Abstract
Glioblastoma (GBM) is the most common primary malignant neoplasm of the central nervous system and, despite the standard therapy; the patients’ prognoses remain dismal. The miRNA expression profiles have been associated with patient prognosis, suggesting that they may be helpful for tumor diagnosis and classification as well as predictive of tumor response to treatment. We described the microRNA expression profile of 29 primary GBM samples (9 pediatric GBMs) and 11 non-neoplastic white matter samples as controls (WM) by microarray analysis and we performed functional in vitro assays on these 2 most differentially expressed miRNAs. Hierarchical clustering analysis showed 3 distinct miRNA profiles, two of them in the GBM samples and a group consisting only of cerebral white matter. When adult and pediatric GBMs were compared to WM, 37 human miRNAs were found to be differentially expressed, with miR-10b-5p being the most overexpressed and miR-630 the most underexpressed. The overexpression of miR-630 was associated with reduced cell proliferation and invasion in the U87 GBM cell line, whereas the inhibition of miR-10b-5p reduced cell proliferation and colony formation in the U251 GBM cell line, suggesting that these miRNAs may act as tumor-suppressive and oncogenic miRNAs, respectively. The present study highlights the distinct epigenetic profiling of adult and pediatric GBMs and underscores the biological importance of mir-10b-5p and miR-630 for the pathobiology of these lethal tumors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM) is the most common primary malignant neoplasm of the central nervous system (CNS), mainly affecting adults. It represents the most aggressive form of malignant gliomas, corresponding to 82% of all cases. Current treatment for GBM is based on maximal surgical resection of the tumor followed by a combination of radiotherapy and temozolomide [1,2,3]. Despite the standard therapy, patients with GBM have an unfavorable prognosis with a 5-year survival rate of 4 to 5% and a median survival of 15 months [1, 4, 5]. Other therapeutic options such as tumor-treating field (TTF) technology in association with temozolomide, target therapy and immunotherapy have been evaluated, but they are not curative [2, 6, 7].
Most GBMs are considered to be primary tumors, occurring de novo and accounting for more than 90% of all cases [8]. Several studies have shown that young patients have a more favorable outcome when compared to older individuals [9, 10], a fact probably related to the molecular and biological differences between pediatric and adult GBM [11, 12]. In view of the lethality of GBM, a better understanding of the biological and molecular nature of these tumors is urgently needed, as well as the recognition of new potential therapeutic targets.
MicroRNAs (miRNAs) are small non-coding RNA molecules approximately 17–22 nucleotides long that act as posttranscriptional regulators of gene expression by direct binding to target mRNAs [13, 14]. miRNAs are involved in diverse biological processes, including cell proliferation and apoptosis. Thus, their expression in non-neoplastic tissues differs from their expression in tumor tissues, suggesting that deregulation of miRNA expression may play a key role in the progression and spread of many neoplasms by causing them to act as oncogenes and/or tumor suppressor genes [13, 15]. Previous studies have shown that the miRNA expression profile in GBM could be associated with prognosis and a distinct response to treatment, which may help to classify, to diagnose and to predict the clinical course of the patients [16,17,18,19,20,21], as well as to distinguish between pediatric and adult GBMs [12]. Moreover, their expression could be modulated and adapted to GBM therapy [7].
Herein, we identified miRNA signatures associated with glioblastoma in children and adults compared to non-neoplastic white matter (WM) samples using the miRNA microarray method. In vitro assays were performed with the two more differentially expressed microRNAs between GBM and non-neoplastic samples.
Materials and methods
Patients and samples
The study comprised primary GBM samples from 40 consecutive patients (31 adults and 9 children). The adult’s median age was 61.5 years (age range 19 to 74 years) and to children the median age was 9 years (range 3 to 18 years-old). Patients were diagnosed and treated at the University Hospital of the Ribeirão Preto Medical School, University of São Paulo (HCFMRP–USP) and at Centro Infantil Boldrini, Campinas, Brazil. The histopathological diagnosis was performed according to WHO criteria [4]. All patients were treated with surgery, radiotherapy and chemotherapy (temozolamide or lomostine based protocols). The 3-year overall survival of the patients enrolled in the study was 4.4 ± 4% for the adults and 22.2 ± 13.9% for the children.
