Abstract
Red rot caused by Colletotrichum falcatum poses a serious threat to sugarcane cultivation in many tropical and sub-tropical countries. Deciphering the molecular network of major defense-signaling pathways in sugarcane cultivars with varying red rot resistance is essential to elucidate the phenomenon of defense priming exerted by resistance inducers. Therefore, in this study, expression pattern of transcripts coding for major defense-signaling pathway regulatory genes was profiled during compatible and incompatible interactions and in response to defense priming using qRT-PCR. Candidate genes that were profiled are involved in or related to hypersensitive response and reactive oxygen species production (HR/ROS), salicylic acid (SA), and jasmonic acid/ethylene (JA/ET) pathways. For compatible and incompatible interactions, susceptible (CoC 671), field tolerant (Co 86032) and resistant (Co 93009) sugarcane cultivars were used, whereas for defense priming, benzothiadiazole (BTH) and the pathogen-associated molecular patterns (PAMPs) of C. falcatum viz., CfEPL1 (eliciting plant response-like) and CfPDIP1 (plant defense inducing protein) were used in CoC 671 cultivar. Results indicated that the master regulator of defense pathways, nonexpressor of pathogenesis-related genes 1 (NPR1) was highly upregulated in incompatible interactions (in both Co 86032 and Co 93009) than the compatible interaction along with SA pathway-associated genes. Similarly, in response to defense priming with BTH, CfEPL1 and CfPDIP1, only the SA pathway-associated genes showed considerable upregulation at 0 h post inoculation (hpi) and other intermittent time points. Overall, this study showed that SA-mediated defense pathway is the most predominant pathway reprogrammed during priming with BTH, CfEPL1 and CfPDIP1 and substantiated the earlier findings that these agents indeed induce systemic resistance against red rot of sugarcane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Red rot caused by the fungus Colletotrichum falcatum is one of the serious problems for sugarcane production in many tropical countries. Owing to the economic significance of this disease, red rot resistance is one of the major and essential criteria for screening sugarcane breeding clones in India. In sugarcane, contribution of multiple alleles to complex traits such as disease resistance is a basic question strangling the breeding efforts, which could only be addressed by the development of novel/new strategies that integrates various biotechnology tools [1,2,3]. However, sugarcane genome sequencing is lagging a long way behind, when compared to other related monocots like sorghum and maize, due to its polyploidy, large size and complexity of chromosomes. As far as the status of molecular basis of sugarcane disease resistance is concerned, the progress is not quite encouraging except for sugarcane smut disease, wherein a substantial progress is reported with the advent of next generation sequencing [4,5,6,7,8].
In these circumstances, any effort to delineate the molecular network of major disease resistance pathways associated with red rot would provide significant insights into the understanding of disease resistance in sugarcane. Infected plants undergo transcriptional reprogramming during local defense and during induction of systemic acquired resistance (SAR) [9, 10]. In sugarcane, with the advent of high-throughput genomic platforms and the availability of sugarcane EST database, it became possible to study the spatial and temporal expression of potential defense-related transcription factors (PlantTFDB), targeted defense-related genes (KEGG pathway) and phenylpropanoid pathway genes, etc. The information thus generated on the differential expression pattern of these genes would possibly elucidate the involvement and regulation of multiple signaling networks during compatible/incompatible interactions and defense priming in sugarcane. Compatible interaction refers to an interaction where a pathogen can successfully invade the host and causes disease, whereas the incompatible interaction refers to an interaction where the host can successfully deploy its defense to contain the pathogen invasion [11].
The strategy of induced resistance is well demonstrated in sugarcane against red rot for many years [12]. Selvaraj et al. [13] reported that few candidate defense-related genes of phenylpropanoid pathway that were upregulated during incompatible interaction were also expressed early during defense priming with benzothiadiazole (BTH) and Cf elicitor. Similarly, temporal expression profiling of five major transcription factor (TF) family genes indicated a significant early induction of few TFs in both incompatible interaction and defense priming. It was suggested that these TFs may play a major role in triggering, coordinating or regulating the expression of defense-related pathway genes in sugarcane against the red rot infection [14].
Recently, Ashwin et al. [15, 16] has reported the identification of two potential molecular signatures viz., CfEPL1 (eliciting plant response-like protein 1, a ceratoplatanin protein) and CfPDIP1 (plant defense inducing protein 1, a novel protein) from C. falcatum. Subsequent functional characterization of CfEPL1 and CfPDIP1 indicated that both of them induce HR in tobacco and systemic resistance against red rot in sugarcane. While, most of the ceratoplatanin family proteins are already reported as defense inducing pathogen-associated molecular patterns (PAMPs), functional assays of the novel CfPDIP1 protein indicated a dichotomous role. It induces host defense when primed by means of foliar spray, while suppresses host defense like an effector during co-infiltration with C. falcatum spores in sugarcane leaves [16]. Comprehensively, these studies have highlighted the potential role of these PAMPs/Effectors of C. falcatum in inducing PAMP-triggered immunity (PTI)/effector-triggered immunity (ETI) in sugarcane.
Profiling and identification of potential candidate defense-related genes encoding major defense-signaling pathways activated during incompatible interactions would portray the holistic view of a complex defense network [17,18,19]. Similarly, the elucidation of defense network activated during the priming with the potential PAMP/effector, CfEPL1 and CfPDIP1 would provide vital insights into the mechanism of defense activation. Therefore, in this study, transcriptional expression of major defense pathway regulatory genes-related/mediated with hypersensitive response and reactive oxygen species production (HR/ROS), salicylic acid (SA) and jasmonic acid/ethylene (JA/ET) was profiled during compatible and incompatible interactions and during priming with BTH, CfEPL1 and CfPDIP1.
