Many species and types of human cells contain circRNAs, which exhibit plurality, evolutionary conservation, stability, and particularity. CircRNAs were found to be involved in multiple biological processes by acting as competitive endogenous RNAs (ceRNAs) to counteract the effects of miRNAs [11]. In recent years, the role of circRNA in regulating transcription or chelating proteins has been gradually recognized and concerned. CircRNAs also play essential roles in the pathogenesis and expansion of breast cancer. For example, circUBR1 was up-regulated in BC, which can trigger BC cell proliferation and metastasis [12]. Silencing it delayed BC tumor growth in vivo, boosted apoptosis, and reduced BC cell proliferation and metastasis in vitro. Circ_0003645's expression was considerably amplified in both breast cancer cell lines and tissues [13]. Elimination of circ_0003645 hindered the growth of breast cancer cells, leading to their programmed cell death. Circ_0003645 had a positive effect on HMGB1 and facilitated the growth of breast cancer cells by attaching miR-139-3p.
Early detection and prompt treatment can significantly reduce breast cancer mortality rates. However, early-stage breast cancer often does not present obvious symptoms and indications. Hence, women tend to neglect the clinical examination of breast cancer, resulting in breast cancer still being diagnosed at a later stage [14]. As a result, early detection of breast cancer is essential for effective therapy [15]. Circular RNAs have been identified to be differentially expressed in the initial stages of breast cancer, which can act as sponges for miRNAs to affect mRNA expression, contributing to the breast cancer development [16]. Therefore, it is hypothesized that circRNA may serve as a potential diagnostic marker and novel therapeutic approach for early-stage breast cancer.
In the following, we will focus on introducing the main functions of circRNAs and their roles in breast cancer cell proliferation, invasion, metastasis, and drug resistance. In addition, the potential of circRNAs as diagnostic and prognostic markers for breast cancer will also be highlighted.
Introduction of CircRNA
Biogenesis of circRNA
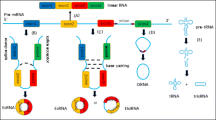
Initially, it was believed that circRNA was generated through the incorrect splicing of exons [17]. With the development of bioinformatics and the maturity of RNA sequencing technology, an increasing number of circRNAs have been discovered in various biological cells [18]. Non-coding RNA of a particular kind called circRNA is commonly expressed in eukaryotic cells. However, unlike the structure of ordinary long non-coding RNA, the circRNA molecule exhibits a covalent bond between its 3′ and 5′ end, resulting in the formation of a covalently closed loop. The absence of a 3′ and 5′ terminus renders it impervious to be digested by RNAase, thereby ensuring its stability in vivo [19]. CircRNAs are spliced in reverse from their mRNA precursors (pre-mRNA). CircRNAs are expressed at low levels in cells [20], but their expression levels appear time and tissue specificity. CircRNAs can be categorized into three subclasses: exon circRNAs (EcRNAs), intron circRNAs (ciRNAs), and exon–intron circRNAs (EIciRNAs), based on their various locations and formation mechanisms [21, 22]. The lasso-driven cyclization model of exon jumping and the intron pairing-driven cyclization model are two hypotheses currently available to explain how exon circular RNAs are generated [23].
As is illustrated in Fig. 1, the lasso-driven cyclization model of exon-skipping begins with the classical splicing of upstream and downstream exons via 3′ and 5′ covalent binding, forming an RNA lasso with exon and intron. Subsequently, the intron breaks down and the exon splices back into exon circRNA [24]. The pairing of complementary sequences on introns outside of the upstream and downstream exons in the intron pairing-driven cyclization model brings the splicing sites near to each other, thereby facilitating the formation of reverse splicing (Fig. 1) [25]. Inverted repeat Alu pairs (IRAlus) were found in the introns upstream and downstream of the ring exon. Its pairing makes the splicing sites outside the upstream and downstream exon to be in a closer position, prompting the formation of reverse splicing. The formation of circular RNA could be facilitated by IRAlus pairing or other complementary sequences [26]. Exon–intron RNA (EIciRNA) is produced by a direct cyclization splicing process after lasso generation without intron degradation [27]. Intron circRNA (ciRNA) is created when introns are partially degraded and then cyclized after lasso formation [28, 29].
In the biogenesis of circRNA, Exonic circRNAs (EcRNAs) are created by back-splicing, which can involve either one or several exons. The lasso-driven cyclization model of exon jumping, and the intron pairing-driven cyclization model, are the other two methods for creating ecRNAs. It is the predominant circRNAs form. Exon–intron circRNAs (EIciRNAs) are primarily found in the nucleus and preserve their intronic sequences between the circularized exons. Intronic lariat precursors escape from the debranching stage of conventional linear splicing to become intronic circRNAs (ciRNAs), which are prevalent in the nucleus
Further investigation has revealed that circRNAs have significant impacts on a range of diseases, including the emergence and progression of tumors [30]. CircRNAs have a wide range of biological functions, most commonly as a miRNA sponge that regulates gene expression to mediate the occurrence and development of cancer [31]. Some circRNAs contain miRNA binding regions, which may function as ceRNAs to block miRNAs, thus boosting the expression of the target gene [32]. The circRNA/miRNA/mRNA regulator axis, which resulted from the adsorption of miRNAs by circRNAs, liberated the target mRNA from functional inhibition [33]. Hansen et al. discovered that CIRS-7 was a special sponge for miR-7 that inhibited the production of multiple oncogenes controlled by miR-7 [34]. In recent years, the role of circRNA and related proteins in controlling protein expression in cancer progression has also been extensively investigated [35]. CircRNAs may adhere to various RNA-binding proteins (RBPs) to perform various functions, including suppressing protein activity, promoting the creation of protein complexes, and enabling cooperation among various proteins [36]. For example, circRNA MBL contains the conserved protein binding sites of muscle blindness (MBL), and the expression levels of MBL strongly influenced circMbl biosynthesis [37]. CircRNA can also regulate gene transcription and influence physiological and pathological processes [38]. CircITCH regulated the expression of ITCH and impeded breast cancer progression by adsorbing miR-214 and miR17 like a sponge [39]. Investigations of circRNA at a translational level uncovered the concealed proteomes and their therapeutic implications for human health [40]. The interaction between circSCRIB and the pre-mRNA of SCRIB hindered the splicing and translation of SCRIB, thus facilitating the advancement of breast cancer [41]. In the last few years, the impact of circRNA on breast cancer has become increasingly well-documented and researched [30].
