Abstract
Background
Breast cancer (BRCA) is the most common and leading cause of cancer-related death in women. MicroRNAs (miRNAs) are short non-coding RNA fragments that play a role in regulating gene expression including the cancer-related pathways. Although dysregulation of miR-223 has been demonstrated in recent studies to have prognostic value in various cancers, its diagnostic and prognostic role in BRCA remains unknown.
Methods
The expression and the prognostic value of miR-223 were evaluated using the TCGA data and verified by qRT-PCR. Subsequently, potential oncogenic targets of miR-223 were identified by using three different miRNA target prediction tools and the GEPIA database. In addition to these databases, protein-protein interaction network, molecular functions, prognostic value, and the expression level of miR-223 targets were included by using several other bioinformatics tools and databases; such as, UALCAN, GeneMANIA and Metascape.
Results
The bioinformatic results demonstrated that miR-223 downregulated in BRCA and associated with poor prognosis of patients. In vitro experiments validated that miR-223 significantly downregulated in BRCA cells, MCF-7, SK-BR3, MDA-MB-231 and HCC1500, compared to normal breast cell line hTERT-HME1. Furthermore, ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes were identified as the potential oncogenic target genes of miR-223 based on their expression and prognosis in BRCA. Additionally, protein-protein interaction network of these target genes was mainly enriched in dynein intermediate chain binding, cell division, regulation of cell cycle process, and positive regulation of cellular component biogenesis.
Conclusions
The results suggests that miR-223 and its targets, ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3, might be reliable potential prognostic biomarkers in BRCA patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BRCA) is the most common malignancy and a leading cause of cancer related deaths among the women due to late diagnosis and insufficient therapeutic means because of heterogeneous nature of breast cancer [1]. The BRCA cells has a high potential of invasion and distant metastasis, which reduces the success of the surgical method in patients diagnosed at advanced stages [2, 3]. Therefore, it is necessary to elucidate the mechanisms underlying BRCA tumorigenesis to identify novel prognostic and diagnostic biomarkers.
MicroRNAs (miRNAs) are short non-coding RNA sequences with about 22 nucleotides in length and they are functional endogenous molecules, which can regulate the expression levels of target genes by binding to 3’ untranslated region of RNA sequences [4, 5]. As with other non-coding RNA family members [6,7,8,9], miRNAs orchestrate many biological processes such as cell cycle progression, immune response, proliferation, apoptosis, and cell differentiation [10, 11]. On the other hand, the bulk of evidence uncovered that dysregulation of miRNAs has been associated with pathogenesis of important diseases including neurological disorders, metabolic diseases, autoimmune diseases, and cancer [12]. miRNAs may serve as molecular markers for cancer prognosis and diagnosis, and that dysregulation of miRNAs responsible for the carcinogenesis of many cancer types [13]. One of these miRNAs, miR-223, is dysregulated in many cancers and it has key roles on cancer hallmarks including metastasis [14], differentiation [15], proliferation [16], and drug sensitivity [17] via regulation of the network of target genes. In a recent study, Citron et al., found that miR-223 was downregulated in ductal carcinoma in situ (DCIS) also they identified the downregulation of miR-223 as an early step in the initiation of luminal BRCA [18]. Another report by Sun et al., demonstrated that miR-223 promotes TRAIL-induced apoptosis in triple-negative breast cancer (TNBC) [19]. Although several direct target genes of miR-223 were reported until now, the majority of these targets still awaits to be characterized. Additionally, miRNAs could regulate several targets expression at same time [20]. Therefore, it is needed to identify potential target network and molecular functions of miR-223 in BRCA.
