Abstract
Background
Fenugreek (Trigonella foenum-graecum L.) is an annual medicinal and spice crop belonging to the family Fabaceae. The occurrence of a yellow vein disease was recorded in fenugreek in Jodhpur (India) in 2022. The infection of begomoviruses in legume crops results in significant yield loss and major economic loss. The current study reports an association of a novel begomovirus species associated with yellow vein disease in Fenugreek.
Methods and results
In symptomatic fenugreek plants, geminivirus-like particles were visible under a transmission electron microscope. Further, nucleotide sequence analysis of the rolling circle amplified product revealed 2743 nucleotide DNA-A genome with close relatedness to French bean leaf curl virus (88.21%) and Senna leaf curl virus (87.63%). It was proposed as a new begomovirus species, Fenugreek yellow vein Rajasthan virus. The genome organization suggested the presence of a typical nonanucleotide sequence along with 7 ORFs in DNA-A. A possible recombination event took place in the coat protein (V1) region with Pedilanthus leaf curl virus and Chilli leaf curl virus as major and minor parents. The recombinant virus poses possible threats to several other legume crops. To the best of our knowledge, this is the first report of the association of FeYVRaV with fenugreek yellow vein disease from northwestern India.
Conclusions
In conclusion, the presence of a novel begomovirus species associated with yellow vein disease in fenugreek is alarming and needs further studies on its infectivity to prevent its spread to legume crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trigonella is a genus of plants from the family Fabaceae. Members of the genus grow in the Canary Islands, southern Europe, non‑tropical Africa, western and central Asia, the Indian subcontinent, and Australia. The best-known member of the genus is the herb fenugreek (Trigonella foenum-graecum L.). Fenugreek (L.) is an annual crop and an important spice used for flavoring food. It is widely cultivated and grown in several countries including India, China, Nepal, North Africa, Australia, and the USA [1]. Fenugreek seeds and leaves are used in traditional medicines, cuisines, cosmetics, and also as a dietary supplement. The medicinal crop is used for the treatment of diabetes, arthritis, hair growth issues, hypothyroidism, hyperlipidemia, and infertility [2].
The incidence of a yellow vein disease in cultivated fenugreek was recorded in a few pockets of Jodhpur (Rajasthan, India) in 2022. Infection of begomoviruses that causes vein yellowing, vein thickening, yellow leaves, mosaic, and stunted growth is common in legume crops. Begomoviruses belong to the genera Begomovirus, family Geminiviridae, and are transmitted by whitefly (Bemisia tabaci, Hemiptera: Aleyrodidae). The family Geminiviridae is classified into fourteen distinct genera and comprises 520 species worldwide [3]. Geminiviruses are small (2.5 to 5.2 kb in size), circular, non-enveloped, and single-stranded DNA (ssDNA) plant viruses. Begomovirus is the largest genus under Geminiviridae and includes viruses with either a single (monopartite genome: DNA-A) or two (bipartite genome: DNA-A and DNA-B) DNA components [4, 5]. The DNA-A virion-sense strand encodes the coat (CP) and pre-coat (precoat) proteins, whereas the complementary-sense strand encodes the replication-associated protein (Rep), transcriptional activator protein (TrAP), replication enhancer protein (REn), and C4 protein in general [4]. DNA-B encodes a nuclear shuttle protein (NSP) on the virion-sense strand and a movement protein (MP) on the complementary-sense strand. The association of a begomovirus molecule causing leaf curl disease in fenugreek (T. corniculata) in India was detected by PCR using degenerate geminivirus primers and by Southern hybridization with a probe specific to tomato leaf curl begomovirus [6]. Infection of Pedilanthus leaf curl virus (PeLCV, MH550115), Ageratum enation virus (AEV), tomato leaf curl Kerala virus (ToLCKeV), and DNA-B molecule of tomato leaf curl New Delhi virus (ToLCNDV) along with Ageratum yellow leaf curl betasatellite in fenugreek was reported earlier [7]. The present study suggests the association of a novel begomovirus species with yellow vein disease in cultivated fenugreek in northern India. The phylogenetic relatedness and recombination analyses suggest its close relatedness with French bean leaf curl virus (FbLCV) and Senna leaf curl virus (SenLCuV) and is proposed as a novel begomovirus species. Further, we also found the recombinational event in the genome. It might pose a potential threat to fenugreek and economically important crops under favorable conditions.