Additionally, 11 non neoplastic white matter CNS tissue samples collected during temporal lobectomy in epilepsy surgeries were used as controls.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by the Ethics Committee of each institution (HC-FMRP-USP protocol number: 4643/2010; Centro Infantil Boldrini protocol number: 1.75-050809) and a signed statement of informed assent and consent was obtained from each adult patient and from the Legal Guardians of children, respectively.
RNA Extraction and cDNA Synthesis
Surgically resected fresh tissues were immediately snap-frozen in liquid nitrogen, stored at – 80 °C and microdissected prior to RNA extraction to exclude necrosis areas or adjacent normal tissue, with at least 80% of neoplastic cells enrichment in the analyzed areas. Extraction of total cellular RNA was made using TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The RNA was reverse transcribed to single-stranded cDNA using the high capacity kit (Applied Biosystems, Foster City, CA, USA).
Microarray analysis of miRNA expression
Initially 29 primary GBM (20 adults and 9 children) and 11 non neoplastic WM samples were used for microarray analysis. To determine the expression prolife of microRNAs the microRNA library was constructed using 100 ng of total RNA and investigated with the Agilent “Human microRNA Microarray Kit (v3)” (G4470C, Agilent Technologies, Santa Clara, CA, USA) that contains 866 human and 89 viral microRNA sequences, according to manufacturer’s instructions. Each miRNA species is printed 20 times with replicate probes on the array. The Feature Extraction Software v10.7.3.1 (Agilent Technologies, Santa Clara, CA, USA) was used to process the spot images. The AgiMicroRna AFE package (Agilent Feature Extraction) from the Bioconductor library was used for standardization, quality evaluation and statistical analysis of microarray data regarding differential expression of miRNA. Each miRNA microarray was analyzed with respect to background signal, signal intensity, as well as internal control blade and spike in controls. To determine the distribution of signals from each spot on each microarray and to check quality and sample integrity a box-plot was generated. To determine differential expression and the false discovery values were used to control the false-positive rate (FDR-False Discovery Rate) an empirical Bayesian approach was used. The 75% quartile method [22] was performed to normalization.
Functional annotation of miRNA targets
The Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was performed using the Genes Union function of the DIANA-mirPath v.3.0 web-server. p-values < 0.001 and validated miRNA interactions from the DIANA-TarBase were considered [23]. All miRNAs found to be differentially expressed in the microarray data (− 2 > fold change (FC) > + 2 and adjusted p < 0.05) were evaluated.
MicroRNA analysis by qRT-PCR
Among the most differentially expressed microRNAs detected by microarray, five were chosen for analysis in 40 GBM samples, including those analyzed by microarray, being with 3 compared between GBM versus non-neoplastic white matter (miR-21-3p, miR 34a-5p and miR-630) and 2 between pediatric versus adult samples GBM (miR-10b-5p and miR-142-5p).
The analyses were made by qRT-PCR using Taqman microRNA assays (Applied Biosystems Inc., Foster City, CA, USA) according to the manufacturer’s recommendations. As references were used the noncoding small nuclear RNA RNU6B and RNU19. Primers were available upon request (Applied Biosystems Inc., Foster City, CA, USA—Supplemental Table 1). The \( 2^{{ - \Delta \Delta {\text{CT}}}} \) method was used to determinate the relative expression using the mean of non-neoplastic white matter control samples as calibrator [24] and the Fold Change expression was calculated by the ratio between the expression values of two groups.
The functional assays were performed for the two miRNAs most differentially expressed in the samples determinate by qRT-PCR, i.e., hsa-miR-630 (the most underexpressed) and hsa-miR-10b-5p (the most overexpressed).
Cell culture
Expression of miR-630 and miR-10b was evaluated in the GBM cell lines KNS42, MO59K, U87, T98G, U138, SF188, U251, LN319 and U343. The cell lines MO59K, U87, T98G, U25 and U343 were purchased from the American Type Culture Collection (ATCC, Rockville, MS, USA). SF188 (pediatric GBM) was kindly provided by Drs. Damien Faury and Nada Jabado (McGill University, Canada) and LN319 by Dr. Frank Furnari (Ludwig Institute for Cancer Research, La Jolla, CA, USA). The GBM pediatric cell line, KNS42, was acquired from the Japan Cell Bank (JCRB Cell Bank—National Institute of Biomedical Innovation). The cell lines were cultured in Ham/F10 medium supplemented with 100 mg/ml streptomycin, 1% glutamine, 100 U/ml penicillin and 10% fetal bovine serum (GIBCO FBS qualified Brazil, ThemoFisher Scientific, Brazil), pH 7.2–7.4, in a humidified atmosphere containing 5% CO2 at 37 °C. All cell lines were authenticated by STR DNA analysis to avoid misidentification or cross-contamination.