The main function of circRNA
The ceRNA notion was first proposed by Salmena et al. [32]. They considered that a single miRNA may control various target genes, while the same target gene may be controlled by several different miRNAs. Because of their competitive connection and regulation by the same miRNA, these RNAs are known as ceRNAs. CircRNAs are competitive endogenous RNAs that control a variety of procedures in biology, including the expression of human genes, the growth and spread of cancer, and many other biological processes (Fig. 2A) [42, 43]. For instance, circDDX21 slowed the progression of triple-negative breast cancer (TNBC) by adsorbing miR-1264 as a sponge to regulate the expression of QKI [44]. Circ_0039960 specifically targeted miR-1178 to up-regulate PRMT7 expression, which helped BC cell grow [45]. Numerous disorders have been demonstrated to be influenced by the circRNA/miRNA/mRNA axis [46, 47].
A CircRNA is a miRNA sponge that indirectly controls mRNA expression and a number of biological processes, including the emergence and advancement of breast cancer. B By binding circACTN4 to FUBP1, the interaction between FUBP1 and FIR is hindered, leading to the regulation of breast cancer progression. C CircIPO11 binds TOP1 to form the CirciPO11-Top1 complex and recruits it to the GLI promoter to trigger its transcription. D circSEMA4B containing IRES can translate the producing protein SEMA4B-211aa in the open reading frame (ORF)
Additionally, circRNA can serve as an RBP sponge. A number of biological functions depend on RNA–protein complexes (RPCs), which are created when circRNAs bind to RBPs [48]. RBPs were responsible for the generation of circRNAs and the regulation of gene expression in transcriptional and post-transcriptional level [49]. For example, RBPs are involved in RNA alternative splicing, which had an impact on the transcription of linear parent genes and controlled the production of affiliated proteins, which were crucial for the development of tumors [50, 51]. CircACTN4 derived from exons 2 to 7 of ACTN4 could interact with far upstream element binding protein 1 (FUBP1) to inhibit the connection between FUBP1 and FIR. Consequently, MYC transcription was triggered to facilitate the emergence and advancement of breast cancer (Fig. 2B) [52]. Circ-Foxo3 was able to attach to several proteins and cooperate with CDK2 and p21 to block the cell cycle and prevent the transition from the G1 stage to the S stage [53]. CircFoxo3 was also found to be able to promote cardiac senescence through binding to as well as inhibiting anti-aging-related proteins (ID1 and E2F1) and anti-stress-related proteins (HIF1a and FAK) in the cytoplasm [54].
CircRNA can also control gene transcription through a variety of methods. CircRNA can manipulate parental genes through the ceRNA mechanism. As an illustration, circGFRA1 might function as a miR-34a sponge to control the production of GFRA1 [55], the parental mRNA, leading to the stimulation of BC cell proliferation and apoptosis blockage. CircIPO11, derived from the IPO11 gene transcript and composed of exons 4 and 5 of the IPO11 gene, recruited TOP1 to the GLI1 (GLI family zinc finger 1) promoter, triggering its transcription and thereby activating the hedgehog signal (Fig. 2C) [56].
For a long time, circRNA has been studied as a non-coding RNA. As a result, its capacity to be translated into proteins has been disregarded. Up until 2017, Legnini et al.'s discovery that circZNF609 could be translated to protein in mouse muscle cells in both a splicing-dependent and a cap-independent way suggested that other eukaryotic circRNAs may encode protein as well [57]. The SEMA4B-211aa protein was translated from an internal ribosome entry site (IRES) sequence of circSEMA4B in a 5′-cap-independent manner. The binding of SEMA4B-211aa to p85 hindered the synthesis of PIP3 and the phosphorylation of AKT, thereby impeding the advancement of breast cancer (Fig. 2D) [58]. By controlling the homeostasis of C-Myc protein, the protein FBXW7 produced by circFBXW7 impeded the onset and progression of glioblastoma [59].
Roles of circRNA in breast cancer
As a new class of epigenetic regulator, circRNA is receiving more and more attention in the process of BC tumor growth, metastasis, chemotherapy resistance, etc. CircRNAs associated with breast cancer mentioned in this paper are listed in Table 1. Because circRNAs have a large number of miRNA binding sites, they can adsorb miRNAs like sponges and prevent them from attaching to target genes, thus affecting tumor formation.