Materials and methods
Sample collection
Yellowing of veins, yellow leaves, and stunting symptoms were recorded in fenugreek (T. foenum-graecum, var. AFg-3) plants. The symptomatic samples were collected in January 2022 from the experimental fields of ICAR-Central Arid Zone Research Institute (CAZRI), Jodhpur (Rajasthan, India). The incidence and symptoms of the disease were recorded. A quadrat of one square meter was selected 10 times randomly in the field. The number of symptomatic plants out of the total plants in each quadrat was counted to record the percent incidence. The whole plant samples were collected in perforated plastic bags and carried to the laboratory at ICAR-Indian Agricultural Research Institute (IARI), New Delhi (India). The samples were cleaned with distilled water and stored at -80 °C for further processing.
Transmission electron microscopy (TEM)
One mm length of symptomatic leaf sample was ground in phosphate buffer (pH 6.5). A drop (3–5 µl) of the extract was adsorbed on the carbon-coated copper grid and washed with double distilled water. The excess extract was manually blotted off the grid using blotting paper. Immediately after blotting the sample, 2% uranyl acetate was added to the grid and excess fluid was drained. The prepared sample grid was examined under the Transmission Electron Microscope (TEM, JEOL-JEM-1011, Japan) at different magnifications for the presence of virus-like particles [8].
DNA extraction and rolling circle amplification (RCA)
Based on the visualization of geminate virus-like particles in TEM, total genomic DNA (gDNA) was extracted from the leaf samples using a modified CTAB method [9, 10]. The isolated gDNA purity and yield were checked in a spectrophotometer (NanoDrop™ 2000, Thermo Fisher Scientific, USA). The isolated gDNA was subjected to rolling circle amplification (RCA). A 20 µl of RCA reaction comprised 7U of φ29 DNA polymerase (Thermo Fisher Scientific), 1X random hexamer primer (Thermo Fisher Scientific), 1X reaction buffer, and 0.4U of pyrophosphatase (Thermo Fisher Scientific) [11].
Viral genome amplification by PCR and cloning
The enriched viral concatamers were used for PCR amplification of the begomovirus molecules. The degenerate universal primer pair, Begomo F (5ʹ-ACGCGTGCCGTGCTGCTGCCCCCATTGTCC-3ʹ) and Begomo R (5ʹ-ACGCGTATGGGCTGYCGAAGTTSAGAC-3ʹ) [12] was used for the amplification of DNA-A in PCR. Degenerate universal primers PBL1v2040/PCRc1 were used for amplification of DNA-B and universal primer pairs beta 01/02 and CLB36F/38R were used to amplify betasatellite in PCR [13, 14]. The PCR was performed in a final volume of 25 µl containing 1X Phusion HF Buffer, 0.2mM dNTPs (Thermo Fisher Scientific), 0.4µM of each forward and reverse primer (GCC Biotech, India), 1U of Phusion™ high-fidelity DNA polymerase (Thermo Fisher Scientific) and ~ 100 ng of template DNA. The thermal cycling was performed in a T100 Thermal Cycler (Bio-Rad, USA) with initial denaturation at 98 °C for 3 min followed by 28 cycles of denaturation at 98 °C for 45 s, annealing at 58 °C for 90 s, extension at 72 °C for 1 min and final extension at 72 °C for 10 min [11]. The amplified product was resolved on 1% agarose gel and visualized in a gel documentation system (MaestroGen Inc, Taiwan) with a 1 kb plus DNA ladder (Thermo Fisher Scientific). The amplified PCR products were purified using the GSure Gel Extraction kit (GCC Biotech) and ligated into the pJET1.2/blunt cloning vector (Thermo Fisher Scientific) using T4 DNA Ligase (Thermo Fisher Scientific) at 4 °C overnight and used further for the transformation in E. coli strain DH5-α competent cells. Randomly selected transformed colonies were allowed to grow in a liquid Luria-Bertani (LB) medium supplemented with ampicillin (50 ug/ml). The bacterial plasmid DNA was isolated using the alkaline-lysis method [15].