Quantitative real time polymerase chain reaction (qRT-PCR)
RNA was extracted with TRIZOL (Invitrogen Inc, Carsdab, CA, USA) and reverse transcribed to single-stranded cDNA using a High Capacity Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. Relative levels of miRNA were determined by real-time quantitative PCR using the primers for hsa-miR-630 and hsa-miR-10-5p in an ABI PRISM™ 7500 Sequence Detection System (Life Biosciences—Applied Biosystems, Carlsbad, CA, USA). Data were normalized using RNU6B and RNU19 as endogenous housekeeping controls and analyzed by the \({2}^{{ - \Delta \Delta {\text{C}}_{{\text{T}}} }}\) method [24]. A human non-neoplastic white matter sample was used as calibrator.
Functional studies
Lentiviral shRNA Transduction, Proliferation and Cell Population Doubling Time, Clonogenic Assay, Invasion Assay, Migration Assay and In silico Analysis were performed as described in Supplemental Method.
Statistical analysis
Differences in miRNA expression levels between sample groups were determined by the Mann–Whitney test. Overall survival (OS) was calculated by Kaplan–Meier curves and log-rank test. Statistical analysis of functional data was performed by the Student t-test and by one-way or two-way ANOVA followed by Bonferroni’s test, as appropriate. Results are presented as means ± SD. Data analysis was carried out using the IBM® SPSS® Statistics software v.20 (SPSS Inc, IL, USA). The level of significance was set at p < 0.05 in all analyses. To validate our results we analyzed the differential expression of miR-10b and miR-630 in a different cohort utilizing a data set from the GSE90603 and GSE42657 studies using the GEO2R program (https://www.ncbi.nlm.nih.gov/geo/geo2r/), Different datasets were also used to compare the expression of miR-10b and miR-630 with their targets as described in the literature (Supplemental Method).
Results
miRNAs signatures distinguish GBM from cerebral WM
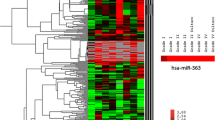
An unsupervised hierarchical clustering analysis distinguished two groups of GBM samples and one group consisting only of cerebral WM (Fig. 1a). We identified a total of 37 miRNAs differentially expressed between GBM and WM samples (fold change range − 5.2 to + 6.7 adjusted p < 0.05), including 13 underexpressed and 24 overexpressed miRNAs (Table 1). Overexpressed miRNAs were closely related to genes of important signaling pathways such as pathways in cancer (hsa05200), transcriptional misregulation in cancer (hsa05202), viral carcinogenesis (hsa05203), regulation of actin cytoskeleton (hsa04810), insulin signaling pathway (hsa04910), and hippo signaling pathway (hsa04390) (Fig. 2a). The underexpressed miRNAs were found to regulate genes involved in pathways in cancer (hsa05200), focal adhesion (hsa04510), transcriptional misregulation in cancer (hsa05202), hippo signaling pathway (hsa04390), signaling pathways regulating pluripotency of stem cells (hsa04550), and cell cycle (hsa04110) (Supplemental Fig. 1a).