Materials and methods
Plant materials
Three sugarcane cultivars, namely, CoC 671 (susceptible to red rot), Co 86032 (field tolerant to red rot) and Co 93009 (resistant to red rot), were planted in 20 feet rows in the institute farm, with three biologically independent replications. Nutritional supplements and proper irrigation were provided to ensure agronomically ideal growing conditions.
Histopathological analysis
For spatio-temporal histopathological analysis of C. falcatum and sugarcane interaction, C. falcatum spores were inoculated on eight months old sugarcane cultivar CoC 671 leaf blade, midrib and stalk tissues, and documented with a light-cum-epifluorescence microscope (Leica, Germany) at different time points. At least five inoculated leaf samples were collected for each time point viz., 0-, 12-, 24-, 48- and 72-h post inoculation (hpi). Similarly, five inoculated cane stalks were collected at each time points (0, 12, 24, 48, 72, 120, 240, 600 and 1080 hpi). For light microscopy, cotton-blue stain was used to stain the C. falcatum spores and hyphal structures. For UV-based epifluorescence microscopy, the tissue sections were bleached with 10% potassium hydroxide and stained with Calcofluor white stain (Cat. No. – 18909, Sigma-Aldrich) and mounted under 50% glycerol for documentation.
Defense priming and pathogen inoculation
For priming, eight months old sugarcane cultivars (CoC 671, Co 86032 and Co 93009) were foliar sprayed (25 mL/cane) separately with three different inducing agents (BTH—125 µM (active ingredient) [12]; CfEPL1—50 µg/mL [15]; CfPDIP1—50 µg/mL [16]) for two times at 10 days interval. After 2 weeks of the second foliar spray, primed canes were inoculated with C. falcatum spores by nodal swabbing method as described by Ashwin et al. [12]. Similarly, 8 months old unprimed sugarcane cv. CoC 671, Co 86032 and Co 93009 were also inoculated to study compatible and incompatible interactions. At least, six canes were inoculated per treatment and per cultivar for pathogen biomass quantitation and gene expression analysis. Deionized water was used for mock priming and mock inoculation (untreated and uninoculated control).
Sample collection
Pathogen challenged and mock inoculated stalk tissue samples of primed and unprimed canes were collected at different time points (0, 12, 24, 48, 72, 120, 600 hpi) for pathogen biomass quantitation and expression analysis, snap frozen and stored at − 80 °C until processing. For phenotypic evaluation of priming effects, at least three canes were separately inoculated per treatment and per cultivar with respective controls and evaluated at 600 hpi (25 days post inoculation).
Nucleic acid extraction
For expression analyses, total RNA was extracted from the nodal region (covering approximately 1 cm above and below the pathogen challenged node) of all the collected samples using TRI Reagent® (Sigma-Aldrich, USA) and then converted into cDNA using RevertAid™ H minus First Strand cDNA Synthesis Kit (Thermo Scientific, USA). RNA samples were quantified using Nanodrop™ (USA). An equimolar pool of good quality RNA samples were used as templates for qRT-PCR analyses. For in planta C. falcatum biomass quantitation, genomic DNA was extracted from the nodal portion of the collected stalk samples using CTAB method.
Primer designing and amplicon identification
Orthologous gene sequences that are putatively involved in the major defense-signaling pathways viz., HR/ROS, SA-mediated and JA/ET-mediated pathways in the model plant pathosystems viz., Arabidopsis, tobacco, sorghum, maize and rice were sourced from NCBI and primers were designed accordingly using Primer BLAST software (https://www.ncbi.nlm.nih.gov/tools/primer-blast) (Supplementary Table 1).
Pathogen biomass quantitation and gene expression analysis using qPCR
C. falcatum biomass was quantified by absolute quantification method using the eukaryotic elongation factor (CfeEF1α) [16, 20]. A standard curve was generated from CfeEF1α amplification from ten-fold dilutions of C. falcatum DNA with a linear relationship (R2 value) of 0.9895 (Supplementary Fig. 1). Cycle threshold (CT) values of the test samples from different time intervals were then substituted in the standard curve equation to determine the quantity of pathogen biomass.
For quantitative reverse transcriptase PCR (qRT-PCR), sugarcane elongation factor (ScEF1α) was identified as the most stable reference gene than glyceraldehyde 3-phosphate dehydrogenase (ScGAPDH) and 25S ribosomal RNA (Sc25S rRNA) by the tool NormFinder (NormFinder Excel add-in v 0.953) [15]. Both qPCR analyses were performed in StepOnePlus™ Real-Time PCR (Applied Biosystems, CA, USA) using Power SYBR Green PCR Master Mix (Applied Biosystems) for three biological replications with two technical replications each. Melt curve analysis was performed to verify amplification specificity of the target genes. Amplicon re-sequencing was performed to verify the identity of the transcript-derived fragments (TDFs) and submitted in Genbank. NCBI accession codes, amplification efficiency of transcripts, primer sequences, amplicon length, and annealing temperature details were listed (Supplementary Table 1). For rapid global comparison of different combination of datasets and to depict the expression levels of differential expression of genes in a simplified way, log2 fold change (∆∆CT) expression data of the genes were represented with various color intensities (heat map profiles) using Microsoft Excel v2013 as described by Slawinska et al. [21]. Also, the datasets of individual genes were statistically analyzed using one-way analysis of variance (ANOVA) and the significant differences among the time points were analyzed with post hoc Tukey’s test (p ≤ 0.05) using IBM SPSS statistics software v 21.0.