Sequence analysis and genome map
Three representative clones of the begomovirus component were sequenced bi-directionally at the DNA Sequencing Facility, University of Delhi South Campus, India. The obtained sequences were manually checked and assembled to construct the full genome of DNA-A. The reconstructed virus sequence was analyzed for coding regions using the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The DNA sequence was further examined using NCBI nucleotide BLAST (blastn) to find out the closest homology sequences. A pairwise sequence alignment was performed using the Multiple Sequence Comparison by Log-Expectation algorithm (MUSCLE) program in Molecular Evolutionary Genetics Analysis (MEGA-X) tool [16]. The full-length genome map of the cloned DNA-A was generated through the Artemis DNA plotter (http://www.sanger.ac.uk/Software/Artemis) analysis tool V18.1.0 [17]. All the ORFs coordinates were manually added to the tool and a schematic map was generated.
Phylogenetic tree and percent nucleotide sequence identity
Based on homology and previous reports of Begomovirus species infecting plants of Fabaceae, Malvaceae, Solanaceae, and some other families, 28 full-length DNA-A sequences of representative virus isolates were retrieved from the NCBI GenBank (Supplementary Table S1). The DNA sequences were analyzed using the MEGA-X tool with Neighbor-Joining (NJ) method with 1000 bootstrap replicates. Percent nucleotide identity was calculated and a three-color coded heat map was generated for DNA-A using the Sequence Demarcation Tool (SDT) version 1.2 [18]. A sequence of polyprotein gene (Accession no. EF445546) of Ranunculus mild mosaic virus (genus Potyvirus) was used as an outgroup to construct the phylogenetic tree.
Recombination analysis
All the representative sequences were retrieved from NCBI GenBank to analyze the genetic recombination. The sequences were aligned first with the MEGA tool. The final aligned sequences were analyzed using RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, and T-3Seq methods implemented in Recombination Detection Programme (RDP) version 4.97 for the detection of the possible recombination events [10, 19].
Results
Symptoms and incidence of yellow vein disease in fenugreek
The incidence of yellow vein disease in fenugreek was observed in January-February, 2022. The diseased plants showed strong yellow vein symptoms (Fig. 1). The fenugreek plants at the pre-flowering to flowering stage (45–51 days) showed initial symptoms on the top and middle leaves. At a later stage, the leaves started drying from the leaf margin towards the middle vein. The infected plants remained stunted and produced fewer seeds. The severe infection resulted in drying and drooping of leaves. About 54 to 68 total plants were grown in each square meter of the sampling quadrat. Out of which, up to one plant in each quadrat was recorded with the typical symptoms. The disease incidence was around 1–2%.
Virus particle structure under TEM
Extract from infected leaf samples was examined under TEM. Geminivirus-like twinned incomplete icosahedral virion particles were observed under TEM. Two isometric structures were fused to form geminate particles. The particles were about 34–36 nm in diameter (Supplementary Fig. 1a). The presence of geminate particles suggested the possible infection of begomovirus in the fenugreek samples.
Amplification of DNA-A and sequencing
The full-length virus genome was amplified by RCA-based ssDNA enrichment followed by PCR amplification. The universal primer pair for DNA-A (Begomo-F and Begomo-R) amplified a ~ 2.8 kb band specific to begomovirus DNA-A (Supplementary Fig. 1b). The results suggested the presence of begomovirus molecules in the collected sample. The PCR amplified molecules were cloned in pJET1.2/blunt-ended cloning vector and the restriction digestion with XbaI and XhoI further confirmed the size of the insert. Restriction digestion suggested the presence of a similar molecule in all the amplified samples. The sequences of three randomly selected clones showed 100% homology. A consensus sequence of the plasmid DNA revealed a 2743 nucleotide-long DNA-A molecule that was closely related to other begomovirus sequences available at NCBI. The nucleotide sequence can be retrieved from the NCBI GenBank using Accession No. ON787805. However, no DNA-B or betasatellite molecules could be detected in multiple attempts either by PCR using universal primer pairs PBL1v2040/PCRc1 (for DNA-B [13]) and beta 01/02; CLB36F/38R (for betasatellite [12]) or by restriction digestion of RCA products using KpnI (for betasatellite), and SalI and BglII (for DNA-B).
