Abstract
Genetic improvement in coconut relies on exploiting the vast existing diversity among coconut accessions. Robust molecular markers are a pre-requisite for efficient characterization of genetic diversity. Microsatellites or simple sequence repeats (SSRs), mined from expressed sequence tags (ESTs), constitute an important resource for analysis of genetic diversity as they are abundant, polymorphic and represent function regions of the genome. We have identified a total of 318,528 putative EST-SSRs from 130,942 unigenes utilizing a leaf transcriptome dataset of coconut. Among the EST-SSRs, dinucleotide repeats were abundant (219,912; 69.04%) followed by trinucleotide (70,722; 22.2%) and tetra-nucleotide repeats (6281; 1.9%). Among the dinucleotide repeat motifs, the dominant repeat was AG/CT (35.87%), followed by AT/AT (18.59%), while the dominant trinucleotide repeat was AAG/CTT (4.59%). One hundred and twenty EST-SSR primer pairs were designed and utilized to amplify six DNA samples of coconut accessions. Fifty primers (41.7%) produced reproducible polymorphic fragments of expected sizes, from which a total of 10 primers were selected for the diversity assessment in 186 palms of 50 coconut accessions, comprising of 25 each of tall and dwarf accessions. A total of 137 alleles were detected with an average of 13.7 alleles per SSR locus. The number of alleles observed at each locus in the data set ranged from 7 to 22. All the loci showed 100% polymorphism with respect to the samples screened. The average observed heterozygosity was 0.46. The PIC values ranged from 0.79 (CnKGDEST129 and CnKGDEST100) to 0.91 (CnKGDEST117 and CnKGDEST122) with a mean value of 0.85, indicating the capacity of the EST-SSR markers to detect high levels of polymorphism. The cluster analysis revealed that accessions were generally clustered based on their relative similarity and irrespective of their geographic origins. The present study demonstrates the usefulness of transcriptome sequencing as a rapid and cost-effective methodology for the development of molecular markers. The EST-SSR markers generated through this study constitute useful and reliable tools for assessment of genetic diversity and marker-assisted selection in coconut.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Popularly regarded as ‘Tree of Life’ or ‘Tree of Heaven’, the coconut palm (Cocos nucifera L.) is the livelihood supporter to millions of people in around 93 tropical countries, comprising mainly of Island nations, where they play an even influential role in sustaining the fragile ecosystem [1]. Although all the parts of the coconut palm are used by humankind, the chief economic part is the edible endosperm of matured fruits, used for fresh consumption as well as for processing to get several oil and milk products, and their derivatives are used either for consumption or industrial purposes. The sweet water and delicious tender kernel from immature fruits are relished for fresh consumption as well as processed to make beverages. The fresh sweet sap tapped from the unopened inflorescence serves as a nutritious drink and is also used to make a range of edible products.
The genus Cocos has only one species nucifera, with two distinct botanical groups viz., talls and dwarfs, mainly distinguished by the palm stature and cross or self-pollination, respectively, and also by several other morphological and qualitative characters [2]. Because coconut is a perennial and mainly a cross-pollinating species, most crop improvement efforts have utilized the diversity existing in natural coconut populations of different regions. Establishment of systematic coconut gene banks in a few countries since the 1920s has paved the way for development and release of several superior varieties through selection and hybridization [3]. Genetic diversity studies have particular importance in crop breeding programs in coconut, especially in the arrangement of accessions into genetic groups, for further use in breeding programs, and in recognition of redundant ecotypes or duplicates in collections [2, 4,5,6]. Genetic diversity analyses in coconut were previously undertaken utilizing various morphological and biochemical features of the germplasm collections, which include foliar traits [7], fruit component analysis [8] and banding profiles of native proteins [9]. As these techniques are prone to environmental influences [10], they have given way to molecular markers for the characterization of crop germplasm and overcome most of the limitations of morphological and biochemical markers.
In coconut, the use of molecular markers like RFLP (Restriction Fragment Length polymorphism) [4], AFLP (Amplified Fragment Length Polymorphism) [11] and SSR (Simple Sequence Repeat) [11,12,13,14, 16,17,18,19] have been reported for genetic characterization and diversity estimation [2, 15]. Among these, SSRs have steadily become the method of choice in population genetics and germplasm characterization studies, construction of linkage maps and mapping of QTLs (quantitative trait loci) in many crops by their co-dominant nature, enhanced polymorphism and reproducibility, and more coverage of the genome. Concerning coconut, SSRs have been used by many researchers in diversity studies [11,12,13,14, 16, 17]. Although widely employed, the development of SSR markers via screening of genomic libraries is time-consuming, labor-intensive and expensive [20].