Results
Histopathological analysis of C. falcatum-sugarcane interaction
Different developmental stages of C. falcatum lifestyle and colonization on leaves and stalks of the red rot susceptible standard cultivar, CoC 671 were examined microscopically and documented at distinct stages. On sugarcane leaf tissues, C. falcatum spores germinated and formed appressorial structures with or without germination tubes by 12 hpi (Fig. 1a, b). By 24 hpi, the pathogen penetrated into the host cell followed by formation of primary hyphal structure (Fig. 1c), which later invaded nearby cells through secondary hyphae by 48 hpi (Fig. 1d) marking the end of biotrophic phase. i.e. invading without killing the cell. However, between 48 and 72 hpi, the secondary hyphae proceeded to damage the cell structure with extensive colonization, thus indicating the transition phase from biotrophic to necrotrophic phase. i.e. killing the colonized cells (Fig. 1e). After 72 hpi, the necrotrophic phase continued to invade and colonize. Mostly, the pathogen spread quickly through and around the nutrient-rich vascular bundles and then colonized the nearby cells (Fig. 1f–h). After intra and intercellular colonization, the pathogen emerged outside through the stomatal pores and formed the sporulating acervuli structures with setae by 72 hpi (Fig. 1i–m).
Histopathology of spatio-temporal interaction of C. falcatum with sugarcane cv. CoC 671 leaf. a C. falcatum spores on leaf surface at 0 hpi. b Spore germination and appressorium formation on leaf surface at 12 hpi. c Penetration of epidermal cell from appressorium and primary hyphae formation (leaf)—24 hpi. d Longitudinal sectioning (LS) showing invasive colonization of adjacent cells through secondary hyphae at 48 hpi. e LS showing extensive colonization of mesophyll cells on leaf blade at 72 hpi. f Cross section (CS) showing progressive colonization of leaf midrib through vascular bundles at 48 hpi. g CS showing hyphal colonization in and around the vascular bundle cells of leaf midrib at 48hpi. h CS showing extensive colonization of leaf midrib at 72 hpi. i, j Pre-emergence of acervuli from stomata at 72 hpi. k CS showing emergence of acervuli after intracellular colonization (midrib)—72 hpi. l Emergence of sporulating acervulus with setae structures at 72 hpi. m Mature sporulating acervulus with setae structure at 72 hpi (bar—50 µm; compound bar—20 µm)
The first 48 h of C. falcatum—sugarcane interaction on sugarcane stalk tissues were similar to the leaf tissues, except the mode of entry. In stalk tissues, the common mode of entry was through the root primordia on the nodal rind portion (Fig. 2a). After 48 h, the secondary hyphal network moved towards the nutrient rich vascular bundles and started to colonize the apoplastic (intercellular) spaces in and around them, in order to colonize fast (Fig. 2b–d). By 120 hpi, the pathogen colonized the cells around the vascular bundles and extended the progression both horizontally and vertically (Fig. 2e–h). By 25 dpi, the entire node and internodes got colonized (Fig. 2i, j) and by 45 dpi, the vascular bundles began to disintegrate and as a result, the entire tissues got macerated (Fig. 2k, l).
Histopathology of spatio-temporal interaction of C. falcatum with sugarcane cv. CoC 671 stalk. a Spore germination and appressorium formation on root primordial surface in nodal portion of cane stalk at 12 hpi. b–d a series of LS showing intercellular (apoplastic) hyphal network colonization in and around the vascular bundles of cane stalk tissues (internode) at 72 hpi. This stage was observed after establishment of primary and secondary hyphae as observed in Fig. 1. e LS showing progressive and intensive intercellular hyphal network around the vascular bundles at 120 hpi. f Colonization of mesophyll cells adjacent to vascular bundles (internode) at 120 hpi. g CS showing colonization of vascular bundle and adjacent mesophyll cells at 120 hpi. h CS showing progressive colonization of vascular bundle and adjacent mesophyll cells at 240 hpi. i CS showing intense colonization of internode tissues at 600 hpi. j CS showing complete colonization of nodal tissues at 600 hpi. k LS showing mycelial colonization in the macerated internal tissues (pith) at 45 dpi. l LS showing complete maceration cellular structures that led to pith formation at 45 dpi (bar—50 µm)
Phenotypic evaluation of compatible and incompatible interactions and defense priming with CfEPL1 and CfPDIP1
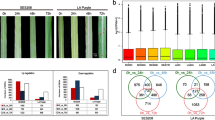
As the name suggests, the compatible interaction between the susceptible cultivar, CoC 671 and C. falcatum resulted in extensive development of red lesions transgressing across more than five nodes (Fig. 3a). In contrast, the incompatible interactions of the field tolerant (resistant) cultivar, Co 86032 and resistant cultivar, Co 93009 with C. falcatum did not produce any visible lesion development.
Representative images showing the extent of disease severity on split-opened canes during different host–pathogen interactions and during defense priming at 25 days post inoculation. a Extent of disease severity during compatible and incompatible interaction of C. falcatum with susceptible, field tolerant and resistant sugarcane cultivars. b Priming efficacy of BTH, CfEPL1 and CfPDIP1 in suppressing the red rot severity in a susceptible sugarcane cultivar
In continuation of the earlier findings on the disease suppressive effects of CfEPL1 and CfPDIP1, their priming efficacy in reducing the severity of red rot was phenotypically evaluated on split-opened canes of CoC 671. BTH, a well demonstrated inducer of systemic resistance, which was shown to reduce the severity of red rot was used as a positive control. Results indicated that CfEPL1 followed by CfPDIP1 priming has significantly reduced the development of red lesion on spilt-opened canes (Fig. 3b). However, the efficacy of both these biotic elicitors in reducing the disease severity was relatively lesser than the efficacy of BTH priming.