In this present study, we evaluated target genes network, functional enrichments, prognostic value, and expression level of miR-223 in BRCA through an integrated approach. First, we assessed expression level of miR-223 in BRCA using TCGA database and validated these expression levels in breast cancer and normal healthy breast cell lines by doing qRT-PCR assay. Further, we used UALCAN database to evaluate relationship between miR-223 expression and clinical pathological parameters such as individual cancer stages, nodal metastasis status, subclasses, and patient’s age. Additionally, we identified potential oncogenic target genes of miR-223 and their interaction network. Subsequently, we revealed their expression level, prognostic value and functional enrichments by using several bioinformatics databases and tools. Taken together, this study highlights the crucial effect of miR-223 on clinical landscape of BRCA as a tumor suppressor. Our findings indicate that miR-223 and its potential targets show promise as therapeutic, diagnostic, and prognostic biomarkers for BRCA. However, due to the study’s exclusive reliance on in silico analysis, it is crucial to recognize the necessity for additional in vitro/vivo investigations. These future studies are essential to verify the clinical translatability of miR-223 and its targets for diagnostic, prognostic, and therapeutic applications.
Materials and methods
Cell culture and cell lines
Estrogen/progesterone receptor positive (ER+/PR+) BRCA cell line MCF-7 (luminal), human epidermal growth factor receptor 2 - enriched (HER2+) BRCA cell lines SK-BR3, hormone receptor positive (HR+) BRCA cell line HCC1500, triple negative breast cancer (TNBC) cell line MDA-MB-231 and hTERT-HME1 mammary cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). MCF-7, MDA-MB-231 cell lines were cultured in Dulbecco’s modified eagle’s medium (DMEM) (Sigma, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma, USA) and 1% penicillin/streptomycin (Sigma, USA) in appropriative conditions (%95 humidified atmosphere, 5% CO2 and 37 oC). Also, SK-BR3, HCC1500 and hTERT-HME1 cell lines were cultured in RPMI 1640 medium (Sigma, USA) supplemented with %10 FBS and 1% penicillin/streptomycin in appropriative conditions. Confluent cells (%80) were re-seeded in 25 cm2 culture flasks to continue passaging, after detached by trypsinization (trypsin 0.25%) (Sigma, USA) and resuspended in culture medium.
Total RNA extraction
Total RNA extraction including miRNA was performed from MCF-7, SK-BR3, MDA-MB-231, hTERT-HME1 and HCC1500 cell lines by using the miRNeasy mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturers’ protocol. Extracted RNA samples were quantified by using NanoDrop ND-1000 (Thermo Scientific, California, USA) at 260/280 nm and stored at -80 oC until their use.
Complementary DNA synthesis
cDNA synthesis was performed with 1 ug of extracted RNA samples using miScript II RT kit (Qiagen GmbH, Hilden, Germany) according to manufacturer’s procedure. For cDNA synthesis reverse transcription PCR was conducted on a Sensoquest Labcycler using the following conditions: incubation for 60 min at 37 oC and inactivation step for 5 min at 95 oC, respectively. Samples were stored at -20 oC until their use.
Quantitative real time PCR
Expression level of the miR-223 in BRCA cells, were evaluated with qRT-PCR by using miScript SYBER Green PCR kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s procedure. qRT-PCR conditions using the kit instructions: an initial activation step 95 oC for 15 min, followed by 40 cycles including 94 oC for 15 s, 55 oC for 30 s and 70 oC for 30 s. The U6 small nuclear RNA (RNU6) (Qiagen GmbH, Hilden, Germany) was used as an endogenous control, and the relative miR-223 expression normalized to RNU6. Comparative threshold cycle method (2−ΔCt) was used to determine the relative expression of miR-223 in BRCA cells (MCF-7, SK-BR3, MDA-MB-231 and HCC1500) vs. normal breast cell line hTERT-HME1. All qRT-PCR experiments were conducted using Rotor Gene Q Thermocycler (Qiagen GmbH, Hilden, Germany) and replicated three times.