CircRNA is involved in breast cancer development and cell proliferation
Some circRNAs feature miRNA binding sites and can function as ceRNAs to suppress the production of miRNA and increase the production of the target genes [32]. As a result, circRNA can operate as a miRNA sponge to control gene expression, thereby influencing the emergence and advancement of cancer. For instance, P21, a mitosis regulator, could attach to the CDK1/cyclinB1 and prevent CDK1 from activating, hence preventing the growth and proliferation of G2/M breast cancer cells [60]. The low expression of circDDX17 in breast cancer cell lines and tissues [61] was linked to poor long-term survival of breast cancer patients. CircDDX17 controlled cyclization factors (CDK1 and P21) by sponging miR-605, which in turn promoted apoptosis and reduced cell proliferation. Circ_103809 prevented breast cancer cell lines from reproducing. By disrupting the EMT signal cascade, low expression of circRNA_103809 could impede the G2/M phase and reduce the growth and spread of BC cells [62]. The amplification of circ_0000526 drastically reduced miR-492 production and increased the production of suppressor of cytokine signaling 2 (SOCS2) through the sponge effect, thereby preventing the proliferation and metastasis of breast cancer cells and encouraging cell death [63]. Circ_001569 expression was significantly up-regulated in BC tissues and cell lines [64]. The elevated expression of circ_001569 was associated with lymph node metastases, increased clinical stage, and decreased overall lifespan. It promoted BC progression through PI3K-AKT pathway. CircUBR1 was also up-regulated in BC [12]. Blocking circUBR1 expression prevented BC tumor growth in vivo, induced apoptosis in vitro, and reduced BC cell proliferation and metastasis. The dual-luciferase reporter gene assay confirmed that circUBR1, potentially acting as a miR-1299 substrate, stimulated the synthesis of its target Cyclin D1 (CCND1) and enhanced the proliferation and metastasis of BC cells. A high-throughput microarray of circRNAs and qRT-PCR research revealed that circGFRA1, derived from the GFRA1 gene, was significantly increased in breast cancer [55], while increased expression of circGFRA1 was associated with a decreased overall survival. CircGFRA1 up-regulated the expression of GFRA1 by adsorbing miR-34a to promote breast cancer cell proliferation and inhibit apoptosis. Silencing circGFRA1 inhibited proliferation and promoted apoptosis by releasing more miR-34a to down-regulate GFRA1 expression in BC. In BC tissues and cell lines, circPTK2 expression was increased [65]. By sponging miR-136 to modulate NFBI and AKT/PI3K pathway, it could dramatically promote the growth, spread, and invasion of TNBC cells. Elevated levels of circ_0005273 in breast cancer cells up-regulated the expression of YAPl (yes-associated protein1) through the adsorption of miR-200a-3p, and then inactivated the Hippo-YAP1 signaling route, which contributes to breast cancer cell multiplication and migration [66]. Besides, the reduction of circ_0005273 inhibited BC cell expansion, migration, cell cycle, and tumor formation in vivo.
The Warburg effect, also known as aerobic glycolysis, is a unique pattern of cell metabolism in cancer cells that shows an escalating rate of glucose uptake and lactic acid fermentation in an aerobic environment. Unlike normal cells, which undergo both oxidative phosphorylation and glycolysis, cancer cells tend to undergo glycolysis in both an anoxic and aerobic microenvironment. CircRNF20 was elevated in BC tissues and cells and served as a sponge for miR-487a, which targeted hypoxia-inducible factor-1 (HIF-1) in BC cells. HIF-1 was a transcription factor targeting the promoter of hexokinase II (HK2) to speed up transcription and promote glycolysis in BC cells. Consequently, circRNF20 could stimulate the proliferation of BC cells by means of Warburg effect (aerobic glycolysis) [67]. In contrast to nearby tissues, Li et al. discovered that circ_0000514 was increased in BC tissues [42]. They found that circ_0000514 could increase the expression of CXCL10 by sponging miR-296-5p to promote BC cell invasion and proliferation. Targeting miR-223-3p, circZFR enhanced the production level of FABP7 in breast cancer, boosting the growth, migration, invasion, and EMT of BC cells while blocking apoptosis [68]. Inhibiting circZFR significantly impeded the growth of tumor cells.
CircRNA is involved in breast cancer invasion and metastasis
One of the major causes of death in breast cancer is metastasis. Breast cancer cell metastasis relies on a mechanism known as epithelial–mesenchymal transformation (EMT), and it was reported that circRNA participated in this process [69]. CircNR3C2 was significantly reduced in BC and was inversely correlated with distant metastasis and mortality in invasive breast cancer [70]. By serving as miR-513a-3p's sponge, circNR3C2 increased the levels of E3 ubiquitin-ligase HRD1. This hindered the expansion, migration, invasion, and EMT of breast cancer cells by generalizing Vimentin and inducing its degradation through the proteasome. Through sponging miR-548o, circ_0047604 directly targeted DACH1 in breast cancer cells, thus preventing the growth and spread of tumor cells [71].
miR-148a-3p and miR-152-3p could be adsorbed by circANKS1B, boosting the production of the transcription element USF1, which then raised the transcription of TGF-1, triggering the TGF-1/Smad signal and encouraging EMT [72]. Circ_0089153 was highly expressed in BC tissues. It increased the expression of E2F6 by serving as the miR-2467-3p sponge to stimulate BC cell growth, migration, invasion, and EMT [73]. Circ_0001955 was also found to be up-regulated in breast cancer cells. It increased the generation of GLUT1 by enhancing miR-1299 and supported angiogenesis, migration, invasion, survival, and glycolytic metabolism in BC cells [74].
Elevated circCD44 expression in BC cells was negatively correlated with patient prognosis [75]. As a competitive endogenous RNA, circCD44 inhibited miR-502-5p-mediated KRAS degradation by directly attaching to miR-502-5p. Additionally, it could also bind to IGF2BP2 in TNBCs to maintain the stability of Myc mRNA, thus promoting the breast cancer cell’s spread, invasion and tumorigenesis. CircHIPK3 was highly expressed in BC cytoplasm, which specifically targeted miR-326 [76]. Down-regulation of circHIPK3 dramatically reduced BC cell proliferation, migration, invasion, and EMT. Endogenous expression of miR-326 partly counteracted the impacts of circHIPK3 on apoptosis and EMT-related proteins.
By targeting miR-326, circ_0000511 controlled the expression of TAZ in BC cells, thus promoting the expansion, migration, and invasion of BC, while inhibiting BC cell death [77]. Circ_0000518 could adsorb endogenous miR-1225-3p and block its biological activity. MiR-1225-3p was able to increase the expression of SOX4 mRNA, thereby promoting the growth, infiltration, and migration of BC cells [78].