In planta quantitation of C. falcatum biomass during compatible and incompatible interactions and defense priming with CfEPL1 and CfPDIP1
To determine the level of colonization of C. falcatum at different developmental stages of host–pathogen interaction, its biomass was quantified during compatible and incompatible interactions, and defense priming with CfEPL1 and CfPDIP1. Quantification profile clearly showed the distinctness in the accumulation of pathogen biomass in compatible interaction with CoC 671, when compared to the incompatible interactions with Co 86032 and Co 93009, as evident by phenotypic evaluation (Fig. 4a). Though the cultivars Co 86032 and Co 93009 were graded as field tolerant and resistant to red rot, respectively, this quantitation profile indicates that C. falcatum colonization is indeed progressing at a relatively very less pace in both cultivars until 120 hpi. However, it almost came down to nil at 600 hpi, in contrast to CoC 671.
In planta quantification of C. falcatum biomass by absolute quantification method. a Quantitation of C. falcatum biomass during compatible and incompatible interactions. a Y-axis values was broken (║) to ensure the small increments in pathogen biomass of Co 93,009 and Co 86,032 are visible. b Quantitation of C. falcatum biomass during priming with SAR inducers in CoC 671. b ‘0 h’ indicates mock inoculated control. Error bars indicate standard deviation of three biological replicates with two technical replicates each
In response to priming with SAR inducers, BTH recorded the least accumulation of C. falcatum biomass, followed by CfEPL1 and CfPDIP1, which were similar to the phenotypic observation (Fig. 4b). Noticeably, all the three inducers showed significant suppression of pathogen colonization since 120 hpi. However, BTH, the positive control performed better than the other two inducers ever since 48 hpi.
Expression profiling of major defense pathway-associated genes during compatible and incompatible interactions
Temporal expression profiling of SA, JA/ET and HR/ROS pathway associated genes showed significant upregulation of most of SA pathway-associated genes (NPR1, ICS, CHI B, CHI 2, GLU D) in few time points of field tolerant cultivar, Co 86032 and most of the time points of resistant cultivar, Co 93009 (Fig. 5) (Supplementary table 2). In addition, few genes associated with JA/ET pathway (DEF 1, DEF 4, DEF 5, and AOS) were also upregulated distinguishably at an earlier timepoint by 12 hpi in the incompatible interaction of Co 93009. However, the HR/ROS pathway genes, except, WRKY 44 did not show considerable changes in their expression pattern in both Co 86032 and Co 93009 cultivars. Interestingly, the expression of NPR1, the master regulator of many defense-signaling pathways was activated early and upregulated in most of the time points in both types of incompatible interactions. Another major regulator gene of JA/ET pathway, ERF3 gene expression was inconsistent and found to be completely absent in some of the time points in all the three interactions. Overall results indicated that SA pathway-associated genes were relatively more active at early time points in response to pathogen challenge during the incompatible interactions in Co 86032 and Co 93009 cultivars than the compatible interaction in CoC 671 cultivar.
Heat map profile showing differential expression of major defense-related pathway genes at different time intervals during compatible and incompatible interaction of C. falcatum with susceptible, field tolerant and resistant sugarcane cultivars. Log2 fold expression (− ∆∆CT) values were represented in this heat map profile. ‘0 hpi’ indicates untreated and uninoculated control. White boxes indicate no detectable expression
Relative expression profiling of major defense-signaling pathway genes in response to BTH, CfEPL1 and CfPDIP1 priming
Expression profiling of defense-signaling pathway-associated genes in response to the priming of BTH, CfEPL1 and CfPDIP1 on the susceptible cultivar, CoC 671 showed significant early upregulation (0 hpi) of most of the SA pathway genes (Fig. 6) (Supplementary table 3). Notably, NPR1 gene expression was significantly upregulated in all the time points of all the primed samples (BTH, CfEPL1 and CfPDIP1) similar to the incompatible interactions. The expression of some of the SA pathway-associated genes (ICS, CHI 7 and GLU D) were upregulated at sporadic time points in primed samples.
Heat map profiles showing temporal expression of major defense-related pathway genes during priming with BTH, CfEPL1 and CfPDIP1 in a susceptible sugarcane cultivar, CoC 671. Log2 fold expression (− ∆∆CT) values were represented in this heat map profile. ‘0 hpi’ indicates treated uninoculated control in primed samples. White boxes indicate no detectable expression
Expression profiling of JA/ET-pathway-associated genes showed that DEF 1, DEF 4, COI 1.1 and COI 1.2 got upregulated at 24 and 48 hpi in response to CfEPL1 priming, whereas the same set of genes were upregulated at 0 hpi due to priming with CfPDIP1. However, there was no considerable uniformity in the expression pattern of JA/ET pathway-associated genes in response to BTH priming. ERF3 gene expression was inconsistent and found to be completely absent in some of the time points of primed samples.
Most of the HR/ROS-associated gene expressions were upregulated at 0 hpi in all the three primed samples, whereas they were completely downregulated at subsequent time points in BTH and CfPDIP1 priming. Except WRKY 44, all the genes of this pathway showed consistent downregulation in CfPDIP1 primed samples. In case of CfEPL1 priming, all the genes of HR/ROS pathway, except HIN1 showed upregulation by 24 and 48 hpi.