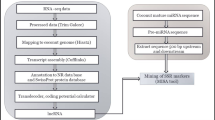
The use of ESTs (Expressed Sequence Tags) to mine for SSRs (EST-SSRs) has largely reduced the dependence on genomic SSRs. The advantages of EST-SSR techniques include hypervariability, suitability for high throughput analysis, high polymorphism and mainly, their high transferability to related species [20]. EST-SSRs have been utilized in diverse spheres, such as marker-assisted selection (MAS), for developing high yielding varieties as well as molecular mapping and quantitative trait loci (QTL) analysis [21]. EST-SSR markers also constitute functional markers as they are derived from expressed regions of the genome [22]. Transcriptome sequencing offers a cost-effective strategy in the identification of microsatellite loci from expressed regions of the genome [23]. The present work was undertaken to screen coconut leaf transcriptome data, generated previously in our laboratory [24], to identify EST-SSRs and to utilize the EST-SSR technique for diversity analysis in coconut utilizing the selected, well known conserved tall and dwarf accessions.
Materials and methods
Mining for EST-SSRs
Leaf transcriptome dataset of Chowghat Green Dwarf cultivar (NCBI SRA: SRX 436961), generated on an Illumina HiSeq 200 platform [24], was used as a sequence source for locating the SSR markers. The identification and localization of perfect as well as compound microsatellites and the frequency of a specific microsatellite type, according to the unit size or individual motifs, were done using MIcroSAtellite (MISA) tool (https://webblast.ipk-gatersleben.de/misa/).
BLASTx analysis
EST-SSR similarity searches were undertaken using the BLASTx program. The sequences were screened with maximum threshold E-value at 0.0001, identity percentage (indicating the similarity between the aligned sequences with respect to the length of the matched region) and coverage percentage (indicating the similarity between the aligned sequences with respect to the size of the query’s sequence). The sequences, which possessed major BLASTx similarity, were further analyzed.
Design of primers flanking EST-SSRs
SSR primers were designed using the flanking sequences by Primer3 software (bioinfo.ut.ee/primer3/). The major primer design parameters were melting temperature (50–60 °C), GC content (40–60%), primer size (18–30 bp) and product size (150–400 bp). Melting temperature (Tm) ranged from 57 to 60 °C, primer GC content ranged from 40 to 60%, primer size ranged from 22 to 27 bp, and product size ranged from 101 to 300 bp. Besides, the following parameters were also used for SSR prediction: di-mers, tri-mers, tetra-mers, penta-mers and hexa-mers, with a minimum of three repeats, and hepta-mers, octa-mers and nona-mers, with a minimum of two repeats. For marker design, we have selected SSR regions with length greater than or equal to 30 bp. The parameters used for primer design are given in Supplementary Table S1. Selected primers were checked for self-annealing and primer dimer formation using the OligoAnalyzer tool (https://www.idtdna.com/pages/tools/oligoanalyzer).
Plant materials and extraction of DNA
Selected conserved coconut germplasm, from the National Coconut Genebank, ICAR-CPCRI, Kasaragod, Kerala State, India and International Genebank for South East Asia and the Middle East (ICG-SAME), Kidu, Karnataka State, India, were used in the study. A total of 186 palms belonging to 50 coconut accessions (comprising of 25 tall and dwarf accessions each; 2–4 palms per accession), representing both exotic and indigenous material (Table 1), were used for diversity analysis. Genomic DNA was isolated from spindle leaves as per the protocol described in [25]. To check DNA for its purity and intactness, the genomic DNA was run in 0.8% agarose gel stained with ethidium bromide and visualized in a gel documentation system (BIO-RAD, USA).
EST-SSR analysis
From the designed EST-SSR primer pairs, 120 primers were selected, from sequences possessing major BLASTx similarities, for screening a subset of germplasm comprising three each of tall and dwarf coconut accessions [West Coast Tall, San Ramon Tall, Philippines Ordinary Tall, Chowghat Green Dwarf, Kenthali Orange Dwarf, and Andaman Yellow Dwarf] for detection of polymorphism. The three tall and three dwarf accessions, used for initial screening, were selected to represent different geographical groups. PCR reactions were conducted in volumes of 20 µl containing 35 ng genomic DNA, 0.2 µM of both forward and reverse primers (Sigma), 0.1 µl of 10 mM of each dNTPs (MBI Fermentas), 2.0 µl of 10 × buffer [10 mM Tris–HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin] and 0.5 µl of Taq DNA polymerase (3 Units/µl) (Genei Laboratories Pvt. Ltd., India). The reaction mixture was given a brief spin for thorough mixing of the cocktail components and aliquoted in 20 µl thin-walled clear PCR tubes. Initially, gradient PCR was carried out to find the optimum annealing temperatures for the designed primers. The PCR conditions were: an initial denaturation step 94 °C for 2 min, followed by 39 cycles at 94 °C for 1 min, annealing (different temperature for each primer pair) for 1 min and 72 °C for 1 min 30 s, terminating with a final extension at 72 °C for 10 min. After gradient PCR, the products were run in a 3% agarose gel and the amplicons were visualized with ethidium bromide staining using a gel documentation system (Bio-Rad, USA). The annealing temperatures, which gave a strong single band of the expected size, were selected and subsequently PCR was carried out at that temperature for the particular pair of primers. Each polymorphic primer was tested at least thrice to ensure the reproducibility of polymorphism and the banding patterns. Based on their ability to detect polymorphism, primers were selected for diversity analysis. PCR products were analyzed, using the DNA 500 kit, using a MultiNA microchip electrophoresis system (MCE-202 MultiNA, Shimadzu). Automated detection of peak sizes on electropherograms was achieved by the MultiNA software (MultiNA Control Software and Data Analysis Software MultiNA Viewer, Shimadzu).