CircRNA is involved in chemotherapy resistance
CircRNAs were found to be expressed in chemotherapy-resistant breast cancer, suggesting that circRNAs may be involved in promoting or reversing chemotherapy resistance in BC cells. Medication resistance-associated circRNAs are promising diagnostic and therapeutic indicators that may improve the clinical management of BC by boosting chemotherapy sensitivity, predicting the efficacy of chemotherapeutic agents in resistant populations, and so on. The most commonly reported drugs associated with circRNAs in recent chemotherapy for BC patients are adriamycin (ADM), tamoxifen (TAM), and paclitaxel (PTX).
BC cells that exhibited resistance to ADM displayed a robust expression of circ_0001667 [79]. The target of circ_0001667, miR-4458, had decreased levels in ADM-resistant carcinoma tissues and cells. Knockdown of circ_0001667 reduced the production of NCOA3 by releasing miR-4458, resulting in a significant decrease in the growth, migration, invasion, and ADM resistance of MCF-7/ADM and MDA-MB-231−/− ADM cells. Circ_0092276 up-regulated ATG7 expression by adsorbing miR-384, thereby enhancing the autophagy of BC cells, augmenting the resistance of MCF-7 and MDA-MB-468 cells to adriamycin (ADM), and preventing apoptosis [80]. In patients with TNBC, elevated circUBE2D2 expression was strongly correlated with progressed TNM staging, lymph node metastases, and worsened prognosis [81]. CircUBE2D2 could protect CDCA3 by working as a miR-512-3p sponge to accelerate the development of BC and adriamycin resistance. The chemo tolerance to ADM in BC cells was decreased by circUBE2D2 knockdown, which also inhibited cell growth, migration, and invasion.
In TAM-resistant BC cells, the expression of circ_0025202 was dramatically reduced [82]. Circ_0025202 could function as a sponge for miR-182-5p, hindering cell proliferation, colony formation, and migration, while also enhancing cell apoptosis and susceptibility to tamoxifen. It could also further stimulate FOXO3a, the direct target gene of miR-182-5p. Meanwhile, circ_0025202 could also target miR-197-3p [83]. Overexpression of miR-197-3p enhanced TAM resistance and promoted the proliferation of BC cells. Circ_0025202 targeted miR-197-3p, thereby enhancing HIPK3 expression, and diminishing TAM resistance and tumorigenesis in BC cells. Both in TAM-resistant breast cancer cells and their exosomes, circUBE2D2 was significantly up-regulated [84]. By adsorbing miR-200a-3p, exosome-mediated circUBE2D2 induced tamoxifen resistance in breast cancer cells.
Circ_0006528 was highly expressed in PTX-resistant breast cancer tissues and cells, which directly targeted miR-1299 and increased CDK8 expression by sponging miR-1299 to promote the proliferation, migration and invasion of PTX-resistant breast cancer cells [85]. Silencing circHIPK3 led to paclitaxel-resistant BC cells more susceptible to treatment through the alteration of HK2 via miR-1286 [86]. In PTX-resistant BC tissues and cells, circHIPK3 expression was in a high level. CircHIPK3 specifically targeted miR-1286 to increase the expression of HK2, which therefore aided in the formation of BC tumors and decreased the susceptibility of BC cells to PTX. LeT-7A-5P/DUSP7 axis was the mechanism through which circABCB10 increased PTX resistance in breast cancer [87]. CircABCB10 knockdown increased PTX susceptibility and death while preventing invasion and autophagy in PTX-resistant BC cells. CircRNF111 levels were also elevated in paclitaxel-resistant BC tissues and cells [88]. CircRNF111 boosted the production of E2F3 by directly aiming at miR-140-5p to induce PTX resistance in breast cancer. Silencing circRNF111BC resulted in a significant decrease in PTX resistance, cell survival, invasion, colony formation, and glycolysis in BC cells.
The miR-153-3p/ANLN axis was controlled by circMMP11 to influence lapatinib resistance in breast cancer cells [89]. CircMMP11 increased the production of ANLN by functioning as a miR-153-3p sponge, causing BC cells more resistance to lapatinib. Knockdown of circMMP11 showed an increased sensitivity to lapatinib, which inhibited the survival, motility and invasion of breast cancer cells, as well as induced their apoptosis. In monastrol-resistant cell lines, circ_0007874 expression levels were down-regulated. Circ_0007874 controlled the TRAF4/Eg5 axis by acting on the Eg5 protein and preventing TRAF4 from interacting with the Eg5 gene [90], thereby reducing BC cell activity and enhancing monastrol-induced cytotoxicity.
Potential of circRNAs as diagnostic and prognostic markers
Prognostic assessment plays a crucial role in prolonging the survival rate of cancer patients. Multiple studies have suggested that circRNAs might be involved in a variety of breast cancer diseases. Circular RNAs have thus drawn further consideration as potential prognostic indicators for breast cancer [91]. CircRNAs are very common and extremely durable molecules that express themselves in particular ways depending on the stage of cell, tissue, and development. CircRNAs are well conserved across species, and resistant to RNaseR action [14]. Therefore, circRNAs have the potential to be utilized as cancer biomarkers because of their distinctive metabolic characteristics.