Discussion
Every plant is being attacked by a wide range of pathogens. During the course of co-evolution, both plants and fungi developed their molecular combat system in a see-saw manner, which ultimately dictates the winner of this arms race. Plants recognize the pathogens by means of PAMPs or elicitors which signals their presence and induce defense in plants. With the recognition of the pathogen, plants often trigger a localized resistance reaction, known as the HR, which is characterized by a rapid cell death at the site of infection [22]. Sometimes, plants do develop an enhanced resistance to further pathogen attack, when primed with PAMPs/inducers and sometimes effectors. This type of enhanced resistance is referred to as systemic acquired resistance (SAR) [23].
At molecular level, SAR is characterized by an increased expression of a large number of pathogenesis-related genes (PR genes), in both local and systemic tissues. Pathogen recognition, downstream signal transduction and activation of plant defense are the critical steps involved in the induction of plant immune system. The perception of a potential pathogen results in the activation of intracellular signalling events including ion fluxes, phosphorylation/dephosphorylation cascades, kinase cascades, and generation of reactive oxygen species (ROS), which in turn activate major defense pathways viz., jasmonic acid (JA), and ethylene (ET) [24, 25]. These signalling events lead to the reinforcement of plant cell wall and the production of defense-related proteins and phytoalexins. These transcriptional events occur during the induction of SAR, and in any compatible or incompatible interactions, however with differential speed and intensity, which ultimately define the outcome of plant immune responses [26, 27].
Spatio-temporal histopathological analysis of C. falcatum interaction with sugarcane provided insights into the biotrophic phase (0–48 hpi), transition phase (48–72 hpi) and necrotrophic phase (> 72 hpi) in both leaves and stalk tissues, as comparable to other hemibiotrophic pathogens belonging to Colletotrichum spp.[28,29,30]. The extent of rapid and aggressive colonization over the vascular bundles by 72 hpi suggests that the intracellular hyphae utilizes the nutrient rich apoplastic spaces of that region to feed and secrete a range of effectors to suppress host defense in that vicinity for further expansion. This is one of the characteristic developments of hemibiotrophic colonization of Colletotrichum spp., where the host defense induced during biotrophic phase of colonization is suppressed by the effectors secreted during the transition phase [31, 32]. Subsequently, the effectors secreted by the secondary necrotrophic hyphae during the necrotrophic phase of infection damage and kill the cells and tissues [33]. Similarly, in our study, the extensive colonization resulted in complete damage and maceration of internal tissues, which led to the formation of pith inside the stalk from 25 to 45 dpi. Further, these observations were substantiated by the degree of pathogen biomass accumulation at respective timepoints. Overall, the analysis depicted the crucial developmental stages/events occurring at different time points, and based on which, the time intervals for drawing samples for transcriptional profiling was determined.
Comparative expression profiling of compatible and incompatible interactions of sugarcane and C. falcatum showed upregulation of some of the SA and JA/ET pathway-associated genes in the field tolerant cultivar—Co 86032 and resistant cultivar, Co 93009 at certain time points. Though, the interaction of C. falcatum with both Co 86032 and Co 93009 was referred to as incompatible interactions based on the outcome of disease phenotyping, Co 86032 is not red rot resistant cultivar, unlike, Co 93009. And, this difference in their resistance level is evident in our pathogen biomass quantitation study. Co 86032 is considered as a field tolerant (resistant) cultivar, because the cultivar exhibits resistant reaction when artificially inoculated by nodal swabbing method, but expresses susceptibility when inoculated by plug inoculation method (an invasive method of inoculation). Plug inoculation method is a rigorous stringent evaluation method for screening red rot resistant varieties. Since, Co 86032 did not succumb to red rot under natural climatic conditions at field trials evaluated at hot spots, the cultivar was released for cultivation as field tolerant (resistant) [34] and presently, it is the predominant cultivar in South India. The differential regulation of these defense-signaling pathways in these two cultivars clearly suggests that SA pathway-associated genes could possibly play a pivotal role in conferring disease resistance, while JA/ET pathway-associated genes might further reinforce the induced defense/resistance as evidenced in Co 93009 and CfEPL1 priming. Recently, Li et al. [35] also showed that priming of tobacco with FocCP1, a ceratoplatanin PAMP similar to CfEPL1, induced the production of both SA and JA molecules while triggering HR and SAR against tobacco mosaic virus and Pseudomonas syringae pv. tabaci 6605 (Pst. 6605) infections. The activation of SAR has also been shown to suppress JA signaling in plants, thereby prioritizing SA-dependent resistance against pathogens over JA-dependent defense against insect herbivory [36, 37].
SA is an important signal molecule in plants, from a disease resistance perspective. Two pathways of SA biosynthesis have been identified in plants. Plants synthesize SA from cinnamate produced by the activity of PAL [38]. Another pathway of SA biosynthesis is facilitated by Isochorismate synthase (ICS) [39]. The transcriptional network of important regulatory genes that govern the major defense-signaling pathways in the model plant systems like Arabidopsis, tobacco, sorghum, maize, rice, etc. are depicted pictorally in Fig. 7. Besides, the depiction also represents the set of defense pathway-associated genes that have been profiled in the present study.