Data analysis
The calculation of genetic diversity values and construction of the unweighted pair group method with arithmetic mean (UPGMA) dendrogram using Nei’s genetic distance [26] was carried out using the GDA software (Genetic Data Analysis) [27].
Results
Mining for EST-SSRs from coconut transcriptome data
A total of 130,942 unigenes, assembled from Illumina sequencing of the leaf of CGD cultivar (SRX 436961), were used for the mining of EST-SSRs. Using the MISA tool, a total of 318,528 potential SSRs were identified. The number of SSR containing sequences was 89,820. The number of sequences containing more than one SSR was 57,578. Di-nucleotide repeats were more (219,912), followed by tri-nucleotide (70,722) and tetra-nucleotide repeats (6281) (Table 2). From Table 2, it is clear that the di-nucleotide SSR is the dominant type (69.04%) followed by tri-nucleotide repeats (22.2%). Among the di-nucleotide repeat motifs, the dominant repeat was AG/CT (35.87%) followed by AT/AT (18.59%) and AC/GT (13.19%). The dominant tri-nucleotide repeat was AAG/CTT (4.59%) (Table 3).
Development of EST-SSR markers
A total of 523 primers were designed from flanking SSR regions using Primer3 software. These primers were named as CnKGDEST01-CnKGDEST523 (to denote Cocos nucifera Kasaragod). The details of the 523 primers are given in Supplementary Table S2. Other sequences were excluded due to very short SSR flanking sequences, which were not suited for primer designing. BLAST analysis revealed that all the primers were novel.
Sequence annotation
For the identification of the putative function of each sequence containing EST-SSRs, BLASTx analysis was done. The 523 EST-SSR sequences were subjected to BLASTx analysis, and no hits were found for 78 EST-SSR sequences. The remaining 445 sequences possessed a putative function and 94 were uncharacterized proteins. The results also showed that 381 of the EST-SSR sequences possessed high homology with Elaeis guineensis (oil palm) (in which 84 were uncharacterized proteins) and the remaining 64 EST-SSR sequences possessed high homology with Phoenix dactylifera (date palm) (in which 10 were uncharacterized proteins). The details are given in Supplementary Table S2.
EST-SSR analysis
From the 523 designed EST-SSR primers, after eliminating the 78 EST-SSR sequences which had no hits in BLASTx analysis and 94 EST-SSR sequences with uncharacterised proteins, a total of 120 primers (NCBI Accession numbers: KX580069 to KX580168; KU999089 to KU999108) were selected and synthesized. Gradient PCR was used to select the optimum annealing temperature for each SSR primer pair. These primers were screened in six coconut accessions comprising of three each of tall and dwarf accessions. A total of 60 primer pairs (50%) produced monomorphic bands in the six accessions. Ten primers (8.3%) did not give amplification or gave weak amplification. Fifty primers (41.7%) produced reproducible polymorphic fragments. From these 50 primer pairs, a total of 10 primers were selected for the diversity assessment. The details of primer sequences, optimum annealing temperature and observed allele sizes of the selected primer pairs are provided in Table 4.
Allele richness of SSR loci
A total of 137 alleles were detected with an average of 13.7 alleles per SSR locus. Digital gel profiles generated by MultiNA Viewer software of tall and dwarf accessions using the primers CnKGDEST142, CnKGDEST129 and CnKGDEST136 are given in Supplementrary Fig. S1 a–c. The number of alleles observed at each locus in the data set ranged from seven (loci CnKGDEST100) to 22 (CnKGDEST136). All the loci showed 100% polymorphism with respect to the samples screened. The average observed heterozygosity was 0.46 (Table 5). The PIC values ranged from 0.79 (CnKGDEST129 and CnKGDEST100) to 0.91 (CnKGDEST117 and CnKGDEST122), with a mean value of 0.85 (Table 5).
Genetic diversity levels
Genetic variation at 10 EST-SSR loci was assessed in 186 palms of 50 coconut accessions (2–4 palms per accession), comprising of 25 each of tall and dwarf accessions. The proportion of polymorphic loci (%), alleles per locus, expected heterozygosity (He), observed heterozygosity (Ho) and fixation index in the 50 accessions overall 14 loci are given in Table 6.