CircRNAs up-regulated in BC and their diagnostic and therapeutic values in BC
Breast tumors exhibit a differential expression of circRNAs. Some circRNAs have been found to be highly expressed in BC and be capable of promoting the development of BC, suggesting that they may one day be served as prognostic indicators and therapeutic targets for BC. For instance, enhanced production of circ_0103552 in BC tissue samples is associated with poor prognosis in BC patients [92]. The results of loss-of-function study on MCF7 cells and gain-of-function experiments on MDA-MB-231 cells revealed that the survival probability of MCF7 cell line was considerably reduced after silencing circ_0103552. Circ_0103552 greatly increased cell viability and metastasis of MDA-MB-231 cells, while decreased their apoptosis by sponging miR-1236. Silencing circUBR1 in BC hindered the proliferation and metastasis of BC cells, triggered apoptosis in vitro, and impeded the expansion of BC cancer in vivo [12]. By controlling TAZ expression, circ_0000511 promoted the expansion, migration, and invasion of BC cells, while inhibiting BC cell autophagy [77]. In BC patients, circCDYL expression levels in serum and tissues were markedly up-regulated, and this up-regulation was closely correlated with medical tumor staging, metastasis, and patient survival. Through adsorbing miR-1275, circCDYL controlled the production of ATG7 and ULK1, and then affected BC cell autophagy and proliferation [93]. BC patients with high circ_0069094 expression appeared a poor prognosis. Circ_0069094, as a suppressor of miR-59, increased the expression of HK2 to promote glycolysis. The increased rate of glycolysis altered the tumor microenvironment and enhanced the aggressiveness of cancer cells [94]. CircYY1 was expressed at an increased level in BC tissues and cells, and individuals with increased circYY1 expression had a poor prognosis. By adsorbing miR769-3p, circYY1 increased the production of the oncogene YY1 to support tumorigenesis and glycolysis in BC cells [95]. Therefore, up-regulation of all circRNAs mentioned heretofore, including circUBR1, circ_0000511, circCDYL, circ_0069094, and circYY1 can be used as prognostic indicators and therapeutic targets for BC.
Increased tumor size, lymph node spread, advanced TNM staging, aggressiveness of tumors, and poor prognosis were all substantially correlated with elevated levels of circUBAP2 [96]. CircUBAP2 could increase the production of the carcinogen MTA1 by absorbing miRNA-661, promoting BC cell migration and expansion. The expression of circSEPT9 was higher in BC tissues than that in normal ones. This finding was positively correlated with a severe disease stage and a poor prognosis [97]. Knockdown of circSEPT9 had a profound impact on the spread, migration, and invasion of BC cells, causing BC cell death and autophagy, and preventing tumor growth and spread in vivo. E2F1 and EIF4A3 promoted BC malignancy and progression via the circSEPT9/miR-637/LIF axis.
In BC tissues and cell lines, circGNB1 was substantially expressed, and its expression was strongly linked with tumor volume size and TNM phase [98]. To facilitate the development of BC cells, circGNB1 elevated the production of the oncogene protein IGF1R by targeting miR-141-5p. Furthermore, a strong correlation was observed between circIFI30 and poor prognoses in BC tissues and cell lines. It might function as an absorbent for miR-520b-3p to increase the level of CD44 and hasten the EMT of BC cells and the emergence of an aggressive phenotype [99]. KIF4A has been shown to be a promising indicator of prognosis and treatment targets for cancer. CircKIF4A was highly expressed in BC and played a regulatory role in controlling BC progression by controlling KIF4A production through adsorbing miR-375 [100]. Therefore, circUBAP2, circSEPT9, circGNB1, circIFI30, and circKIF4A are all excellent candidates for targeted therapies and diagnostic and prognostic biomarkers for BC patients.
CircRNAs down-regulated in BC and their values in the treatment of BC
Some circRNAs are down-regulated in BC and prevent breast cancer from growing and progressing, suggesting that these circRNAs could be new BC therapeutic agents. For instance, BC patients with low circDDX17 expression usually had poor long-term survival [61]. By sponging miR-605, it altered the cell cyclization factors (CDK1 and P21) to reduce BC cell proliferation and increase apoptosis. By increasing the production of ubiquitin-specific protease 4 (USP4) through the absorption of miR-553 [101], circBMPR2 prevented the evolution of BC and TAM resistance. BC patients with decreased DACH1 expression had a poor survival rate [71]. By functioning as the sponge for miR-548o, circ_0047604 specifically targeted DACH1 in BC, thus preventing cancer cell proliferation and migration. Therefore, the down-regulation of circDDX17, circBMPR2, and circ_0047604 can be employed as possible prognostic indicators for BC while they could also be investigated as new treatment medicines for BC.
CircNR3C2 was considerably decreased in BC and showed an inverse linkage with the spread and death of invasive breast cancer [70]. By sponging miR-513A-3p, circNR3C2 increased HRD1 production, which in turn prevented migration, invasion and EMT progression of breast cancer cells. In the meantime, it could generalize Vimentin in breast cancer and cause its degradation through the proteasome, thereby impeding the progression of BC. The expression of circ_0006220 was significantly reduced in BC [102]. It was a miR-197-5p absorbent and effectively controlled CDH19 expression, thereby inhibiting the development of BC development. Consequently, circPTK2, circNR3C2, and circ_0006220 are considered to be promising biomarkers for the diagnosis and therapy of BC.
Although numerous circRNAs possess potential for BC diagnosis and treatment, how to utilize them clinically remains unclear, and still needs further research.
Summary and prospects
Because of their distinctive characteristics, circRNAs have garnered a lot of interest and have recently emerged as a new research hotspot. CircRNAs are important regulators of many physiological and pathological processes. They are uniquely expressed in tissues, and are expressed in different ways in both tumor and non-tumor tissues. Due to their abundance in bloodstream fluid, saliva, and exosomes, circRNAs can be used as potential diagnostic or predictive biomarkers for diseases, particularly in the emergence, progression, and prognosis of malignant tumors [103]. CircRNAs are now new options for the early identification of BC and development of prognostic indicators, owing to their aberrant expression in BC and high specificity and sensitivity in detection.