Genome organization
The full-length DNA-A exhibited seven ORFs (Fig. 2), five in the complementary-sense strand (C1, C2, C3, C4, and C5) and two in the virion-sense strand (V1, and V2). The stem-loop structure contained a typical nonanucleotide sequence (TAATATTAC) at the common region (CR). In complementary-sense strand, C1 (2594 − 1506 nucleotide), C2 (1609 − 1211 nucleotide), C3 (1470 − 1087 nucleotide), and C4 (2440 − 2138 nucleotide) encoded replication-associated protein, transcriptional activator protein, replication enhancer protein, and silencing suppressor, respectively. The presence of C5 (1106 − 600 nucleotide) was also predicted in the complementary-sense strand. In the virion-sense strand, V1 (294–1070 nucleotide) encoded coat protein, and V2 (134–478 nucleotide) encoded pre-coat protein.
Genome organization of the DNA-A component. The scale represents the nucleotide position in the genome. C1 (red encodes Rep), C2 (blue encodes TrAP), C3 (green encodes REn), C4 (violet encodes silencing suppressor), C5 (aqua), V1 (orange encodes coat-protein), and V2 (olive encodes pre-coat protein). A yellow circle represents the hairpin-like structure at the common region (CRA) of DNA-A.
Evolutionary relationship of the cloned DNA-A
The begomovirus sequence isolated from T. foenum-graecum showed 88.21% and 87.63% sequence homology to its closest relatives FbLCV (Accession no. JQ866297) and SenLCuV (Accession no. KU852742), respectively. Since the International Committee on Taxonomy of Viruses (ICTV) describes a < 91% similarity threshold for begomovirus species delimitation [20], the present isolate appeared to be a novel begomovirus species, Fenugreek yellow vein Rajasthan virus. In phylogenetic analysis, fenugreek yellow vein Rajasthan virus (FeYVRaV) showed close evolutionary relatedness with the SenLCuV (Accession no. KU852742) which were previously reported to infect Senna occidentalis (family Fabaceae) (Fig. 3). FeYVRaV clustered in the same clade with SenLCuV, FbLCV, and Corchorus yellow vein mosaic virus (CoYV) infecting Malvaceae and Fabaceae plants.
Phylogenetic relationships of the cloned DNA-A component of Fenugreek yellow vein Rajasthan virus. Begomoviruses associated with Fabaceae, Malvaceae, Solanaceae, Apiaceae, and Piperaceae plants were retrieved from NCBI GenBank, and the tree was generated using the MEGA-X tool with 1000 bootstrap replicates. For each sequence, the accession number with the isolates name, country code, place of isolation, and year of sequence submitted have been mentioned. Each of the distinct nodes is also represented in a color-coded circle. The distance scale bar represents the rate of nucleotide substitution/per site/ year. Ranunculus mild mosaic virus (Potyvirus, Accession no. EF445546) sequence encoding polyprotein was used as an outgroup
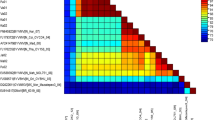
To further understand the nucleotide sequence identity, a three-color coded matrix of the full-length sequence of FeYVRaV along with other begomovirus sequences was generated using SDT (Fig. 4a). The results confirmed that the DNA-A sequence with 88.21% pairwise identity, belonged to a novel begomovirus species. In SDT analysis, FeYVRaV exhibited high nucleotide identity with PeLCV (MN566097) and FbLCV (JQ866297) infecting Fabaceae plants.