a Heatmap of unsupervised hierarchical clustering analysis showing all samples evaluated by microarray and the significant differentially expressed miRNAs between GBM (adult and pediatric) and non-neoplastic white matter. Twenty four GBMs were arranged in a distinct cluster of the white matter samples. Five GBMs were arranged in the same cluster of WM samples, but in a distinct subgroup. b Heatmap of unsupervised hierarchical clustering analysis was able in distinguish two groups of GBM samples, one composed preferentially by pediatric patients and another one by adult patients
a The miR-630 relative expression in glioblastoma cell lines KNS42, LN319, U343, SF198, T98G, MO59K, U138, U251 and U87 and miR-630 expression after lentiviral overexpression in U87 cell line. MiR-630 expression in glioblastoma cell lines was determined by qRT-PCR. The dashed line represents the average of the white matter (WM) sample used as the calibrator. b miR-630 overexpression was confirmed by qRT-PCR in U87 cell line and was used as control the respective cell line transfected with the miRNA negative control (miR-NC)
miRNA profiles differ between adult and pediatric GBMs
We also sought to investigate whether the miRNA expression profile differs between adult and pediatric GBM samples. An unsupervised hierarchical clustering analysis was able in distinguish two groups of GBM samples, one composed preferentially by pediatric patients and another one by adult patients (Fig. 1b). In a supervised analysis we detected a total of 16 miRNAs differentially expressed between these two groups including 14 overexpressed and only 2 underexpressed miRNAs (FC range − 2.4 to 7.2, adjusted p < 0.05) (Supplemental Table 2). Functional analysis by KEGG annotations of the differentially expressed miRNAs showed that the upregulated ones are involved in pathways such as TGF-beta signaling pathway (hsa04350), p53 signaling pathway (hsa04115), adherens junction (hsa04520), and hippo signaling pathway (hsa04390). In contrast, the only two underexpressed miRNAs were associated with cell cycle (hsa04110), hippo signaling pathway (hsa04390), fatty acid metabolism (hsa01212), fatty acid biosynthesis (hsa00061), and steroid biosynthesis (hsa00100) (Supplemental Fig. 1b).
qRT-PCR analysis
In order to analyze the expression profile detected in the microarray analysis we selected five miRNAs found to be differentially expressed in order to evaluate their expression by qRT-PCR in 40 samples of GBM, including the specimens studied by microarray. With the exception of miR-21-3p, GBM samples showed significant differential expression of miR-10b-5p, miR-34a-5p and miR-630 when compared to non-neoplastic WM. Among them, miR-10b-5p and miR-630 were the most overexpressed and underexpressed miRNAs, respectively (Supplemental Table 3 and Supplemental Fig. 2a). Similar results were observed when the miR-10b-5p and miR-630 expression were analyzed in the in different cohort utilizing the data set from the GSE90603 e GSE42657 studies (Supplemental Fig. 3). Adult GBM samples showed significant overexpression of miR-10b-5p and miR-142-5p when compared to children’s GBM (Supplemental Fig. 2b).
In silico analysis
Although molecular GBM subgroups analysis were not performed in the present study, we analyzed the expression profile of miR-10b and miR-630 using the TCGA-GBM dataset composed by 563 GBM cases (553 adults and 10 pediatrics) according to molecular classification and gene mutational status (https://doi.org/10.7908/C17080T1). The miR-10b showed a significant lower expression level in the mesenchymal subgroup and showed also differences according the mutation status of the genes TP53, IDH1, PTEN, EGFR and RB1. The miR-630 showed a significant higher expression level in the mesenchymal subgroup, with significant differences according the mutation status of the genes PTEN and EGFR (Supplemental Fig. 4).
Two-years overall survival (OS) was calculated according to the median values of expression to miR-10b and miR-630. We observed significant lower OS to the patients that presented a miR-10b expression higher the median compared with those with lower expression: 4.8 ± 4.6% versus 22.7 ± 4.6% (p = 0.023). No differences in OS was observed to miR-630 (p = 0.914). Using the TCGA-GBM dataset (https://doi.org/10.7908/C17080T1), a lower expression of miR-10b and a higher expression of miR-630 were associated with lower OS (p = 0.049 and p = 0.027, respectively) (Supplemental Fig. 5).
miR-630 and miR-10b-5p expression profiles in GBM cell lines
According to the profiles observed in GBM samples, the majority of cell lines (7 out of 9) showed low miR-630 expression compared to the control sample (WM) (Fig. 2a). The U87 cell line showed the lowest expression level and was selected for lentiviral transfection to induce miR-630 expression. After selection with puromycin, the levels of miR-630 was increased by > 6000 × (Fig. 2b).
Additionally, miR-10b-5p expression was evaluated in the 9 GBM cell lines, which showed high expression compared to control (Fig. 3a). The U251 cell line showed the highest level and was selected for lentiviral transfection in order to inhibit the miR-10b-5p. After selection with puromycin, the levels of miR-10b-5p was decreased by > 80% (Fig. 3b).