For all the accessions, the observed heterozygosity was less than expected heterozygosity indicating a tendency towards inbreeding within a population (Table 6). The mean fixation index across the 50 accessions was 0.17. Negative fixation index was noticed for three accessions viz., Katchal Green Dwarf, Andaman Orange Dwarf and Sri Lankan Yellow Dwarf. The fixation index was the highest for the accession East Coast Tall (0.88). The fixation index value was zero for the accessions Kulashekharan Green Tall, Kenthali Orange Dwarf, Gangabondam Green Dwarf, Chowghat Green Dwarf, Andaman Yellow Dwarf, Andaman Green Dwarf, Sri Lankan Green Dwarf, Hari Papua Orange Dwarf, Coco Bleu Dwarf and Pemba Green Dwarf (Table 6).
Clusters based on the dendrogram
The clustering pattern, based on the analyses of the EST-SSR profiles, revealed the formation of two main clusters among the selected 25 tall accessions, representing the diversity among tall accessions from different geographical regions. The grouping was observed, generally, based on geographical affinity (Fig. 1). Cluster 1, which was a major cluster with 21 accessions, included accessions from South East Asia, most of the Indian and African accessions and one accession from the Pacific region. Within the sub-clusters, most of the large-fruited accessions including Andaman Giant Tall, San Ramon Tall, Borneo Tall, and Cochin China Tall were grouped whereas the majority of the small and medium fruited types formed separate sub-cluster. West Coast Tall Spicata, which is a sub-population of West Coast Tall, clustered together indicating their close relatedness whereas Tiptur Tall, a coconut population from Karnataka state of India showing greater extent of morphological similarity to West Coast Tall, grouped with Philippines Lono Tall.
UPGMA cluster phenogram showing the genetic relationships among the 25 tall coconut accessions, based upon Nei’s genetic distance [26]
Interestingly, the two native coconut populations from Andaman Islands viz., Andaman Ordinary Tall and Andaman Giant Tall were positioned at two extremities. While Andaman Ordinary Tall has normal sized nuts, Andaman Giant Tall has giant sized nuts. Both the populations were collected from South Andaman Island. The two populations from the Konkan region in the West Coast of India, viz., Benaulim Tall and shared similarity with East Coast Tall (a population from the state of Tamil Nadu, on the East Coast of India), and separated from other coconut populations from the West Coast of India. Cluster 2 grouped only four accessions, comprising two from the Pacific Ocean region (British Solomon Island Tall and Guam Tall) and one each from the African region (Zanzibar Tall) and Andaman Islands (Andaman Ordinary Tall) (Fig. 1).
The dendrogram constructed for dwarf accessions in the study revealed the unique nature of Niu Leka Green Dwarf which has formed a single accession in Cluster 1. In contrast, the rest of the dwarfs (24 accessions) grouped together into Cluster 2 (Fig. 2). Within Cluster 2, Malayan Orange Dwarf and Andaman Yellow Dwarf formed a separate sub-cluster. The two Green Dwarfs from Andaman and Nicobar Islands, India, viz. Andaman Green Dwarf and Katchal Green Dwarf clustered together. A few relatively similar (morphologically) dwarfs clustered together, viz. the Kenthali Orange Dwarf with Chowghat Orange Dwarf, while the Chowghat Green Dwarf clustered with Laccadive Green Dwarf. The Kulasekharam Yellow Dwarf was grouped with Malayan Yellow Dwarf. Also, the dwarf accessions from the Pacific Region, namely, Hari Papua and Nikkore, both having orange-coloured fruits, are seen closely related. Among the remaining accessions, the clustering pattern does not show any definite geographical affinity or any specific trait, although there are some exceptions (Fig. 2).
UPGMA cluster phenogram showing the genetic relationships among the 25 dwarf coconut accessions, based upon Nei’s genetic distance [26]
Discussion
A large set of clean reads of high quality, obtained from RNA-sequencing of coconut leaf in an Illumina platform were assembled. A total of 130,942 non-redundant unigenes were obtained, providing a substantial basis for EST-SSR identification and characterization. Our study helped to identify a total of 3,18,528 potential SSRs which is testimony to the high frequency of SSRs in transcribed regions of the coconut genome (one SSR per 2.7 kb). A comparative study of the SSR densities between different species showed that Glycine max (one SSR per 1.77 kb) has a higher SSR density when compared to coconut (one SSR per 7.4 kb), whereas Zea mays (one SSR per 8.1 kb), Arabidopsis thaliana (one SSR per 13.83 kb), Solanum lycopersicum (one SSR per 11.1 kb), Populus spp. (one SSR per 14 kb) and Gossypium spp. (one SSR per 20 kb) demonstrated lower SSR densities in their EST sequences [28]. This difference in densities could be attributed to the number/length of EST sequences, the criteria used for identification, size of the database as well as the various bioinformatics tools used for the research [28].