(a) Pairwise nucleotide sequence similarity matrix score of FeYVRaV. The sequence data were aligned by the MUSCLE program using the sequence demarcation tool (SDT) version 1.2. (b) Schematic representation of recombinational events in DNA-A genome of FeYVRaV. PeLCV might act as major and ChiLCV as minor parent donors in the recombination. The recombination breakpoint is marked by highlighted background. @Recombination breakpoints indicate the position in the genome where the recombination event took place. *Location of the corresponding recombination breakpoint positions in the viral genome. $R-RDP; G-GENCONV; B-BOOTSCAN; M-MAXCHI; C-CHIMERA; S-SISCAN; T-3SEQ are the methods used to identify the recombination breakpoints. #The lowest p-value detected by the underlined method has been mentioned in the previous column
Recombination analysis
The DNA-A sequence of FeYVRaV was recombinant and possible recombination events took place in the coat protein (V1) region. The recombinant analysis revealed that PeLCV (Accession no. JQ012790) and ChiLCV (Accession no. KX499526) were the major and minor parents of FeYVRaV, respectively. The p-value of the analyzed component was 6.656 × 10− 05 with the CHIMERA method in the RDP tool, suggesting the recombinant significance of the cloned DNA-A component. A pictorial representation of the predicted recombination event using RDP has been shown in Fig. 4b. The expected breakpoint position lies between 613 and 1346 in multiple sequence alignment with other begomovirus species. The detailed analyses of recombination events and phylogenetic relatedness suggested that FeYVRaV is a novel virus species and it might have been introduced from other cultivated and non-cultivated plants.
Discussion
Fabaceae is the third largest plant family in the world [21] that includes legume crops. A wide range of begomoviruses affects the production and quality of legume crops [22]. Infection of AEV, ChiLCV, Dolichos yellow mosaic virus (DoYMV), mung bean yellow mosaic virus (MYMV), mung bean yellow mosaic India virus (MYMIV), ToLCNDV, FbLCV, SenLCuV, PeLCV, and cotton leaf curl Kokhran virus (CLCuKoV) in several legume crops suggests their susceptibility to begomoviruses. Fenugreek is cultivated worldwide as a semiarid crop. Its seeds and leaves are used as herbs, spices, and vegetables. India is a major producer of fenugreek, and over 80% of India’s output is from Rajasthan. The incidence of yellow vein disease has been recorded that reduces the seed yield and makes the leaves unacceptable to consumers [10]. In the present study, an association of a novel begomovirus with yellow vein disease of fenugreek was confirmed by TEM, nucleotide sequencing, and phylogenetic analyses. Geminate particles were visible under TEM indicating possible begomovirus infection. A 2743 nucleotide sequence showed homology to DNA-A of other begomoviruses. The genome organization suggests the presence of a typical nonanucleotide sequence along with seven ORFs in DNA-A. The occurrence of a leaf curl disease of fenugreek caused by AEV was reported in India, Pakistan, and Nepal [23, 24]. Infection of PeLCV (Accession no. MH550115) in fenugreek was also reported from Pakistan. Association of ToLCKeV, DNA-B molecule of ToLCNDV, and a novel betasatellite with severe leaf curl disease of fenugreek have been reported recently [7]. However, the disease complex and its components were not fully understood. In the present study, the DNA-A amplified from the symptomatic fenugreek samples shared a maximum sequence identity of 88.21% with FbLCV DNA-A followed by SenLCuV (87.63%). The study does not overrule the coinfection of other begomovirus species and satellite molecules. The pairwise percent nucleotide identity of DNA-A is showing below the ≥ 91% threshold level for begomovirus species delimitation as per the ICTV guidelines [3]. Therefore, we proposed a new begomovirus species, Fenugreek yellow vein Rajasthan virus infecting fenugreek in India. Further studies are anticipated to understand its infectivity, satellite molecules, transmission biology, host range, and epidemiology.