a The miR-10b expression in glioblastoma cell lines KNS42, LN319, U343, SF198, T98G, MO59K, U138, U251 and U87 and miR-10b expression after lentiviral knockdown in U251 cell line. The miR-10b expression in glioblastoma cell lines was determined by qRT-PCR. The dashed line represents the average of the white matter (WM) sample used as the calibrator. b miR-10b inhibition was confirmed by qRT-PCR in U251 cell line and was used as control the respective cell line transfected with the miRNA negative control (miR-NC)
Overexpression of miR-630 and Inhibition of miR-10b-5p reduced cell functional roles associated with tumor progression
In order to evaluate the functional roles of miR-630 and miR-10b-5p in GBM, in vitro assays were performed after the induction of miR-630 overexpression in the U87 cell line and the inhibition of miR-10b-5p in the U251 cell line. Overexpression of miR-630 reduced cell proliferation and increased cellular doubling time compared to negative controls (Fig. 4a). In addition, the mir-630 overexpressed cell line showed lower rates of cell invasion (p < 0.05) and migration (non-significant), (Fig. 4b, c). Interestingly, in silico analysis showed that miR-630 was mainly involved in pathways related to proliferation, migration and invasion of tumor cells (Supplemental Fig. 6a), as confirmed by the cellular processes regulated by the most relevant miR-630 target genes (Supplemental Fig. 6b). Using again the TCGA-GBM (https://doi.org/10.7908/C17080T1) we correlate the expression profile of the target genes described to this miR and observed a significant correlation with the genes BCL2, CD200, CCN1, MTDH, SNAI2, TGFBR2 and YAP1 (Supplemental Fig. 7). To correlate the expression profile of the described miR-630 target genes in the pediatric GBM, we use the Pfister dataset (https://doi.org/10.1016/j.ccr.2012.08.024) with 44 GBM samples (27 pediatric GBM and 17 adult GBM samples). Only the CD200 gene presented a significant correlation in this dataset.
Effects of miR-630 overexpression in U87 cell line determined by functional assays. a Proliferation and doubling time of population cells after miR-630 overexpression in U87. b Number of invaded cells comparing U87 mir-630 overexpressed to U87 negative control (miR-NC). c Migration rate of U87 cells after miR-630 overexpression compared to time 0 h. *p < 0.05, **p < 0.01, ***p < 0.001
The miR-10b inhibition in the U251 cell line decreased cell proliferation and increased cellular doubling time (Fig. 5a) and reduced the number of colonies formed compared to control (Fig. 5b). A lower, though non-significant, cell invasion rate was also observed (Fig. 5c). In silico analysis showed that miR-10b-5p is mainly involved in cancer pathways (Supplemental Fig. 8a) and the most relevant miR-10b-5p target genes are related to cell cycle, proliferation and apoptosis (Supplemental Fig. 8b). Using the TCGA-GBM dataset (https://doi.org/10.7908/C17080T1) we correlate the expression profile of the target genes described to this miR and observed a significant correlation with the genes ACLY, AXL, CDK2, CDKN1A, FHL2, NOTCH1, PAX6, SLC2A3, STAT6 and TFP1 (Supplemental Fig. 9). To correlate the expression profile of the target gene in the pGBM, we use the Pfister database (https://doi.org/10.1016/j.ccr.2012.08.024). Only the CDKN2A gene presented a significant correlation.
Effects of miR-10b inhibition in U251 cell line determined by functional assays. a Cell proliferation after miR-10b inhibition in U251. b Number of colonies comparing U251 shRNA miR-10b to negative control (miR-NC). c Number of invaded U251 cells after miR-10b inhibition compared to control. *p < 0.05, **p < 0.01
Discussion
GBMs are the major representatives of high grade gliomas, being considered grade IV neoplasms according to the WHO [4], and still have a very dismal prognosis [25]. Described in the early 1990s, miRNAs form a relatively new group of non-coding molecules that play an essential role in the regulation of various physiological and pathological processes, acting as post-transcriptional regulators [13, 25, 26]. Their expression has the potential to be modulated and shows a promising therapeutic application in GBM [27].
In the present study we observed a differential pattern of microRNA expression in GBM when compared to non-neoplastic white matter controls, aligned with a different expression profile between GBM in adult patients versus children. Differential microRNA expression between GBM and non-neoplastic brain controls has been described [28,29,30] and, although few studies have analyzed microRNA expression in pediatric high grade gliomas [12, 31, 32], a distinct microRNA expression profile between GBM of children and adults has also been shown [12]. Around 50% of the miRNAs differentially expressed between GBM versus WM and between adult versus pediatric GBM and found in the present study were also described in the above studies.