Expression and survival analysis of miR-223 and target genes
UALCAN is an online analysis and mining web tool based on TCGA database [21]. By using this database, we assessed the relative expression of miR-223 in BRCA tissue and normal healthy breast tissue. Next, we evaluated the relative expression level of miR-223 in various subgroups of BRCA, encompassing individual cancer stages, patient’s race, patient’s gender, patient’s age, nodal metastasis status, menopause status, major subclasses, and TP53 mutation status. Furthermore, expression levels of miR-223 targets, ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes, were verified by using the GEPIA2 tool (http://gepia2.cancer-pku.cn/). Additionally, we used the Kaplan-Meier plotter (http://kmplot.com/analysis/index.php) to determine prognostic value of miR-223 and target genes in BRCA.
Prediction of miR-223 targets
miRDB (miRDB (http://mirdb.org/), DIANAmT (http://diana.imis.athenainnovation.gr/DianaTools/) and TargetScan (http://www.targetscan.org) are online databases to predict potential miRNA targets. Results from these three algorithms and upregulated genes in BRCA from the GEPIA were intersected by using the Venny 2.1 online tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html) in order to detect upregulated targets of miR-223 in BRCA.
PPI network construction and functional enrichment analysis
Protein-protein interaction (PPI) of target genes of miR-223 was constructed through the GeneMANIA (www.genemania.org) database. Interactions was determined according to physical interactions, shared protein domains and common pathways, co-expressions, co-localizations, genetic interactions, and predicted interactions. Furthermore, gene ontology terms enrichment analysis was carried out using the Metascape (http://metascape.org) web tool [22] based on PPI network of miR-223 targets.
Statistics
All quantitative data were expressed as the mean ± SD and were analyzed by the GraphPad Prism Software Version 7.0 (GraphPad Software Inc., LaJolla, CA). Student’s t-test and Mann Whitney U test were applied to compare differences in gene expression between two groups depending on the data distribution (normally or non-normally). Kaplan-Meier method was used for survival analysis through log-rank tests. Less than 0.05 p-values were considered statistically significant.
Results
Expression and prognostic analysis of miR-223 in BRCA
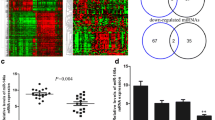
We determined the miR-223 expression levels in four breast cancer cell lines including MCF-7, SK-BR3, MDA-MB-231, and HCC1500 and mammary cell line, hTERT-HME1, by using qRT-PCR. The results showed that miR-223 expression level was significantly lower in MCF-7, SK-BR3, MDA-MB-231, and HCC1500 cell lines than hTERT-HME1 cells (Fig. 1a). The expression level of miR-223 in BRCA was screened by using UALCAN database which showed that, miR-223 was significantly downregulated in 749 tumor samples compared to 76 normal healthy samples (Fig. 1b). Additionally, the association analysis between miR-223 and overall survival rate of BRCA patients was done by using the Kaplan Meier plotter online tool. Regarding the Kaplan Meier plotter analysis, high expression level of miR-223 was significantly correlated with favorable survival rates (Fig. 1c).
miR-223 was downregulated in BRCA cell lines and in vivo samples. (A) Comparison of miR-223 expression level between BRCA and normal breast samples. (B) Comparison of miR-223 expression level between BRCA cell lines MCF-7, SK-BR3, MDA-MB-231, and HCC1500 and hTERT-HME1 normal breast cell line. (C) Kaplan-Meier analysis of the effect of the miR-223 expression level on BRCA patient overall survival (n = 1076)
Clinicopathological value of miR-223 in BRCA
The expression level of miR-223 in BRCA based on clinicopathological characteristics including, individual cancer stages, nodal metastasis status, subclasses, menopause status, TP3 mutation status, gender, age, and race was analyzed by using the UALCAN database. There was a meaningful decrease in the expression level of miR-223 in cancer stages and nodal metastasis status compared to the normal samples, but there was not a meaningful difference between nodal and cancer stages (Fig. 2a-b). Moreover, miR-223 was significantly downregulated in subclasses of BRCA including luminal and HER2 + compared to TNBC and normal healthy samples (Fig. 2c) and also its expression level was significantly lower in pre-menopause and post-menopause BRCA patients (Fig. 2d). According to TP53 mutation status analysis, miR-223 expression was decreased in TP53-non mutant samples compared the normal samples and TP53 mutant samples (Fig. 2e). On the other hand, gender-based analysis revealed that miR-223 expression was significantly downregulated in both genders, but its expression level varied in male and female patients, with male patients having lower expression than females (Fig. 2f). Furthermore, patient’s age analysis indicated that miR-223 was significantly downregulated in 21–40, 41–60 and 61–80 years of age groups (Fig. 2g). Race based analysis showed that miR-223 was significantly downregulated in Caucasian, African-American and Asian patients, but downregulation of miR-223 between the races did not show statistically significant difference (Fig. 2h).