Despite the progress made in the field of circRNA, there are still numerous issues that require further investigation. Our current knowledge of circRNA is still relatively shallow compared to coding RNA, miRNAs, and lncRNAs. The biological function of most circRNAs in physiological and pathological processes and how to apply them clinically still need further research. Additionally, there is still a lack of knowledge regarding the regulatory processes and functions of BC circRNA-miRNA-mRNA regulatory system. Considering that there are multiple subtypes of breast cancer, each with different clinical treatments and disease prognosis, exploring the distinctively expressed circRNAs of each subtype and targeting them will undoubtedly become a hotspot of research in the diagnosis, treatment and prognosis of breast cancer. Target investigation and early mechanism validation are the main areas of concern in current research. Therefore, it will be important to focus on other mechanisms in-depth, combine laboratory and clinical research, and endeavor to translate experimental findings into clinical application and practice in addition to the exploration and preliminary mechanism verification of circRNA in the future.
Data availability
Not applicable.
Code availability
Not applicable.
Consent for publication
The authors all consent for publication.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ (2011) Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol Off J Eur Soc Med Oncol 22(8):1736–1747
Liang Y, Zhang H, Song X, Yang Q (2020) Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol 60:14–27
Liu H, Li X, Li H, Feng L, Sun G, Sun G et al (2022) Potential molecular mechanisms and clinical progress in liver metastasis of breast cancer. Biomed Pharmacother 149:112824
Akram M, Iqbal M, Daniyal M, Khan AU (2017) Awareness and current knowledge of breast cancer. Biol Res 50(1):33
Wu J, Cheng J, Zhang F, Luo X, Zhang Z, Chen S (2020) Estrogen receptor α is involved in the regulation of ITGA8 methylation in estrogen receptor-positive breast cancer. Ann Transl Med 8(16):993
Venkatesh J, Wasson MD, Brown JM, Fernando W, Marcato P (2021) LncRNA-miRNA axes in breast cancer: novel points of interaction for strategic attack. Cancer Lett 509:81–88
Xu J, Wu KJ, Jia QJ, Ding XF (2020) Roles of miRNA and lncRNA in triple-negative breast cancer. J Zhejiang Univ Sci B 21(9):673–689
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73(11):3852–3856
Xie R, Zhang Y, Zhang J, Li J, Zhou X (2020) The role of circular RNAs in immune-related diseases. Front Immunol 11:545
Han TS, Hur K, Cho HS, Ban HS (2020) Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers 12(9):2622
Zhang L, Sun D, Zhang J, Tian Y (2021) Circ-UBR1 facilitates proliferation, metastasis, and inhibits apoptosis in breast cancer by regulating the miR-1299/CCND1 axis. Life Sci 266:118829
Zhang J, Ke S, Zheng W, Zhu Z, Wu Y (2020) Hsa_circ_0003645 promotes breast cancer progression by regulating miR-139-3p/HMGB1 axis. OncoTargets Ther 13:10361–10372
Milosevic M, Jankovic D, Milenkovic A, Stojanov D (2018) Early diagnosis and detection of breast cancer. Technol Health Care Off J Eur Soc Eng Med 26(4):729–759
DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A et al (2019) Breast cancer statistics, 2019. CA Cancer J Clin 69(6):438–451
Rao A, Arvinden VR, Ramasamy D, Patel K, Meenakumari B, Ramanathan P et al (2021) Identification of novel dysregulated circular RNAs in early-stage breast cancer. J Cell Mol Med 25(8):3912–3921
Cocquerelle C, Mascrez B, Hétuin D, Bailleul B (1993) Mis-splicing yields circular RNA molecules. FASEB J Off Publ Fed Am Soc Exp Biol 7(1):155–160
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777
Harland R, Misher L (1988) Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development (Camb Engl) 102(4):837–852
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15(7):409
Shang Q, Yang Z, Jia R, Ge S (2019) The novel roles of circRNAs in human cancer. Mol Cancer 18(1):6
Chen L, Shan G (2021) CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 505:49–57
Chen J, Li Y, Zheng Q, Bao C, He J, Chen B et al (2017) Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 388:208–219
Barrett SP, Wang PL, Salzman J (2015) Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 4:e07540
Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28(20):2233–2247
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–147
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al (2015) Exon–intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264
Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y et al (2015) Metazoan tRNA introns generate stable circular RNAs in vivo. RNA (New York NY) 21(9):1554–1565
Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG (2019) Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47(12):6452–6465
Zhou B, Mo Z, Lai G, Chen X, Li R, Wu R et al (2023) Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF-. J Exp Clin Cancer Res 42(1):48
Fu B, Liu W, Zhu C, Li P, Wang L, Pan L et al (2021) Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. Int J Biol Sci 17(12):3104–3117
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358
Yan L, Zheng M, Wang H (2019) Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res 11:1033–1041
Hansen TB, Kjems J, Damgaard CK (2013) Circular RNA and miR-7 in cancer. Cancer Res 73(18):5609–5612
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH (2020) Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 19(1):172
Tran AM, Chalbatani GM, Berland L, De Los Santos CM, Raj P, Jalali SA et al (2020) A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular RNAs. Front Cell Dev Biol 8:50
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66
Sun Z, Chen C, Su Y, Wang W, Yang S, Zhou Q et al (2019) Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms. J Cancer 10(13):2885–2891
Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q et al (2019) Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. Neoplasma 66(2):232–239
Sinha T, Panigrahi C, Das D, Chandra PA (2022) Circular RNA translation, a path to hidden proteome. Wiley Interdiscip Rev RNA 13(1):e1685
Ma J, Du WW, Zeng K, Wu N, Fang L, Lyu J et al (2021) An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther J Am Soc Gene Ther 29(9):2754–2768
Li L, Feng G, Chen T, Zhang L (2022) Circ_0000514 promotes breast cancer progression by regulating the miR-296-5p/CXCL10 axis. J Biochem 170(6):753–761
Liang Y, Song X, Li Y, Su P, Han D, Ma T et al (2019) circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene 38(42):6850–6866
Huang R, Yang Z, Liu Q, Liu B, Ding X, Wang Z (2022) CircRNA DDX21 acts as a prognostic factor and sponge of miR-1264/QKI axis to weaken the progression of triple-negative breast cancer. Clin Transl Med 12(5):e768
Ma W, Sun X, Zhang S, Chen Z, Yu J (2022) Circ_0039960 regulates growth and Warburg effect of breast cancer cells via modulating miR-1178/PRMT7 axis. Mol Cell Probes 64:101829
Zhou J, Ge Y, Hu Y, Rong D, Fu K, Wang H et al (2018) Circular RNAs as novel rising stars with huge potentials in development and disease. Cancer Biomark A 22(4):597–610
Wu Z, Liu P, Zhang G (2022) Identification of circRNA-miRNA-immune-related mRNA regulatory network in gastric cancer. Front Oncol 12:816884
Zheng S, Zhang X, Odame E, Xu X, Chen Y, Ye J et al (2021) CircRNA-protein interactions in muscle development and diseases. Int J Mol Sci 22(6):3262
Lyu D, Huang S (2017) The emerging role and clinical implication of human exonic circular RNA. RNA Biol 14(8):1000–1006
Zhang H, Shen Y, Li Z, Ruan Y, Li T, Xiao B et al (2020) The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal 34(1):e23049
Wang Z, Lei X, Wu FX (2019) Identifying cancer-specific circRNA-RBP binding sites based on deep learning. Molecules (Basel Switz) 24(22):4035
Wang X, Xing L, Yang R, Chen H, Wang M, Jiang R et al (2021) The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. Mol Cancer 20(1):91
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44(6):2846–2858
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z et al (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38(18):1402–1412
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W et al (2017) circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res 36(1):145
Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N et al (2021) Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer 20(1):132
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O et al (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66(1):22-37.e9
Wang X, Jian W, Luo Q, Fang L (2022) CircSEMA4B inhibits the progression of breast cancer by encoding a novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell Death Dis 13(9):794
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F et al (2018) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 110(3):304–315
He J, Chen Y, Cai L, Li Z, Guo X (2018) UBAP2L silencing inhibits cell proliferation and G2/M phase transition in breast cancer. Breast Cancer (Tokyo Jpn) 25(2):224–232
Peng HH, Wen YG (2020) CircDDX17 acts as a competing endogenous RNA for miR-605 in breast cancer progression. Eur Rev Med Pharmacol Sci 24(12):6794–6801
Qiu X, Wang Q, Song H, Shao D, Xue J (2020) circ_103809 promotes breast cancer progression by regulating the PI3K/AKT signaling pathway. Oncol Lett 19(6):3725–3730
Wang WB, Ren P, Ren FH, Huang M, Cheng X (2021) Circ_0000526 blocks the progression of breast cancer by sponging miR-492. Cancer Biother Radiopharm 36(6):467–476
Xu JH, Wang Y, Xu D (2019) Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomark A 25(2):193–201
Wang M, Chen D, Zhang H, Luo C (2022) Circular RNA circPTK2 modulates migration and invasion via miR-136/NFIB signaling on triple-negative breast cancer cells in vitro. Inflamm Res Off J Eur Histamine Res Soc 71(4):409–421
Wang X, Ji C, Hu J, Deng X, Zheng W, Yu Y et al (2021) Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1-hippo signaling pathway. J Exp Clin Cancer Res 40(1):29
Cao L, Wang M, Dong Y, Xu B, Chen J, Ding Y et al (2020) Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis 11(2):145
Tian X, Yang H, Fang Q, Quan H, Lu H, Wang X (2022) Circ_ZFR affects FABP7 expression to regulate breast cancer progression by acting as a sponge for miR-223-3p. Thoracic Cancer 13(9):1369–1380
Haynes B, Sarma A, Nangia-Makker P, Shekhar MP (2017) Breast cancer complexity: implications of intratumoral heterogeneity in clinical management. Cancer Metastasis Rev 36(3):547–555
Fan Y, Wang J, Jin W, Sun Y, Xu Y, Wang Y et al (2021) CircNR3C2 promotes HRD1-mediated tumor-suppressive effect via sponging miR-513a-3p in triple-negative breast cancer. Mol Cancer 20(1):25
Zhao B, Zhou R, Ji C, Liu D, Wu T, Xu H et al (2022) The function of circRNA-0047604 in regulating the tumor suppressor gene DACH1 in breast cancer. BioMed Res Int 2022:6589651
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M et al (2018) The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer 17(1):160
Gao SL, Fan Y, Liu XD, Liu W, Zhao M (2022) circ_0089153 exacerbates breast cancer cells proliferation and metastasis via sponging miR-2467-3p/E2F6. Environ Toxicol 37(6):1458–1471
Cheng H, Kuang S, Tan L, Sun S (2022) Circ_0001955 plays a carcinogenic role in breast cancer via positively regulating GLUT1 via decoying miR-1299. Thoracic Cancer 13(7):913–924
Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B et al (2021) CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer 20(1):138
Qi L, Sun B, Yang B, Lu S (2021) circHIPK3 (hsa_circ_0000284) promotes proliferation, migration and invasion of breast cancer cells via miR-326. OncoTargets Ther 14:3671–3685