Mono-nucleotide repeats were not considered in our study. Di-nucleotide SSRs were determined to be more abundant (69.03%) when compared to tri-nucleotide repeats (22.2%) which are in line with a previous study conducted in coconut [18]. Among di-nucleotide repeat motifs, the dominant repeat was found to be AG/CT (35.87%) followed by AT/AT (18.59%) and AC/GT (13.19%) which corroborated findings from previous studies in many species including Hevea brasiliensis and Elaeis guineensis [29, 30]. In E. guineensis (oil palm), di-nucleotide repeats formed the largest group (45.6%), consisting of AG/CT (66.9%), AT/AT (21.9%), AC/GT (10.9%) and CG/CG (0.3%) motifs followed by tri-nucleotide repeats (34.5%) [30], which was similar to the result obtained in this study. The repeat CCG/CGG was reported as the abundant tri-nucleotide repeats in monocots, which might be due to the high G+C content and consequent codon usage in monocot ESTs [31, 32]. In this study, the dominant tri-nucleotide repeat was AAG/CTT (4.59%), which was different from the previous report [18]. The frequent occurrence of tri-nucleotide repeats in coding regions is likely because changes in these repeats are unlikely to cause frameshift mutations [33, 34]. Among di-nucleotide repeats, GA/TC repeat motifs were more abundant than AG/CT in plants like Hordeum vulgare, Zea mays, Oryza sativa, Sorghum bicolor and Triticum spp. [35]. The higher number of AG/CT or GA/TC repeats may be correlated with the frequent occurrence of four types of amino acid products in translated proteins: GAG (Glu), AGA (Arg), CUC (Leu) and CUC (Ser) [5]. Tri-nucleotide repeats were found to be the most dominant SSRs in legume crops like Vicia faba, Glycine max and Glycine soja [36, 37]. The di, tetra or penta-nucleotide variations produce a shift in the reading frame which results in negative selection and a lower degree of polymorphism [34]. The sequence annotation was done using the BLASTx program against oil palm and date palm sequences. The results showed coconut unigenes shared high homology with oil palm.
We designed 523 primers through Primer3 software; all these primers were found to be novel compared to the earlier study [18]. A total of 60 primer pairs (50%) produced monomorphic bands in the six accessions used in the screening process. A possible reason could be that since these SSRs are present in expressed regions of the genome, they could be highly conserved among members of a species causing a lower level of polymorphism [38]. Ten primers did not give amplification, which might be due to the presence of large introns within the primer binding regions, preventing primer annealing [39]. Also, amplicons could be sometimes smaller or larger than the expected sizes, which could be attributed to mutations, like large insertions or deletions, occurring within the region bounded by the primers [39]. A total of 50 primers (41.7%) produced reproducible polymorphic fragments, from which 10 primers were selected at random for diversity studies. The 10 primer pairs could detect a total of 137 alleles, with an average of 13.7 alleles per SSR locus. The number of alleles observed at each locus in the data set was high, ranging from seven (loci CnKGDEST100) to 22 (CnKGDEST136), indicating the capacity of the EST-SSRs to detect high polymorphism. Also, all the 10 loci showed 100% polymorphism with respect to the samples screened. The average fixation index detected was 0.47, while the average observed heterozygosity was 0.46. Earlier studies using SSR markers also detected higher heterozygosity values for tall populations of coconut [12,13,14, 16, 17, 40]. The PIC values ranged from 0.79 to 0.91 with a mean value of 0.85, indicating the capacity of the EST-SSR markers to detect high levels of polymorphism.
For all the accessions, the observed heterozygosity was less than expected heterozygosity indicating a tendency towards inbreeding within a population, with the mean fixation index across the 50 accessions being 0.17. This could be due to the fact that the conserved accessions are developed from a few selected palms of the original population. While a higher level of population differentiation (FST = 0.36) was observed in Mexican coconut populations analyzed using 15 enzymatic systems [41], a moderate level of population differentiation (FST = 0.054) was observed in Sri Lankan coconut populations analyzed using eight SSR primer pairs [14]. Three accessions viz., Katchal Green Dwarf, Andaman Orange Dwarf and Sri Lankan Yellow Dwarf displayed a negative fixation index, indicating an excess of heterozygotes. The highest fixation index was recorded for East Coast Tall (0.88) which is a highly selected cultivar predominantly grown in the eastern coast of the Indian mainland. The fixation index value was zero for 10 dwarf accessions (Kulashekharam Green Tall, Kenthali Orange Dwarf, Gangabondam Green Dwarf, Chowghat Green Dwarf, Andaman Yellow Dwarf, Andaman Green Dwarf, Sri Lankan Green Dwarf, Hari Papua Orange Dwarf, Coco Bleu Dwarf and Pemba Green Dwarf) which indicated that these sub-populations were identical in allele frequencies. These accessions are selected and collected or from original populations, conserved, multiplied with other selection and maintained for at least two generations and hence could have attained the identical allele frequencies in the studied palms. The initial outcome of autogamy in dwarfs was the random fixation of one allele to each locus and the subsequent conservation of a major part of its genetic structure. The ensuing evolution of dwarfs might have encompassed extended periods of relative stability, interspersed with sporadic gene exchanges with local tall populations or other dwarfs, which would have helped to differentiate between these cultivars [4].