Expression of miR-223 in various clinicopathological parameters in BRCA patients. (A) Expression of miR-223 in individual cancer stages of BRCA. (B) Expression of miR-223 in nodal metastasis status of BRCA. (C) Expression of miR-223 in different subclasses of BRCA. (D) Expression of miR-223 in menopause status of BRCA. (E) Expression of miR-223 in TP53 mutation status of BRCA. (F) Expression of miR-223 in BRCA patient’s gender. (G) Expression of miR-223 in BRCA patient’s age. (H) Expression of miR-223 in BRCA patient’s race
Upregulated target genes of miR-223 in BRCA
The common targets of miR-223 were identified by using 3 different miRNA target prediction tools (TargetScan, miRDB and DianaMT) and common targets were reduced to 9 genes by the intersect regions of Venn diagram (Fig. 3a and b). According to the outcomes of these databases, we identified 279 common target genes of miR-223 (Fig. 3a). Subsequently, we used the GEPIA database and obtained the significantly upregulated genes in BRCA. Intersection between upregulated genes and common targets of miR-223 revealed that ANLN, DYNLT1, C9orf152, LRRC59, RAB3D, SLC12A8, SLC2A1, TMEM209 and, TPM3 were the upregulated target genes of miR-223 in BRCA (Fig. 3b).
Expression and prognostic analysis of miR-223 target genes in BRCA
Identification of potential oncogenic targets of miR-223 were done by the validated expression levels of common target genes and later their prognostic value analysis in BRCA patients. According to these results ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes were the upregulated genes associated with poor prognosis of BRCA patients (Fig. 4). Hence, we continued our study by selecting these hub genes (ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3). Expression levels ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes that were obtained from GEPIA database were significantly overexpressed in BRCA tissues compared to normal breast healthy tissues (Fig. 4). Next, we evaluated clinical significance of these overexpressed targets of miR-223 in BRCA by using the Kaplan-Meier plotter web tool. Consequently, high expression levels of ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes were associated with poor prognosis of BRCA patients (Fig. 5). These results suggested that ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 might have oncogenic functions in initiation and progression of BRCA.
Functional enrichment analysis of miR-223 target genes network
We generated PPI network of miR-223 target genes via using the GeneMANIA database. According to PPI network, there were 30 proteins that were associated with ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 (Fig. 6a). Subsequently, we used these interaction network to reveal their potential enrichments in gene ontology terms. The enriched terms of miR-223 targets network in both molecular functions and biological processes were colored and visualized according to cluster ID of enriched terms (Fig. 6b). The results of gene ontology terms analysis demonstrated that ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 network was particularly enriched in dynein intermediate chain binding, cell division, regulation of cell cycle process, and positive regulation of cellular component biogenesis (Fig. 6c).