According to microarray analysis, the subset of the most overexpressed miRNAs found in GBM samples suggests hyper-regulation of important pathways associated with cancer such as pathways in cancer, transcriptional misregulation, viral carcinogenesis, regulation of actin cytoskeleton, insulin signaling pathway, and hippo signaling pathway. Similarly, the underexpressed miRNAs were also found to regulate pathways such as pathways in cancer, focal adhesion, transcriptional misregulation in cancer, hippo signaling pathway, pathways regulating pluripotency of stem cells, and cell cycle. Comparison of KEGG pathways between GBM and non-neoplastic brain tissue have been described, with pathway in cancer being found to be the most frequently deregulated one [33].
In this study, we found low expression of miR-630 in GBM tumor samples and GBM cell lines when compared to WM. A lower expression of this miR was also observed in silico analysis using the data of the GSE90603 e GSE42657 studies. Functional assays showed that its overexpression by lentiviral shRNA transduction reduces cell proliferation and invasion. miR-630 plays dual roles in tumor behavior, acting as an oncogene by promoting tumor progression, with a consequent poor prognosis in renal cell, gastric, colorectal and ovarian cancers [34,35,36,37,38]. In the present study, miR-630 was found to have a tumor suppressor role in GBM, in agreement with other studies showing that miR-630 inhibits tumor progression and metastasis in breast, esophageal squamous cell and lung cancer [36, 39,40,41]. Moreover, it was shown that anti-miR-630 enhances the radio-resistance of human glioblastoma cells by targeting CDC14, suggesting that miR-630 can be involved in the response to radiation treatment in GBM [42]. Among the various target genes, miR-630 has been linked to important signaling pathways such as TGFβ [35], Wnt [43], ERK [36], IGF1R [44, 45] and BCL-2 [36], that regulate relevant cell mechanisms such as proliferation, apoptosis and epithelial-mesenchymal transition. We also performed in silico analysis using the TCGA-GBM dataset and we found significant correlation between the miR-630 and some genes of the pathways described above including the genes BCL2, CD200, CCN1, MTDH, SNAI2, TGFBR2 and YAP1.
Regarding miR-10b-5p, we found significant high levels in patients’ samples with GBM and also in GBM cell lines, in agreement with other studies on GBM [46,47,48]. A higher expression of this miR in GBM when compared to WM was also observed in silico analysis using the data of the GSE90603 and GSE42657 studies. This microRNA is not expressed in human brain and has been found to be overexpressed not only in high grade gliomas but also in low grade gliomas such as pilocytic astrocytoma [47,48,49]. Furthermore, miR-10b expression levels seem to be directly associated with the degree of malignancy, being much higher in GBMs than in other gliomas [47, 50]. In agreement with the present study, which showed that miR-10b inhibition decreases cell proliferation and colony formation, cellular and murine models have associated miR-10 inhibition with the reduction of tumor growth and invasion [48, 50]. Additionally, miR-10b has been reported to be a regulator of invasion and metastasis processes by targeting the genes PLAUR, RHOC, HOXD10, MMP14, alpha- and beta-integrin [47, 50], and to regulate proliferation and apoptosis by direct targets BCL2L11, CDKN1A, CDKN2A, TFAP2C [48]. Recently, El Fatimy et al. (2017) performed miR-10b ablation using CRISP-Cas9 and showed that this process can be lethal to glioma cell cultures and intracranial GBM models [51]. In silico analysis using the TCGA-GBM dataset showed significant correlation between the miR-10 with the genes ACLY, AXL, CDK2, CDKN1A, FHL2, NOTCH1, PAX6, SLC2A3, STAT6 and TFP1, some of them involved in the biological processes described above.
The main limitation of the present study was the lack of molecular classification of the analyzed samples. Despite the overexpression of miR-10b was observed in all of the GBM cell lines analyzed and the lower expression of miR-630 in 7 of 9 GBM cell lines, that represent different pediatric and adults GBM molecular subgroups, it is important to point that is not possible to confirm that these differential expression could be act as a general mechanism of gene regulation in GBM and this data needs to be viewed with caution. It is interesting to point that in silico analysis using TCGA-GBM dataset showed a differential expression in both miR-10b-5p and miR-630 in the mesenchymal subgroup. In this data set it was not possible to compare these expression levels with non-neoplastic brain white matter.
In summary, the present study highlights distinct miR signatures of adult and pediatric GBM, as well as important biological and functional roles of miR-10b-5p and miR-630 for the pathobiology of these lethal tumors.