With respect to clustering of tall accessions, the position of Andaman Giant Tall and Andaman Ordinary Tall, both from coconut populations of the Andaman Islands in India, in two different clusters highlights the effectiveness of the selection based on fruit component traits (such as size, shape, copra content and husk proportion) and palm morphology for successful varietal improvement. Two high yielding selections, made from Andaman Giant Tall and Andaman Ordinary Tall, have been released as distinct varieties for commercial exploitation, namely ‘Kalpa Dhenu’ and ‘VPM3’, respectively [3]. While Andaman Ordinary Tall showed a greater affinity with Zanzibar Tall in the present study, an earlier study by Devakumar et al. [42] using SSR markers had indicated a close association of Zanzibar Tall with Andaman Ranguchan Tall, implying a possible gene flow between coconut populations of Andamans and Africa and also highlighting the diversity present in coconut populations of the Andaman & Nicobar region. The Kulasekharam Green Tall, a tall selection from naturally introgressed progenies of Kulasekharam Green Dwarf, a secondary collection of Malayan Green Dwarf, introduced from Sri Lanka to Kulashekaram in the Tamil Nadu State of India, grouped with other tall accessions indicating the fixation of tall traits in this selection. This selection, with high yield and showing a lesser incidence of eriophyid mite damage, has been released as a variety (Kalpa Haritha) and is observed to produce vigorous and more uniform progenies [43]. The small-fruited accessions such as Ayiramkachi Tall and Laccadive Micro Tall clustered with West Coast Tall indicating their close relation with medium fruited types. This is in conformation with the report on the production of medium-sized fruits in alternate years in some palms of the micro-fruited (small fruited) populations [44]. The two coconut populations from Konkan region in the West Coast of India, Ratnagiri Tall and Benaulim Tall, shared greater similarity with East Coast Tall than coconut populations from the West Coast of India, indicating that the accessions neither grouped as per geographical affinity nor clustered as per few morphological traits. Both main clusters and most sub-clusters have the parental populations of improved varieties which have been released for different purposes having distinct morphological features, level of tolerance/resistance to biotic and abiotic stress. Hence, further observations on the accessions based on clustering through EST-SSR marker analysis will be useful in fine-tuning the selection process and utilization of truly diverse genotypes. The clustering of these genotypes could be useful in selecting diverse types for use in coconut improvement where the selection could be based on desirable morphological and yield features.
The dendrogram constructed for dwarf accessions in the study revealed the unique nature of Niu Leka Dwarf, which has formed a single accession cluster, whereas the remaining grouped together (Fig. 2). The Niu Leka Dwarf is well known to be distinct from all other dwarfs for many morphological, reproductive and fruit component traits [45]. Earlier studies using molecular markers have also revealed the uniqueness of this accession [12, 30]. Considering the unique traits of dwarfness coupled with high copra content observed in tall accessions, Niu Leka Dwarf has been registered as unique germplasm under the Indian germplasm registry [46]. Among the remaining accessions, the clustering pattern has not shown any definite geographical affinity or any specific trait-based clustering with some exceptions. The clustering of differently colored dwarf accessions indicates the importance of palm stature manifested by combinations of many other morphological features such as earliness for flowering, stem girth variation, leaf length, pollination behavior etc. and in case of dwarf, the entire dwarf populations from different geographical origins, except Niu Leka Dwarf (from Fiji), were grouped together into a single cluster. Within Cluster 2 possessing 24 dwarf accessions, the Indian Ocean collections viz., Pemba Green Dwarf and Coco Bleu Dwarf (a dwarf selection from the original Coco Bleu Tall collection) grouped together and were distinct in comparison to the remaining accessions. The Cameroon Red Dwarf, known for its relatively high copra content, grouped with Sri Lankan Orange Dwarf, a similar type with bold orange fruits. The Pacific accessions Hari Papua and Nikkore, both bearing orange fruits, of medium to small size, respectively, clustered together, indicating they could be closely related. Chowghat Green Dwarf and Laccadive Green Dwarf were grouped together; these two accessions with a slender trunk and early flowering, possess many similar traits except for fruit shape at the time of maturity. While Laccadive Green Dwarf produces round fruits, Chowghat Green Dwarf produces elongated fruits with a characteristic beak at the time of maturity.
Interestingly, two yellow dwarfs with dropping fronds, the Malayan Yellow Dwarf and Kulashekaram Yellow Dwarf clustered together indicating that they are more closely related and distinct from the Andaman Yellow Dwarf. Further, historical reports indicate that the Kulashekaram Yellow Dwarf is evolved from the secondary collection of Malayan Yellow Dwarf, introduced from Sri Lanka to Kulashekaram in the Tamil Nadu State of India. The Kenthali Orange Dwarf was seen grouped with Chowghat Orange Dwarf, where the former is considered morphologically similar and possibly derived from the latter ecotype. These two are orange dwarf accessions of mainland India; the orange dwarf palms of the West Coast region of Kerala State in India are referred to as Chowghat Orange Dwarf whereas it is popularly termed as Kenthali Dwarf in the adjoining Karnataka State. Nair et al. [3] also reported Kenthali Orange Dwarf as a subpopulation of Chowghat Orange Dwarf. Interesting relationships of Laccadive Yellow Dwarf with Sri Lankan Yellow Dwarf and Gangabondam Green Dwarf with Sri Lankan Green Dwarf also could be seen from the clustering. The Malayan Green Dwarf grouped with Laccadive Orange Dwarf, both exhibiting robust growth characters and these two were closely related to Gudanjali Dwarf, another green dwarf collected from Gujarat State in India and known for early flowering at knee height of palms.