A. Protein interaction network of ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 was created based on top interacted proteins. B. Enriched terms of miR-223 targets network in both molecular functions and biological processes based on cluster ID of enriched terms. C. Functional enrichment analysis of miR-223 targets interaction network was evaluated based on cluster ID of enriched terms
Discussion
BRCA is the leading cause of cancer-related mortality and most common invasive malignancy in female population [23]. The main factor that complicates the determination of effective therapeutic strategies for the treatment of BRCA is the variability of the phenotypic characteristics of subtypes of BRCA and it is generally classified into 4 major subtypes including luminal A, luminal B, HER2 and TNBC [24]. The ER positive/luminal is the most diagnosed subtype of the BRCA, and the more aggressive subtype is called luminal B, while luminal A has a better survival potential. In addition, TP53 mutations are less frequent in luminal A than in luminal B [25]. TNBC (including ER-negative, PR-negative, and HER2-negative) is the clinically most aggressive type of breast malignancies [26]. Therefore, better understanding of clinicopathological mechanisms of BRCA is required to define more effective methods and targetable new molecules. miRNAs are important epigenetic regulators and therapeutic targets and their dysregulated expressions is associated with prognosis of many cancers including BRCA [27,28,29]. One of these miRNAs is miR-223 that is downregulated in many tumors including lung cancer [30], osteosarcoma [31], and hepatocellular carcinoma [32]. Yang et al., reported that miR-223 inhibits proliferation and migration of BRCA cell lines [33]. Further enlightening of the role of miR-223 in the clinicopathology of BRCA and identification of its targets in BRCA might contribute to the improvement of new approaches for the treatment and prognosis of BRCA.
In this study, we revealed that miR-223 was downregulated in BRCA tissues compared to normal breast tissues according to the UALCAN database analysis and verified it by qRT- PCR analysis. Consistent with our results, Citron et al., found that miR-223 was downregulated in BRCA [18]. It has been reported that low expression level of miR-223 was correlated with poor survival rates in many cancers including acute myeloid leukemia (Yu et al. 2020), osteosarcoma (Dong et al. 2016) and oral cancer (Li et al. 2020). Likewise we demonstrated that miR-223 expression was significantly associated with poor overall survival rates of BRCA patients.
The expression level of the miRNAs are important indicators for clinopathological characteristics of the BRCA patients [34, 35]. Therefore, we evaluated clinicopathological role of miR-223 in BRCA patients by using UALCAN database. According to these results, low miR-223 expression was significantly related with individual cancer stages and nodal metastasis status in BRCA samples compared to normal healthy breast samples (Fig. 2a-b). Additionally, miR-223 was downregulated in luminal, HER2 positive and TNBC subtypes of BRCA (Fig. 2c). Consistent with our results Citron et al., found that miR-223 was downregulated in luminal and HER2 positive BRCA patients [18]. In addition, Sun et al., demonstrated that miR-223 has low expression in TNBC cell lines [19]. Low expression of miR-223 was significantly correlated with pre-menopause and post-menopause status of BRCA patients compared the normal healthy samples, although, there was no significant correlation between each other of individual menopause status (Fig. 2d). Kangas et al., reported that hormone replacement treatment (HRT) causing the downregulation of miR-223-3p expression in post-menopause patients, however, they found miR-223 expression is higher in post-menopause patients then pre-menopause patients [36]. On the other hand, Olivieri et al., used only estrogen treatment to HRT caused a decrease in miR-223 expression in MCF-7 cells [37]. We combined the results of both studies [36, 37] and using the UALCAN database (Fig. 2d), we concluded that estrogen treatment in menopause may trigger BRCA via downregulation of miR-223. miR-223 expression in relation to mutant and nonmutant TP53 in BRCA based on UALCAN database bioinformatic analysis was done in which miR-223 was significantly downregulated in TP53-non mutant and mutant samples compared to normal healthy samples (Fig. 2e). Masciarelli et al., report supports partly our findings that mutant p53 suppresses transcriptional activity of miR-223 by targeting promoter region of miR-223 in SK-BR3 and MDA-MB-231 cell lines [38].