Wu D, Jia H, Zhang Z, Li S (2021) Circ_0000511 accelerates the proliferation, migration and invasion, and restrains the apoptosis of breast cancer cells through the miR-326/TAZ axis. Int J Oncol 58(4):1
Shujuan K, Zhongxin L, Jingfang M, Zhili C, Wei W, Liu Q et al (2022) Circular RNA circ_0000518 promotes breast cancer progression through the microRNA-1225-3p/SRY-box transcription factor 4 pathway. Bioengineered 13(2):2611–2622
Cui Y, Fan J, Shi W, Zhou Z (2022) Circ_0001667 knockdown blocks cancer progression and attenuates adriamycin resistance by depleting NCOA3 via releasing miR-4458 in breast cancer. Drug Dev Res 83(1):75–87
Wang Q, Liang D, Shen P, Yu Y, Yan Y, You W (2021) Hsa_circ_0092276 promotes doxorubicin resistance in breast cancer cells by regulating autophagy via miR-348/ATG7 axis. Transl Oncol 14(8):101045
Dou D, Ren X, Han M, Xu X, Ge X, Gu Y et al (2020) CircUBE2D2 (hsa_circ_0005728) promotes cell proliferation, metastasis and chemoresistance in triple-negative breast cancer by regulating miR-512-3p/CDCA3 axis. Cancer Cell Int 20:454
Sang Y, Chen B, Song X, Li Y, Liang Y, Han D et al (2019) circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther J Am Soc Gene Ther 27(9):1638–1652
Li H, Li Q, He S (2021) Hsa_circ_0025202 suppresses cell tumorigenesis and tamoxifen resistance via miR-197-3p/HIPK3 axis in breast cancer. World J Surg Oncol 19(1):39
Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W et al (2020) Exosomes mediated transfer of circ_UBE2D2 enhances the resistance of breast cancer to tamoxifen by binding to miR-200a-3p. Med Sci Monit Int Med J Exp Clin Res 26:e922253
Liu G, Zhang Z, Song Q, Guo Y, Bao P, Shui H (2020) Circ_0006528 contributes to paclitaxel resistance of breast cancer cells by regulating miR-1299/CDK8 axis. OncoTargets Ther 13:9497–9511
Ni J, Xi X, Xiao S, Xiao X (2021) Silencing of circHIPK3 sensitizes paclitaxel-resistant breast cancer cells to chemotherapy by regulating HK2 through targeting miR-1286. Cancer Manag Res 13:5573–5585
Yang W, Gong P, Yang Y, Yang C, Yang B, Ren L (2020) Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through Let-7a-5p/DUSP7 axis. Cancer Manag Res 12:2327–2337
Zang H, Li Y, Zhang X, Huang G (2020) Circ-RNF111 contributes to paclitaxel resistance in breast cancer by elevating E2F3 expression via miR-140-5p. Thoracic Cancer 11(7):1891–1903
Wu X, Ren Y, Yao R, Zhou L, Fan R (2021) Circular RNA circ-MMP11 contributes to lapatinib resistance of breast cancer cells by regulating the miR-153-3p/ANLN axis. Front Oncol 11:639961
Liu Y, Dong Y, Zhao L, Su L, Luo J (2018) Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol 53(4):1752–1762
Deng G, Sui G (2013) Noncoding RNA in oncogenesis: a new era of identifying key players. Int J Mol Sci 14(9):18319–18349
Yang L, Song C, Chen Y, Jing G, Sun J (2019) Circular RNA circ_0103552 forecasts dismal prognosis and promotes breast cancer cell proliferation and invasion by sponging miR-1236. J Cell Biochem 120(9):15553–15560
Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W et al (2020) Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer 19(1):65
Xing Z, Wang X, Liu J, Zhang M, Feng K, Wang X (2021) Hsa_circ_0069094 accelerates cell malignancy and glycolysis through regulating the miR-591/HK2 axis in breast cancer. Cell Signal 79:109878
Zhang X, Li J, Feng Q (2021) CircRNA circYY1 (hsa_circ_0101187) modulates cell glycolysis and malignancy through regulating YY1 expression by sponging miR-769-3p in breast cancer. Cancer Manag Res 13:1145–1158
Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y et al (2018) Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun 505(4):996–1002
Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R et al (2020) The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer 19(1):73
Liu P, Zou Y, Li X, Yang A, Ye F, Zhang J et al (2020) circGNB1 facilitates triple-negative breast cancer progression by regulating miR-141-5p-IGF1R axis. Front Genet 11:193
Xing L, Yang R, Wang X, Zheng X, Yang X, Zhang L et al (2020) The circRNA circIFI30 promotes progression of triple-negative breast cancer and correlates with prognosis. Aging 12(11):10983–11003
Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G et al (2019) circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer 18(1):23
Liang Y, Song X, Li Y, Ma T, Su P, Guo R et al (2019) Targeting the circBMPR2/miR-553/USP4 axis as a potent therapeutic approach for breast cancer. Mol Ther Nucleic Acids 17:347–361
Shi Y, Han T, Liu C (2021) CircRNA hsa_circ_0006220 acts as a tumor suppressor gene by regulating miR-197-5p/CDH19 in triple-negative breast cancer. Ann Transl Med 9(15):1236
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J et al (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25(8):981–984
Acknowledgements
We would like to thank everyone in our study group for their assistance. We appreciate Biorender for contributing the expertly created figure elements to the article.
Funding
This research was funded by Grants from the Natural Science Foundation of Jiangsu Province, Grant Number BK20191429, and the Professional Research Foundation for Advanced Talents of Jiangsu University, Grant Number 11JDG063.
Author information
Authors and Affiliations
Contributions
Giving idea: XS; Reference preparation: CC, HZ, and YJ; Original draft preparation: DG; Review and editing: ML and JW; Supervision: XS. The manuscript's published form was approved by all authors after they had read it.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Informed consent
The authors all consent to participate.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, D., Cui, C., Jiao, Y. et al. Circular RNA and its potential diagnostic and therapeutic values in breast cancer. Mol Biol Rep 51, 258 (2024). https://doi.org/10.1007/s11033-023-09172-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-09172-z