Based on the pattern of diversity indicated in the present study and taking into consideration desirable morphological traits, the different dwarf as well as tall accessions could be selected and used in the breeding program for exploitation of heterosis and development of inter varietal hybrids, not only Dwarf × Tall and Tall × Dwarf hybrids, but also Dwarf × Dwarf as well as Tall × Tall hybrids with desirable traits.
Conclusion
This study reports comprehensive characterization of specific co-dominant EST-SSR markers for coconut. As proof of concept, we have also shown that these markers can be used to genotype the accessions to assess the diversity in germplasm collections and group coconut accessions based on their similarity levels. Therefore, EST-SSRs offer great potential for identification of functional markers and also facilitating marker-assisted selection in coconut.
References
Rajesh MK, Sabana AA, Rachana KE, Rahman S, Jerard BA, Karun A (2015) Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT analysis. 3 Biotech 5(6):999-1006
Arunachalam V, Rajesh MK (2017) Coconut genetic diversity, conservation and utilization. In: Ahuja MR, Jain SM (eds) Biodiversity and conservation of woody plants. Springer, Cham, pp 3–36
Nair RV, Jerard BA, Thomas RJ (2016) Coconut breeding in India. In: Al-Khayari JM, Jain SM (eds) Advances in plant breeding strategies: agronomic, abiotic and biotic stress traits. Springer, Cham, pp 257–279
Lebrun P, N’Cho YP, Seguin M, Grivet L, Baudouin L (1998) Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers. Euphytica 101(1):103–108
Qiu L, Yang C, Tian B, Yang JB, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10(278):1–10
Rao VR, Hodgkin T, Bourdeix R (2005) Locating coconut genetic diversity. In: Batugal P, RamananthaRao V, Oliver J (eds) Coconut genetic resources. International Plant Genetic Resources Institute, Regional Office for Asia, the Pacific and Oceania (IPGRI-APO), Serdang, pp 13–31
Arunachalam V, Jerard BA, Damodaran V, Ratnambal MJ, Kumaran PM (2005) Phenotypic diversity of foliar traits in coconut germplasm. Genet Resour Crop Ev 52(8):1031–1037
Sugimura Y, Itano M, Salud CD, Otsuji K, Yamaguchi H (1997) Biometric analysis on diversity of coconut palm: cultivar classification by botanical and agronomical traits. Euphytica 98:29–35
Geethalakshmi P, Parthasarathy VA, Niral V (2005) Genetic diversity among coconut (Cocos nucifera L.) genotypes using isozymes. Asian J Plant Sci 4:678–683
Kumar P, Gupta VK, Misra AK, Modi DR, Pandey BK (2009) Potential of molecular markers in plant biotechnology. Plant Omics 2(4):141–162
Teulat B, Aldam C, Trehin R, Lebrun P, Barker JHA, Arnold GM, Karp A, Baudouin L, Rognon F (2000) An analysis of genetic diversity in coconut (Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. TheorAppl Genet 100(5):764–771
Merrow AW, Wisser RJ, Brown SJ, Kuhn DN, Schnell RJ, Broschat TK (2003) Analysis of genetic diversity and population structure within Florida coconut (Cocos nucifera L.) germplasm using microsatellite DNA, with special emphasis on the Fiji Dwarf cultivar. Theor Appl Genet 106(4):715–726
Perera L, Russell JR, Provan J, Powell W (2000) Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.). Genome 43(1):15–21
Perera L, Russell JR, Provan J, Powell W (2001) Levels and distribution of genetic diversity of coconut (Cocos nucifera L. var Typica form typica) from Sri Lanka assessed by microsatellite markers. Euphytica 122(2):381–389
Rajesh MK, Karun A, Parthasarathy VA (2019) Coconut biotechnology. In: Nampoothiri KUK, Krishnakumar V, Thampan PK, Nair MA (eds) The coconut palm (Cocos nucifera L.)-research and development perspectives. Springer, Singapore, pp 191–226
Rajesh MK, Arunachalam V, Nagarajan P, Lebrun P, Samsudeen K, Thamban C (2008) Genetic survey of 10 Indian coconut landraces by simple sequence repeats (SSRs). Sci Hortic 118(4):282–287
Rivera R, Edwards KJ, Barker JHA, Arnold GM, Ayad G, Hodgkin T, Karp A (1999) Isolation and characterization of polymorphic microsatellites in Cocos nucifera L. Genome 42(4):668–675
Xia W, Xiao Y, Liu Z, Luo Y, Mason AS, Fan H, Yang Y, Zhao S, Peng M (2014) Development of gene-based simple sequence repeat markers for association analysis in Cocos nucifera. Mol Breed 34(2):525–535
Xiao Y, Luo Y, Yang Y, Fan H, Xia W, Mason AS, Zhao S, Sager R, Qiao F (2013) Development of microsatellite markers in Cocos nucifera and their application in evaluating the level of genetic diversity of Cocos nucifera. Plant Omics 6(3):193–200
Senan S, Kizhakayil D, Sasikumar B, Sheeja TE (2014) Methods for development of microsatellite markers: an overview. Not Sci Biol 6(1):1–13
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Casu R, Dimmock C, Thomas M, Bower N, Knight D, Grof C (2001) Genetic and expression profiling in sugarcane. Proc Int Soc Sugarcane Technol 24:626–627
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Gene 12(7):499–510
Rajesh MK, Rachana KE, Kulkarni K, Sahu BB, Thomas RJ, Karun A (2018) Comparative transcriptome profiling of healthy and diseased Chowghat Green Dwarf coconut palms from root (wilt) disease hot spots. Eur J Plant Pathol 151(1):173–193
Rajesh MK, Jerard BA, Preethi P, Thomas RJ, Fayas TP, Rachana KE, Karun A (2013) Development of a RAPD-derived SCAR marker associated with tall-type palm trait in coconut. Sci Hortic 150:312–316
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19(2):153–170
Lewis PO, Zaykin D (2001) Genetic Data Analysis: computer program for the analysis of allelic data. Version 1.0.