In our study, BTH, an analog of SA was used as a positive control to compare the effects of CfEPL1 and CfPDIP1 priming, because, it has been a well demonstrated SAR inducer that operates through SA pathway and its priming efficacy against red rot in sugarcane has been proven [12, 40]. Here, the pathogen biomass quantitation study also clearly demonstrated the suppression effects of these inducers. Especially, the suppression of pathogen biomass in response to the priming of these biotic inducers suggested that these inducers have indeed activated the defense-signaling pathways and intensified the same upon pathogen perception. NPR1, the master regulator of SA and JA/ET pathway showed early upregulation in both incompatible interaction and in response to SAR priming response. Systemic resistance is associated with the expression of the master regulator, NPR1 and other PR proteins [41]. Transduction of the SA signal requires the function of NPR1. It interacts with the TGA subclass of basic leucine-zipper (bZIP) transcription factors [42]. The presence of two protein–protein interaction domains in NPR1 suggests that it might regulate SAR-related gene expression through interaction with transcription factors. Expression of the NPR1 gene is constitutive, but moderately influenced by SA. Overexpression of NPR1 does not lead to constitutive PR gene expression in the absence of SAR induction, indicating that the NPR1 protein requires SA activation to be functional [43].
This activation would have been accomplished through the redox changes induced as a result of SA accumulation during SAR, which leads to conformational changes in NPR1 from the inactive oligomer to an active monomer. The above phenomenon was also observed in our study, in response to CfEPL1 and CfPDIP1 priming, wherein which the PR proteins, chitinases and glucanases did not express considerably in line with the expression of NPR1 at many time points, unlike the case in Co 93009. Chen et al. [38] reported upregulation of sugarcane NPR1 at early time intervals and the expression was found to be positively regulated in response to SA application, thus indicating the likely involvement of NPR1 in the induction of systemic resistance. Over the years, many reports have established the potential role of PR proteins in conferring disease resistance during sugarcane-pathogen interaction in sugarcane. Differential expression analysis of compatible and incompatible interactions involving C. falcatum and sugarcane using DD-RT-PCR and suppression subtractive hybridization (SSH) revealed two-fold higher expression of β-1,3-glucanase, chitinase and PR10 genes at early intervals (0 to 24 hpi) in incompatible interaction [19, 44]. On the other hand, the pre-activation of NPR1 in response to inducer applications (BTH, CfEPL1 and CfPDIP1), even before the perception of the pathogen have induced the expression of PR genes at the early hours of host–pathogen interaction (0 hpi). Thus, NPR1 was asserted to be involved in downstream regulation of genes involved in SAR [41, 45].
In the meanwhile, the primary and foremost defense response-associated genes related with HR/ROS pathways did not show considerable differences in expression during both compatible and incompatible interactions. However, some of the genes of this pathway got upregulated in response to priming with BTH, CfEPL1 and CfPDIP1 at 0 hpi. This phenomenon suggests a completely different mode of defense activation or induction might have been operated during incompatible interaction and during priming. Nevertheless, many JA/ET pathway genes were also activated during incompatible interaction, which are otherwise antagonistic to the expression of SA pathway-associated genes [46, 47]. These transcriptional reprogramming have substantiated the fact that signalling crosstalks indeed occur between these pathways during dynamic interaction with pathogens and they are mutually antagonistic [48]. Generally, the SA pathway associated genes were found to positively regulate plant defense against biotrophic pathogens, whereas, the JA/ET pathway-associated genes were prominently regulated against necrotrophic pathogens [36]. Since, C. falcatum is a hemibiotrophic pathogen, here, in this study, both SA and JA/ET pathway-associated genes might have been differentially regulated during incompatible interactions and during defense priming. Nevertheless, the temporal and quantitative variations in the basal mechanism of disease resistance operated during incompatible interaction and defense priming might be due to the cultivar specific innate immune mechanisms.
Comprehensively, this study has suggested that the PAMP/effector molecules viz., CfEPL1 and CfPDIP1 indeed suppress pathogen colonization by activating systemic resistance with upregulation of the master regulator NPR1 and PR genes, a similar phenomenon demonstrated for the induction of systemic resistance by BTH. For the first time, this study has indicated that SA-mediated defense pathway is one of the most active defense-signaling pathways that are reprogrammed during priming with BTH, CfEPL1 and CfPDIP1 and substantiated the earlier findings that these agents induce systemic resistance against red rot of sugarcane. The results have opened up new vistas for the identification of interacting partners of these potential PAMPs/Effectors viz. CfEPL1 and CfPDIP1 to elucidate the series of molecular signaling events occurring since perception to defense induction, which thereby could help us to delineate the mechanism of PTI/ETI response in sugarcane.