Data availability
If the data used in this study are required, they can be available from the corresponding author.
References
Stupp R, Mason WP, van den Bent MJ et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996
Stupp R, Taillibert S, Kanner A et al (2017) Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 318(23):2306–2316
Orringer D, Lau D, Khatri S et al (2012) Extent of resection in patients with glioblastoma: limiting factors, perception of resectability, and effect on survival. J Neurosurg 117(5):851–859
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Ostrom QT, Gittleman H, Liao P et al (2014) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol 16(Suppl 4):iv1–iv63
Seystahl K, Wick W, Weller M (2016) Therapeutic options in recurrent glioblastoma—an update. Crit Rev Oncol Hematol 99:389–408
Anthiya S, Griveau A, Loussouarn C et al (2018) MicroRNA-based drugs for brain tumors. Trends Cancer 4(3):222–238
Areeb Z, Stylli SS, Koldej R et al (2015) MicroRNA as potential biomarkers in glioblastoma. J Neurooncol 125(2):237–248
Filippini G, Falcone C, Boiardi A et al (2008) Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol 10(1):79–87
Krex D, Klink B, Hartmann C et al (2007) Long-term survival with glioblastoma multiforme. Brain 130(Pt 10):2596–2606
Paugh BS, Qu C, Jones C et al (2010) Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol 28(18):3061–3068
Miele E, Buttarelli FR, Arcella A et al (2014) High-throughput microRNA profiling of pediatric high-grade gliomas. Neuro Oncol 16(2):228–240
Catania A, Maira F, Skarmoutsou E, D'Amico F, Abounader R, Mazzarino MC (2012) Insight into the role of microRNAs in brain tumors (review). Int J Oncol 40(3):605–624
Lu J, Getz G, Miska EA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev Biol 302(1):1–12
Kim TM, Huang W, Park R, Park PJ, Johnson MD (2011) A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 71(9):3387–3399
Parker NR, Correia N, Crossley B, Buckland ME, Howell VM, Wheeler HR (2013) Correlation of microRNA 132 up-regulation with an unfavorable clinical outcome in patients with primary glioblastoma multiforme treated with radiotherapy plus concomitant and adjuvant temozolomide chemotherapy. Transl Oncol 6(6):742–748
Henriksen M, Johnsen KB, Olesen P, Pilgaard L, Duroux M (2014) MicroRNA expression signatures and their correlation with clinicopathological features in glioblastoma multiforme. Neuromolecular Med 16(3):565–577
Malzkorn B, Wolter M, Liesenberg F et al (2010) Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 20(3):539–550
Low SY, Ho YK, Too HP, Yap CT, Ng WH (2014) MicroRNA as potential modulators in chemoresistant high-grade gliomas. J Clin Neurosci 21(3):395–400
Huang SW, Ali ND, Zhong L, Shi J (2018) MicroRNAs as biomarkers for human glioblastoma: progress and potential. Acta Pharmacol Sin 39(9):1405–1413
Meyer SU, Kaiser S, Wagner C, Thirion C, Pfaffl MW (2012) Profound effect of profiling platform and normalization strategy on detection of differentially expressed microRNAs—a comparative study. PLoS ONE 7(6):e38946
Vlachos IS, Zagganas K, Paraskevopoulou MD et al (2015) DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43(W1):W460–466
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408
Wang Y, Jiang T (2013) Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett 331(2):139–146
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Hummel R, Maurer J, Haier J (2011) MicroRNAs in brain tumors: a new diagnostic and therapeutic perspective? Mol Neurobiol 44(3):223–234
Ciafrè SA, Galardi S, Mangiola A et al (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334(4):1351–1358
Rao SA, Santosh V, Somasundaram K (2010) Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol 23(10):1404–1417
Chen W, Yu Q, Chen B, Lu X, Li Q (2016) The prognostic value of a seven-microRNA classifier as a novel biomarker for the prediction and detection of recurrence in glioma patients. Oncotarget 7(33):53392–53413
Jha P, Agrawal R, Pathak P et al (2015) Genome-wide small noncoding RNA profiling of pediatric high-grade gliomas reveals deregulation of several miRNAs, identifies downregulation of snoRNA cluster HBII-52 and delineates H3F3A and TP53 mutant-specific miRNAs and snoRNAs. Int J Cancer 137(10):2343–2353