In this study, it has been found that miR-223 expression has strong relationship with BRCA patient’s gender, whereas, it is downregulated in male and female, but miR-223 had significantly lower expression in male compared to females (Fig. 2f). According to age-based analysis miR-223 significantly downregulated in 21–80 years old patients but there was no significant difference in 81–100 years old patients (Fig. 2g).
In many studies based on US populations, the majority of BRCA patients participating in the trials represent the Asian population and this reduces the success of treatment methods to be applied to patients from different populations [39]. In order to improve BRCA treatment efficacy should considerate the genomic make up of different race and ethnicity of BRCA patients. In this study, the Caucasian and the African-American patients had significantly low miR-223 levels compared to normal healthy samples and the Asian patients had the lowest miR-223 levels (Fig. 2h).
To further understanding the molecular mechanism of miR-223 in BRCA, we identified the potential oncogenic target genes of miR-223 by using computational tools and databases. Our results revealed that ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes are potential oncogenic targets of miR-223 in BRCA. Consistent with our results Magnusson et al., found that high expression of ANLN correlated with poor survival rates in BRCA [40]. Li et al. reported that SLC12A8 promotes unfavorable prognosis of BRCA [41]. Yao et al., indicated that platelet derived-TPM3 may be useful biomarker for metastatic BRCA [42]. Terp et al., found that LRRC59 expression associated with poor prognosis of BRCA. In addition, Li et al., demonstrated that the overexpression of LRRC59 correlated with poor prognosis of lung adenocarcinoma [43]. However, we couldn’t find any paper on the expression level of DYNLT1 in literature. Moreover, we found that high expression levels of ANLN, DYNLT1, LRRC59, SLC12A8 and TPM3 genes were significantly correlated with poor overall survival rates of BRCA patients.
This study build a single miRNA and multiple mRNA network that is significantly associated with poor prognosis of BRCA patients. However, there were some limitations in our study, the further research is needed to clarify physical or indirect interaction between targets of miR-223 and miR-223 level in BRCA. To clarify underlaying mechanisms of BRCA progression we need to determine these potential interactions with more solid experiments.
Conclusion
In conclusion, the findings of this study, conducted entirely in silico, strongly suggest that ANLN, DYNLT1, LRRC59, SLC12A8, and TPM3 are potential oncogenic targets of miR-223. Moreover, the downregulation of miR-223 is implicated in poor clinicopathological outcomes in BRCA. These results highlight the significance of in silico analyses in identifying potential molecular targets and unraveling the mechanistic implications of miR-223 in breast cancer. However, further in vitro and in vivo investigations are essential to validate these findings and assess the clinical relevance of miR-223 and its targets.
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article.
Code Availability
Not applicable.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33. https://doi.org/10.3322/CAAC.21708
Jin X, Mu P (2015) Targeting breast Cancer metastasis. Breast Cancer (Auckl) 9:23. https://doi.org/10.4137/BCBCR.S25460
Cole M, Parajuli S, Laske D et al (2014) Peripheral primitive neuroectodermal tumor of the dura in a 51-year-old woman following intensive treatment for breast cancer. Am J Case Rep 15:294–299. https://doi.org/10.12659/AJCR.890656