References
Dal-Bianco M, Carneiro MS, Hotta CT, Chapola RG, Hoffmann HP, Garcia AAF, Souza GM (2012) Sugarcane improvement: how far can we go? Curr Opin Biotechnol 23(2):265–270. https://doi.org/10.1016/j.copbio.2011.09.002
Thirugnanasambandam PP, Hoang NV, Henry RJ (2018) The Challenge of analyzing the sugarcane genome. Front Plant Sci 9(May):616. https://doi.org/10.3389/fpls.2018.00616
Ali A, Khan M, Sharif R, Mujtaba M, Gao SJ (2019) Sugarcane omics: an update on the current status of research and crop improvement. Plants 8(9):344. https://doi.org/10.3390/plants8090344
Wu Q, Xu L, Guo J, Su Y, Que Y (2013) Transcriptome profile analysis of sugarcane responses to Sporisorium scitaminea infection using Solexa sequencing technology. Biomed Res Int 2013:298920. https://doi.org/10.1155/2013/298920
Que Y, Su Y, Guo J, Wu Q, Xu L (2014) A global view of transcriptome dynamics during Sporisorium scitamineum challenge in sugarcane by RNA-seq. PLoS ONE 9(8):e106476. https://doi.org/10.1371/journal.pone.0106476
Taniguti LM, Schaker PDC, Benevenuto J, Peters LP, Carvalho G, Palhares A et al (2015) Complete genome sequence of Sporisorium scitamineum and biotrophic interaction transcriptome with sugarcane. PLoS ONE 10(6):e0129318. https://doi.org/10.1371/journal.pone.0129318
Schaker PDC, Palhares AC, Taniguti LM, Peters LP, Creste S, Aitken KS et al (2016) RNAseq transcriptional profiling following whip development in sugarcane smut disease. PLoS ONE 11(9):e0162237. https://doi.org/10.1371/journal.pone.0162237
Rody HVS, Bombardelli RGH, Creste S, Camargo LEA, Van Sluys MA, Monteiro-Vitorello CB (2019) Genome survey of resistance gene analogs in sugarcane: Genomic features and differential expression of the innate immune system from a smut-resistant genotype. BMC Genomics 20(1):809. https://doi.org/10.1186/s12864-019-6207-y
Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA et al (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26(4):403–410. https://doi.org/10.1038/82521
van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162. https://doi.org/10.1146/annurev.phyto.44.070505.143425
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923
Ashwin NMR, Barnabas EL, Ramesh Sundar A, Muthumeena M, Malathi P, Viswanathan R (2017) Disease suppressive effects of resistance-inducing agents against red rot of sugarcane. Eur J Plant Pathol 149(2):285–297. https://doi.org/10.1007/s10658-017-1181-1
Selvaraj N, Ramadass A, Amalraj RS, Palaniyandi M, Rasappa V (2014) Molecular profiling of systemic acquired resistance (SAR)-responsive transcripts in sugarcane challenged with Colletotrichum falcatum. Appl Biochem Biotechnol 174(8):2839–2850. https://doi.org/10.1007/s12010-014-1230-6
Muthiah M, Ramadass A, Amalraj RS, Palaniyandi M, Rasappa V (2013) Expression profiling of transcription factors (TFs) in sugarcane X Colletotrichum falcatum interaction. J Plant Biochem Biotechnol 22(3):286–294. https://doi.org/10.1007/s13562-012-0157-7
Ashwin NMR, Barnabas L, Ramesh Sundar A, Malathi P, Viswanathan R, Masi A et al (2017) Comparative secretome analysis of Colletotrichum falcatum identifies a cerato-platanin protein (EPL1) as a potential pathogen-associated molecular pattern (PAMP) inducing systemic resistance in sugarcane. J Proteomics 169:2–20. https://doi.org/10.1016/j.jprot.2017.05.020
Ashwin NMR, Barnabas L, Ramesh Sundar A, Malathi P, Viswanathan R, Masi A et al (2018) CfPDIP1, a novel secreted protein of Colletotrichum falcatum, elicits defense responses in sugarcane and triggers hypersensitive response in tobacco. Appl Microbiol Biotechnol 102(14):6001–6021. https://doi.org/10.1007/s00253-018-9009-2
Oloriz MI, Gil V, Rojas L, Portal O, Izquierdo Y, Jiménez E, Höfte M (2012) Sugarcane genes differentially expressed in response to Puccinia melanocephala infection: identification and transcript profiling. Plant Cell Rep 31(5):955–969. https://doi.org/10.1007/s00299-011-1216-6
Kawahara Y, Oono Y, Kanamori H, Matsumoto T, Itoh T, Minami E (2012) Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE 7(11):e49423. https://doi.org/10.1371/journal.pone.0049423
Sathyabhama M, Viswanathan R, Nandakumar M, Malathi P, Ramesh Sundar A (2015) Understanding sugarcane defence responses during the initial phase of Colletotrichum falcatum pathogenesis by suppression subtractive hybridization (SSH). Physiol Mol Plant Pathol 91:131–140. https://doi.org/10.1016/j.pmpp.2015.07.003
Diguta CF, Rousseaux S, Weidmann S, Bretin N, Vincent B, Guilloux-Benatier M, Alexandre H (2010) Development of a qPCR assay for specific quantification of Botrytis cinerea on grapes. FEMS Microbiol Lett 313(1):81–87. https://doi.org/10.1111/j.1574-6968.2010.02127.x
Slawinska A, Hsieh JC, Schmidt CJ, Lamont SJ (2016) Heat stress and lipopolysaccharide stimulation of chicken macrophage-like cell line activates expression of distinct sets of genes. PLoS ONE 11(10):e0164575. https://doi.org/10.1371/journal.pone.0164575
Doehlemann G, Requena N, Schaefer P, Brunner F, O’Connell R, Parker JE (2014) Reprogramming of plant cells by filamentous plant-colonizing microbes. New Phytol 204(4):803–814. https://doi.org/10.1111/nph.12938
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209. https://doi.org/10.1146/annurev.phyto.42.040803.140421
Panstruga R, Parker JE, Schulze-Lefert P (2009) Plant immune response pathways. Cell 136(5):6–8. https://doi.org/10.1016/j.cell.2009.02.020
Garner CM, Kim SH, Spears BJ, Gassmann W (2016) Express yourself: Transcriptional regulation of plant innate immunity. Semin Cell Dev Biol 56:150–162. https://doi.org/10.1016/j.semcdb.2016.05.002
Mauch-Mani B, Baccelli I, Luna E, Flors V (2017) Defense priming: an adaptive part of induced resistance. Annu Rev Plant Biol 68(1):485–512. https://doi.org/10.1146/annurev-arplant-042916-041132