Virus evolution is generally found to be linked with inter- and intra-species recombinations. Geminiviruses exhibit a complex evolutionary process and they commonly initiate genetic recombination to adapt and survive in suitable host plants [25]. Our data suggested that FeYVRaV genome is recombinant. PeLCV (JQ012790) might act as a major and ChiLCV (KX499526) as a minor parent donor for the recombinational events. The coat protein (V1) region of DNA-A has been identified as the hot spot for recombination. Phylogenetic relatedness further revealed the close association of FeYVRaV with other begomoviruses isolated from legumes such as SenLCuV, and FbLCV. FeYVRaV also demonstrated genetic relatedness with CoYV. The diverse association of FeYVRaV along with other known begomovirus species in the phylogenetic study and possible inter- or intra-species recombination events that occurred among the DNA-A molecules suggests it might be a potential threat to the cultivation of legumes and other crops [26, 27]. In-depth studies need to be undertaken to understand its host range and the likelihood of economic losses in important field crops. The diverse begomovirus species in a common host gives a huge scope for virus molecules to evolve and also allows the species to acclimatize to a new host and ecological niche [28, 29]. However, the role of insect-vector (whitefly) [30], mutation, pseudo-recombinations [31], national and international trades of non-symptomatic infected plants, and other anthropogenic activities cannot be ruled out.
References
Zandi P (2017) In: Basu SK (ed) Fenugreek (Trigonella foenum-graecum L.): an important Medicinal and aromatic crop. IntechOpen, Rijeka. Ch. 12
Bahmani M, Shirzad H, Mirhosseini M et al (2016) A review on Ethnobotanical and Therapeutic uses of Fenugreek (Trigonella foenum-graceum L). J Evid Based Complementary Altern Med 21:53–62. https://doi.org/10.1177/2156587215583405
Fiallo Olive E, Lett JM, Martin DP et al (2021) ICTV Virus Taxonomy Profile: Geminiviridae 2021. J Gen Virol 102:001696. https://doi.org/10.1099/jgv.0.001696
Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788. https://doi.org/10.1038/nrmicro3117
Devendran R, Namgial T, Reddy KK et al (2022) Insights into the multifunctional roles of geminivirus-encoded proteins in pathogenesis. Arch Virol 167:307–326. https://doi.org/10.1007/s00705-021-05338-x
Raj SK, Pandey SK, Chandra G et al (2001) First report of a geminivirus causing leaf curl disease on Trigonella corniculata evidenced by PCR using degenerate primers. EPPO Bull 31:115–117. https://doi.org/10.1111/j.1365-2338.2001.tb00980.x
Ashwathappa KV, Venkataravanappa V, Hiremath S et al (2022) Fenugreek plants showing the severe leaf curl disease are associated with tomato leaf curl Kerala virus, DNA-B molecule of tomato leaf curl New Delhi virus and a novel betasatellite. Australas Plant Dis Notes 17. https://doi.org/10.1007/s13314-022-00472-0
Kumar A, Rout BM, Choudhary S et al (2022) First Report of Cucurbit Chlorotic Yellows Virus Infecting Pumpkin in India. Plant Dis 106:null–1767. https://doi.org/10.1094/PDIS-07-21-1473-PDN
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Report 1:19–21. https://doi.org/10.1007/BF02712670
Yogindran S, Kumar M, Sahoo L et al (2021) Occurrence of cotton leaf curl Multan virus and associated betasatellites with leaf curl disease of Bhut-Jolokia chillies (Capsicum chinense Jacq.) In India. Mol Biol Rep 48:2143–2152. https://doi.org/10.1007/s11033-021-06223-1
Roy B, Chakraborty P, Ghosh A (2021) How many begomovirus copies are acquired and inoculated by its vector, whitefly (Bemisia tabaci) during feeding? PLoS ONE 16:e0258933
Akhter A, Qazi J, Saeed M, Mansoor S (2009) A severe Leaf Curl Disease on Chilies in Pakistan is Associated with multiple Begomovirus Components. Plant Dis 93:962. https://doi.org/10.1094/PDIS-93-9-0962B