de Moor CH, Meijer H, Lissenden S (2005) Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol 16:49–58. https://doi.org/10.1016/J.SEMCDB.2004.11.007
Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, mechanism, and function. Cell 116:281–297. https://doi.org/10.1016/S0092-8674(04)00045-5
Sahin Y (2022) LncRNA H19 is a potential biomarker and correlated with immune infiltration in thyroid carcinoma. Clin Exp Med. https://doi.org/10.1007/S10238-022-00853-W
Liu H-T, Liu S, Liu L et al (2018) EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res 78:5877–5890
Liu B, Xiang W, Liu J et al (2021) The regulatory role of antisense lncRNAs in cancer. Cancer Cell International 2021 21:1 21:1–15. https://doi.org/10.1186/S12935-021-02168-4
Mathieu E-L, Belhocine M, Dao LT et al (2014) Functions of lncRNA in development and diseases. Med Sci (Paris) 30:790–796
Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH (2019) An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 234:5451–5465. https://doi.org/10.1002/JCP.27486
Kabekkodu SP, Shukla V, Varghese VK et al (2018) Clustered miRNAs and their role in biological functions and diseases. Biol Rev 93:1955–1986. https://doi.org/10.1111/BRV.12428
Ardekani AM, Naeini MM (2010) The role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol 2:161
Seven M, Karatas OF, Duz MB, Ozen M (2014) The role of miRNAs in cancer: from pathogenesis to therapeutic implications. Future Oncol 10:1027–1048. https://doi.org/10.2217/FON.13.259
Li X, Zhang Y, Zhang H et al (2011) miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 9:824–833. https://doi.org/10.1158/1541-7786.MCR-10-0529
Fazi F, Racanicchi S, Zardo G et al (2007) Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 12:457–466. https://doi.org/10.1016/J.CCR.2007.09.020
Qu H, Zhu F, Dong H et al (2020) Upregulation of CCT-3 induces breast Cancer Cell Proliferation through miR-223 competition and Wnt/β-Catenin signaling pathway activation. https://doi.org/10.3389/FONC.2020.533176. Front Oncol 10:
Streppel MM, Pai S, Campbell NR et al (2013) MicroRNA 223 is upregulated in the multistep progression of Barrett’s esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res 19:4067–4078. https://doi.org/10.1158/1078-0432.CCR-13-0601
Citron F, Segatto I, Vinciguerra GLR et al (2020) Downregulation of miR-223 expression is an early event during Mammary Transformation and confers resistance to CDK4/6 inhibitors in luminal breast Cancer. Cancer Res 80:1064–1077. https://doi.org/10.1158/0008-5472.CAN-19-1793
Sun X, Li Y, Zheng M et al (2016) MicroRNA-223 increases the sensitivity of Triple-Negative breast Cancer stem cells to TRAIL-Induced apoptosis by Targeting HAX-1. PLoS ONE 11:e0162754. https://doi.org/10.1371/JOURNAL.PONE.0162754
Catalanotto C, Cogoni C, Zardo G (2016) MicroRNA in Control of Gene expression: an overview of Nuclear Functions. Int J Mol Sci 17. https://doi.org/10.3390/IJMS17101712
Chandrashekar DS, Bashel B, Balasubramanya SAH et al (2017) UALCAN: a portal for facilitating Tumor Subgroup Gene expression and survival analyses. Neoplasia 19:649–658. https://doi.org/10.1016/J.NEO.2017.05.002
Zhou Y, Zhou B, Pache L et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications 2019 10:1 10:1–10. https://doi.org/10.1038/s41467-019-09234-6
Ferlay J, Colombet M, Soerjomataram I et al (2021) Cancer statistics for the year 2020: an overview. Int J Cancer 149:778–789. https://doi.org/10.1002/IJC.33588
Carey LA, Perou CM, Livasy CA et al (2006) Race, breast cancer subtypes, and survival in the Carolina breast Cancer Study. JAMA 295:2492–2502. https://doi.org/10.1001/JAMA.295.21.2492
Koboldt DC, Fulton RS, McLellan MD et al (2012) Comprehensive molecular portraits of human breast tumors. Nature 490:61. https://doi.org/10.1038/NATURE11412