Birkenbihl RP, Liu S, Somssich IE (2017) Transcriptional events defining plant immune responses. Curr Opin Plant Biol 38:1–9. https://doi.org/10.1016/j.pbi.2017.04.004
Munch S, Lingner U, Floss DS, Ludwig N, Sauer N, Deising HB (2008) The hemibiotrophic lifestyle of Colletotrichum species. J Plant Physiol 165(1):41–51. https://doi.org/10.1016/j.jplph.2007.06.008
O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF et al (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44(9):1060–1065. https://doi.org/10.1038/ng.2372
Gan P, Ikeda K, Irieda H, Narusaka M, O’Connell RJ, Narusaka Y et al (2012) Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol 197(4):1236–1249. https://doi.org/10.1111/nph.12085
Kleemann J, Rincon-Rivera LJ, Takahara H, Neumann U, van Themaat EVL, van der Does HC et al (2012) Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog 8(4):e1002643. https://doi.org/10.1371/journal.ppat.1002643
Vargas W, Martín JMS, Rech GE, Rivera LP, Benito EP, Díaz-Mínguez JM et al (2012) Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol 158(3):1342–1358. https://doi.org/10.1104/pp.111.190397
De Silva DD, Crous PW, Ades PK, Hyde KD, Taylor PWJ (2017) Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biol Rev 31:155–168. https://doi.org/10.1016/j.fbr.2017.05.001
Mohanraj D, Kaverinathan K (2011) The concept of field tolerance and its relevance in screening for red rot of sugarcane. J Sugarcane Res 1(2):16–22
Li S, Dong Y, Li L, Zhang Y, Yang X, Zeng H et al (2019) The novel cerato-platanin-like protein FocCP1 from Fusarium oxysporum triggers an immune response in plants. Int J Mol Sci 20(11):2849. https://doi.org/10.3390/ijms20112849
Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5(5):308–316. https://doi.org/10.1038/nchembio.164
Gimenez-Ibanez S, Solano R (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Frontiers in Plant Science 4:72. https://doi.org/10.3389/fpls.2013.00072
Chen Z, Zheng Z, Huang J, Lai Z, Fan B (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4(6):493–496. https://doi.org/10.4161/psb.4.6.8392
Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562–565. https://doi.org/10.1038/35107108
Tripathi D, Raikhy G, Kumar D (2019) Chemical elicitors of systemic acquired resistance—Salicylic acid and its functional analogs. Curr Plant Biol 17(March):48–59. https://doi.org/10.1016/j.cpb.2019.03.002
Pajerowska-Mukhtar KM, Emerine DK, Mukhtar MS (2013) Tell me more: roles of NPRs in plant immunity. Trends Plant Sci 18(7):402–411. https://doi.org/10.1016/j.tplants.2013.04.004
Kinkema M, Fan W, Dong X (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12:2339–2350. https://doi.org/10.1105/tpc.12.12.2339
Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot 62(10):3321–3338. https://doi.org/10.1093/jxb/err031
Prathima PT, Raveendran M, Kumar KK, Rahul PR, Kumar VG, Viswanathan R et al (2013) Differential regulation of defense-related gene expression in response to red rot pathogen Colletotrichum falcatum infection in sugarcane. Appl Biochem Biotechnol 171(2):488–503. https://doi.org/10.1007/s12010-013-0346-4
Pieterse CMJ, Van Loon LC (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7(4):456–464. https://doi.org/10.1016/j.pbi.2004.05.006
Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet. https://doi.org/10.1371/journal.pgen.1000772
Yang Y-X, Ahammed G, Wu C, Fan S, Zhou Y-H (2015) Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr Protein Pept Sci 16(5):450–461. https://doi.org/10.2174/1389203716666150330141638
Li N, Han X, Feng D, Yuan D, Huang L (2019) Signaling crosstalk between salicylic acid and ethylene / jasmonate in plant defense : do we understand what they are whispering ? Int J Mol Sci 20:671. https://doi.org/10.3390/ijms20030671
Acknowledgements
The authors are grateful to The Director, ICAR-Sugarcane Breeding Institute for providing facilities and continuous encouragement. The financial support received from Department of Science and Technology (DST), Department of Biotechnology (DBT) and Indian Council of Agricultural Research (ICAR), New Delhi are greatly acknowledged.
Funding
This work was financially supported in part by the Department of Science and Technology (Sanction F.No. SR/SO/PS-22/10/1 dated 04/01/2011), Department of Biotechnology (DBT) (Sanction no. BT/PR23621/BPA/118/297/2017 dated 31/08/2018) and Indian Council of Agricultural Research in the form of research grants to the corresponding author. Besides, the work was supported by Council of Scientific and Industrial Research (CSIR), India (IN) in the form of CSIR-UGC NET JRF fellowship (S. No. 313505) awarded to the first author.
Author information
Authors and Affiliations
Contributions
NMRA and ARS conceived and designed the experiments. NMRA performed the experiments and wrote the first draft of the manuscript. DA and KVL were involved in statistical analysis of expression data and in revising the manuscript. ARS, LB, PM and RV shared their expertise in analysis and interpretation of the data, and were involved in revising and improving the intellectual content of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Ethical approval
The present research work did not involve Human Participants and/or Animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2020_5944_MOESM1_ESM.tif
Electronic supplementary material 1 (TIF 8622 kb) Supplementary fig. 1 Generation of standard curve of cycle threshold values of CfEF1α gene amplification from different concentration of C. falcatum DNA extracted from an axenic culture.
Rights and permissions
About this article
Cite this article
Ashwin, N.M.R., Barnabas, L., Amalamol, D. et al. Transcriptional reprogramming of major defense-signaling pathways during defense priming and sugarcane-Colletotrichum falcatum interaction. Mol Biol Rep 47, 8911–8923 (2020). https://doi.org/10.1007/s11033-020-05944-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05944-z