Kumar R, Palicherla SR, Mandal B, Kadiri S (2018) PCR based detection of betasatellite associated with the begomoviruses using improved universal primers. Australas Plant Pathol 47:115–118. https://doi.org/10.1007/s13313-017-0537-5
Rojas R (1993) Use of degenerate primers in the polymerase chain reaction to detect Whitefly-Transmitted geminiviruses. Plant Dis 77:340–347
Kumar M, Kumar RV, Chakraborty S (2020) Association of a begomovirussatellite complex with yellow vein.pdf. Arch Virol 165:2099–2103
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Carver T, Thomson N, Bleasby A et al (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. https://doi.org/10.1093/bioinformatics/btn578
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9. https://doi.org/10.1371/journal.pone.0108277
Martin DP, Murrell B, Golden M et al (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:1–5. https://doi.org/10.1093/ve/vev003
Zerbini FM, Briddon RW, Idris A et al (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98:131–133. https://doi.org/10.1099/jgv.0.000738
Tekdal D (2021) In: Singh VP, Singh S, Tripathi DK et al (eds) 13 - plant genes for abiotic stress in legumes. Academic Press, pp 291–301
Malathi VG, Renukadevi P, Chakraborty S et al (2017) Begomoviruses and Their Satellites Occurring in India: Distribution, Diversity and Pathogenesis
Swarnalatha P, Venkataravanappa V, Lakshminarayana Reddy CN et al (2019) Molecular characterization of ageratum enation virus and beta satellite associated with leaf curl disease of fenugreek in India. Indones J Biotechnol 24:74–81. https://doi.org/10.22146/ijbiotech.49939
Tahir M, Amin I, Haider MS et al (2015) Ageratum enation virus—A begomovirus of weeds with the potential to infect crops. Viruses 7
Vinoth Kumar R, Singh AK, Singh AK et al (2015) Complexity of begomovirus and betasatellite populations associated with chilli leaf curl disease in India. J Gen Virol 96:3143–3158. https://doi.org/10.1099/jgv.0.000254
Lefeuvre P, Moriones E (2015) Recombination as a motor of host switches and virus emergence: geminiviruses as case studies. Curr Opin Virol 10:14–19. https://doi.org/10.1016/j.coviro.2014.12.005
Devendran R, Kumar M, Ghosh D et al (2021) Capsicum-infecting begomoviruses as global pathogens: host–virus interplay, pathogenesis, and management. https://doi.org/10.1016/j.tim.2021.05.007. Trends Microbiol
Seal SE, vandenBosch F, Jeger MJ (2006) Factors influencing Begomovirus Evolution and their increasing global significance: implications for sustainable control. CRC Crit Rev Plant Sci 25:23–46. https://doi.org/10.1080/07352680500365257
Chakraborty S, Kumar M (2020) Tomato Leaf Curl New Delhi Virus (Geminiviridae). Elsevier Ltd.
Ghosh A, Roy B, Nekkanti A, et al (2021) Transovarial transmission of dolichos yellow mosaic virus by its vector bemisia tabaci Asia II 1. Front Microbiol 12 https://doi.org/10.3389/fmicb.2021.755155
Garcia Arenal F, Zerbini FM (2019) Life on the Edge: Geminiviruses at the interface between crops and wild plant hosts. Annu Rev Virol 6:411–433. https://doi.org/10.1146/annurev-virology-092818-015536
Acknowledgements
We gratefully acknowledge the support received from the IARI, New Delhi, CAZRI, Jodhpur, and Department of Biotechnology, Government of India, New Delhi, India.
Funding
DBT (Grant no. BT/PR40767/AGIII/103/12777/2020).
Author information
Authors and Affiliations
Contributions
KSJ, RKK, and RKS performed the field observation and collection of the infected samples. MK and AG conceived the experiments and MK performed the experiments and generated scientific data. BK performed the TEM analysis. MK and AG analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have agreed to publish in the journal.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, M., Ghosh, A., Jadon, K.S. et al. Association of a novel begomovirus species with fenugreek yellow vein disease in India. Mol Biol Rep 50, 9203–9211 (2023). https://doi.org/10.1007/s11033-023-08806-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08806-6