Lehmann BD, Bauer JA, Chen X et al (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121:2750–2767. https://doi.org/10.1172/JCI45014
Chang JTH, Wang F, Chapin W, Huang RS (2016) Identification of MicroRNAs as breast Cancer prognosis markers through the Cancer Genome Atlas. PLoS ONE 11:e0168284. https://doi.org/10.1371/JOURNAL.PONE.0168284
Ding HX, Lv Z, Yuan Y, Xu Q (2018) MiRNA polymorphisms and cancer prognosis: a systematic review and meta-analysis. Front Oncol 8:596. https://doi.org/10.3389/FONC.2018.00596/BIBTEX
Altan Z, Sahin Y (2022) miR-203 suppresses pancreatic cancer cell proliferation and migration by modulating DUSP5 expression. Mol Cell Probes 66:101866. https://doi.org/10.1016/J.MCP.2022.101866
Jia CY, Li HH, Zhu XC et al (2011) MiR-223 suppresses cell proliferation by targeting IGF-1R. https://doi.org/10.1371/JOURNAL.PONE.0027008. PLoS One 6:
Ji Q, Xu X, Song Q et al (2018) Mir-223-3p inhibits human Osteosarcoma Metastasis and Progression by directly targeting CDH6. Mol Ther 26:1299–1312. https://doi.org/10.1016/J.YMTHE.2018.03.009
Karakatsanis A, Papaconstantinou I, Gazouli M et al (2013) Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog 52:297–303. https://doi.org/10.1002/MC.21864
Yang Y, Jiang Z, Ma N et al (2018) MicroRNA-223 targeting STIM1 inhibits the Biological behavior of breast Cancer. Cell Physiol Biochem 45:856–866. https://doi.org/10.1159/000487180
Lv P, Zhang Z, Hou L et al (2020) Meta-analysis of the clinicopathological significance of miRNA-145 in breast cancer. Biosci Rep 40. https://doi.org/10.1042/BSR20193974
Nadeem F, Hanif M, Ahmed A et al (2017) Clinicopathological features associated to MiRNA-195 expression in patients with breast cancer: evidence of a potential biomarker. Pak J Med Sci 33:1242. https://doi.org/10.12669/PJMS.335.13008
Kangas R, Morsiani C, Pizza G et al (2018) Menopause and adipose tissue: miR-19a-3p is sensitive to hormonal replacement. Oncotarget 9:2279. https://doi.org/10.18632/ONCOTARGET.23406
Olivieri F, Ahtiainen M, Lazzarini R et al (2014) Hormone replacement therapy enhances IGF-1 signaling in skeletal muscle by diminishing miR-182 and miR-223 expressions: a study on postmenopausal monozygotic twin pairs. Aging Cell 13:850–861. https://doi.org/10.1111/ACEL.12245
Masciarelli S, Fontemaggi G, di Agostino S et al (2013) Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 2014 33(12):1601–1608. https://doi.org/10.1038/onc.2013.106
Hirko KA, Rocque G, Reasor E et al (2022) The impact of race and ethnicity in breast cancer—disparities and implications for precision oncology. BMC Med 20. https://doi.org/10.1186/S12916-022-02260-0
Magnusson K, Gremel G, Rydén L et al (2016) ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer 16. https://doi.org/10.1186/S12885-016-2923-8
Li LW, Xia J, Cui RT, Kong B (2021) Solute carrier family 12 member 8 impacts the biological behaviors of breast carcinoma cells by activating TLR/NLR signaling pathway. Cytotechnology 73:23–34. https://doi.org/10.1007/S10616-020-00439-Y
Yao B, Qu S, Hu R et al (2019) Delivery of platelet TPM3 mRNA into breast cancer cells via microvesicles enhances metastasis. FEBS Open Bio 9:2159. https://doi.org/10.1002/2211-5463.12759
Li D, Xing Y, Tian T et al (2020) Overexpression of LRRC59 is Associated with poor prognosis and promotes Cell Proliferation and Invasion in Lung Adenocarcinoma. Onco Targets Ther 13:6453. https://doi.org/10.2147/OTT.S245336
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sahin, Y., Altan, Z., Karabulut, A. et al. The role of miR-223 in breast cancer; an integrated analysis. Mol Biol Rep 50, 10179–10188 (2023). https://doi.org/10.1007/s11033-023-08850-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08850-2