Liang ML, Hsieh TH, Ng KH et al (2016) Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget 7(15):19723–19737
Zhu XP, Mou KJ, Xu QF et al (2015) Microarray analysis of the aberrant microRNA expression pattern in gliomas of different grades. Oncol Rep 34(1):318–324
Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y (2016) miR-630 functions as a tumor oncogene in renal cell carcinoma. Arch Med Sci 12(3):473–478
Chen WX, Zhang ZG, Ding ZY et al (2016) MicroRNA-630 suppresses tumor metastasis through the TGF-β- miR-630-Slug signaling pathway and correlates inversely with poor prognosis in hepatocellular carcinoma. Oncotarget 7(16):22674–22686
Chen MJ, Wu DW, Wang GC, Wang YC, Chen CY, Lee H (2018) MicroRNA-630 may confer favorable cisplatin-based chemotherapy and clinical outcomes in non-small cell lung cancer by targeting Bcl-2. Oncotarget 9(17):13758–13767
Chu D, Zhao Z, Li Y et al (2014) Increased microRNA-630 expression in gastric cancer is associated with poor overall survival. PLoS ONE 9(3):e90526
Chu D, Zheng J, Li J et al (2014) MicroRNA-630 is a prognostic marker for patients with colorectal cancer. Tumour Biol 35(10):9787–9792
Jin L, Yi J, Gao Y et al (2016) MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 48(9):810–819
Zhou CX, Wang CL, Yu AL et al (2016) MiR-630 suppresses breast cancer progression by targeting metadherin. Oncotarget 7(2):1288–1299
Song YF, Hong JF, Liu DL, Lin QA, Lan XP, Lai GX (2015) miR-630 targets LMO3 to regulate cell growth and metastasis in lung cancer. Am J Transl Res 7(7):1271–1279
Zhang L, Wang C, Xue ZX (2017) Inhibition of miR-630 enhances the cell resistance to radiation by directly targeting CDC14A in human glioma. Am J Transl Res 9(3):1255–1265
Li D, Tian B, Jin X (2018) miR-630 inhibits epithelial-to-mesenchymal transition (EMT) by regulating the Wnt/β-catenin pathway in gastric cancer cells. Oncol Res 27(1):9–17
Corcoran C, Rani S, Breslin S et al (2014) miR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol Cancer 13:71
Farhana L, Dawson MI, Murshed F, Das JK, Rishi AK, Fontana JA (2013) Upregulation of miR-150* and miR-630 induces apoptosis in pancreatic cancer cells by targeting IGF-1R. PLoS ONE 8(5):e61015
Teplyuk NM, Uhlmann EJ, Wong AH et al (2015) MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget 6(6):3770–3783
Gabriely G, Yi M, Narayan RS et al (2011) Human glioma growth is controlled by microRNA-10b. Cancer Res 71(10):3563–3572
Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E (2009) MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 125(6):1407–1413
Darrigo Júnior LG, Lira RCP, Fedatto PF et al (2019) MicroRNA profile of pediatric pilocytic astrocytomas identifies two tumor-specific signatures when compared to non-neoplastic white matter. J Neurooncol 141(2):373–382
Sun L, Yan W, Wang Y et al (2011) MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res 1389:9–18
El Fatimy R, Subramanian S, Uhlmann EJ, Krichevsky AM (2017) Genome editing reveals glioblastoma addiction to microRNA-10b. Mol Ther 25(2):368–378
Funding
This study was supported by the Brazilian Public Research Agencies: Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Grant Number: 2010/07020-9) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Grant Number: 471885/2013-4).
Author information
Authors and Affiliations
Contributions
LGDJ, MB, LGT and CAS designed the project; LGDJ, MB, RCPL, ST, PFF, VSS, VKS, LCV, RAP and DSMA conducted the experiments and microarray analysis; LGDJ, MB, MB, JAY, SRB, SSA, LN, RSO, HRM, CGC, LGT, ETV, CAS were responsible for acquisition, analysis, or interpretation of data; LGDJ, MB, ETV and CAS wrote the manuscript. All authors revised and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Junior, L.G.D., Baroni, M., Lira, R.C.P. et al. High-throughput microRNA profile in adult and pediatric primary glioblastomas: the role of miR-10b-5p and miR-630 in the tumor aggressiveness. Mol Biol Rep 47, 6949–6959 (2020). https://doi.org/10.1007/s11033-020-05754-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05754-3