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9(1):6
Feng SP, Li WG, Huang HS, Wang JY, Wu YT (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23(1):85–97
Ting NC, Noorhariza MZ, Rozana R, Low ET, Ithnin M, Cheah SC, Tan SG (2010) SSR mining in oil palm EST database: application in oil palm germplasm diversity studies. J Genet 89:135–145
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7(2):537–546
Wang YW, Samuels TD, Wu YQ (2011) Development of 1,030 genomic SSR markers in switch grass. Theor Appl Genet 122(4):677–686
Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10(1):72–80
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48(5–6):501–510
Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li ZY (2010) Generation and characterization of 11 novel EST derived microsatellites from Vicia faba (Fabaceae). Am J Bot 97(7):e69–e71
Kuroda Y, Tomooka N, Kaga A, Wanigadeva SMSW, Vaughan DA (2009) Genetic diversity of wild soybean and Japanese cultivated soybeans based on microsatellite (SSR) analysis and the selection of a core collection. Genet Resour Crop Ev 56(8):1045–1055
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177(3):309–334
Wang Z, Yu G, Shi B, Wang X, Qiang H, Gao H (2014) Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS ONE 9(3):e92029
Rajesh MK, Nagarajan P, Jerard BA, Arunachalam V, Dhanapal R (2008) Microsatellite variability of coconut accessions from Andaman and Nicobar Islands. Curr Sci 94(12):1627–1631
Zizumbo-Villarreal D, Colunga-GarcíaMarín P (2001) Morpho-physiological variation and phenotypic plasticity in Mexican populations of coconut (Cocos nucifera L.). Genet Resour Crop Evol 48(6):547–554
Devakumar K, Neema BTS, Uma M, Naganeeswaran S, Niral V, Jerard BA (2016) Analysis of genetic diversity among Indian Ocean coconut accessions through microsatellite markers. Indian J Hort 73(1):13–18
Niral V, Samsudeen K, Sudha R, Ranjini TN (2019) Genetic resource management and improved varieties of coconut. Ind Cocon J 28:10–14
Samsudeen K, Jacob PM, Rajesh MK, Jerard BA, Kumaran PM (2006) Origin and evolution and evolution of Laccadive Micro Tall, a coconut cultivar from Lakshadweep Islands of India. J Plant Crops 34(3):220–225
Harries HC (1978) The evolution, dissemination and classification of Cocos nucifera L. Bot Rev 44:205–315
Jerard BA, Damodaran V, Niral V, Samsudeen K, Rajesh MK, Sankaran M (2013) Conservation and utilization of soft endosperm coconut accession from Andaman Islands. J Plant Crops 41(1):14–21
Funding
This research was funded by Indian Council of Agricultural Research (ICAR-CPCRI Institute Project Code No. 1000761030).
Author information
Authors and Affiliations
Contributions
MKR designed the study; BAJ and VN provided the samples; PP, SR and SN performed the experiments; PP, AAS, KPG and MKR analyzed the data; PP wrote the paper; MKR, VN and BAJ checked the paper. The decision to submit the manuscript for publication was made by all the authors. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None of the authors have any financial or non-financial conflict of interest.
Research involving human and/or animals rights
This research did not involve Human Participants and/or Animals.
Informed consent
Not applicable as this research did not involve human participants or clinical trials.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Preethi, P., Rahman, S., Naganeeswaran, S. et al. Development of EST-SSR markers for genetic diversity analysis in coconut (Cocos nucifera L.). Mol Biol Rep 47, 9385–9397 (2020). https://doi.org/10.1007/s11033-020-05981-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05981-8