Abstract
Over 10% of genetic diseases are caused by mutations that introduce a premature termination codon in protein-coding mRNA. Nonsense-mediated mRNA decay (NMD) is an essential cellular pathway that degrades these mRNAs to prevent the accumulation of harmful partial protein products. NMD machinery is also increasingly appreciated to play a role in other essential cellular functions, including telomere homeostasis and the regulation of normal mRNA turnover, and is misregulated in numerous cancers. Hence, understanding and designing therapeutics targeting NMD is an important goal in biomedical science. The central regulator of NMD, the Upf1 protein, interacts with translation termination factors and contextual factors to initiate NMD specifically on mRNAs containing PTCs. The molecular details of how these contextual factors affect Upf1 function remain poorly understood. Here, we review plausible models for the NMD pathway and the evidence for the variety of roles NMD machinery may play in different cellular processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eukaryotic cell complexity has increasingly become more intricate as the latest research proposes many new models and mechanisms for a variety of cellular functions. With approximately 21,000 protein-encoding genes and up to around one million possible proteins, much research has been focused on the functions and structures of these proteins [1]. There are a variety of different types of proteins, including, but not limited to, enzymes, transport proteins, and motor proteins. Each of these proteins has an essential role in ensuring each living thing and all of its cells are healthy and functioning properly. What may seem to be a minor defect or mutation in a single protein, can actually lead to a disease state.
Lots of cytoplasmic surveillance mechanisms for monitoring mRNA translation exist in eukaryotes that maintain the quality of gene expression, including nonsense-mediated mRNA decay (NMD), no-go decay (NGD), and non-stop decay (NSD). NMD works on a multitude of mRNAs, including those newly synthesized and already existing, and degrades those containing a premature termination codon (PTC), thereby stopping the production of truncated proteins that could result in human diseases [2]. NGD works on mRNAs containing modified nucleobases and those in which the codon-anticodon interaction is interfered with. These mRNA modifications damage the decoding process and cause the ribosome to ‘not go’ [3]. Certain abnormal mRNA structures including hairpins and pseudoknots also stall normal translation [4]. NSD works on truncated mRNA transcripts that lack a stop codon, which causes the ribosomes to consistently move to the end of the mRNA and stall [5].
The “nonsense” part of NMD refers to a codon that causes termination of translation. It was originally thought that the sole purpose of NMD was to degrade messenger RNAs (mRNAs) that contain a premature translation–termination codon [6]. These mRNAs with PTCs arise from failure to remove exons with nonsense mutations in coding regions, pseudogenes, alternative splicing events, and more [7,8,9]. However, several other cellular targets have been found, including transcripts that have no functional value, such as noncoding RNAs, intergenic transcripts, and even bicistronic mRNAs and certain transposable elements’ transcripts [7, 10,11,12]. Furthermore, the steady state levels of approximately 10% of mRNAs are regulated by NMD [13]. Depending on the conditions of the cell, NMD can downregulate different transcripts in order to meet the demands of the cell.
A translation termination codon is usually recognized to be premature if it is located between 50 and 55 nucleotides upstream of the last exon–exon junction, since that is when translation termination directs NMD instead of normal termination [14,15,16]. If a protein is produced from an mRNA transcript with a PTC, then there is an increased likelihood that the protein will be shortened and not function properly [17]. Therefore, mRNAs that are able to escape from NMD can lead to the production of aberrant proteins that contribute to a variety of diseases and illnesses in humans.
During the process of NMD, three proteins that play critical roles are the up-frameshift protein 1, 2 and 3 (Upf1, 2 and 3). The Upf proteins were initially discovered in Saccharomyces cerevisiae [18] and later in higher eukaryotes including Caenorhabditis elegans [19], Drosophila melanogaster [20] and humans [21]. Upf1, also known as regulator of nonsense transcripts 1 (RENT1) or suppressor with morphogenetic defects in genitalia 2 (SMG2), is a highly conserved phosphoprotein possessing nucleic acid-dependent ATPase and 5′-to-3′ helicase activities [19, 21]. The ATP-binding and hydrolysis abilities of Upf1 are critical for assembling with the 40S ribosomal subunit [22] (Min et al., 2013), disassociating from a terminating mRNP [23], releasing a peptide, and the recycling of protein synthesis components and NMD machineries [24]. Upf2 is a molecular bridge between Upf1 and Upf3 [25], and its binding to Upf1 changes Upf1 from RNA clamping to RNA unwinding and stimulates Upf1’s RNA helicase ability [26]. The central RRM (RNA recognition motif) domain of Upf3 links to Upf2 and its C-terminal domain connects to the exon–junction complex (EJC) at the 3′UTR [27, 28]. Upf1 and Upf2 concentrate in the cell cytoplasm, while Upf3 shuttles between cytoplasm and nucleus [29]. The Upf proteins interact with each other and other NMD-related factors, including Smg proteins, release factors, and exon junction complex proteins, for eliciting NMD [30]. However, their exact mechanistic roles in NMD still remain to be clarified.
Previous researchers have showed that mutations of human NMD-related genes, especially the Upf genes, can lead to serious neurodevelopmental disorders and tumors. The somatic mutations in the Upf1 gene is the unique molecular hallmark of pancreatic adenosquamous carcinoma (ASC) [31]. Loss-of-function mutations in Upf3B cause variable clinical symptoms such as intellectual disability (ID) [32,33,34], autism [35], attention deficit hyperactivity disorder (ADHD) [36] and schizophrenia [35, 37, 38]. Moreover, nonsense mutations in human genes results in disease phenotypes. Daar and Maquat showed an anemia-inducing mutation, creating a nonsense codon, in the gene for triosephosphate isomerase (TPI) led to the initiation of NMD and formation of mRNA with a reduced cytoplasmic half-life [39]. It was thus concluded that premature translation termination could mediate TPI deficiency. A nonsense mutation of the fibrillin-1 (FBN1) gene induces NMD and disrupts in-frame exon skipping of FBN1 exon 51, which mediate Marfan syndrome occurrence [40].
Due to these resulting disease states, much research has been done to understand the biology of NMD, with the eventual goal of bringing a platform therapy to patients. Though much progress has been made, many unanswered questions remain.
Upf proteins
Upf1
Upf1’s role in the cell was initially looked at in the 1980s; however, it wasn’t until the 1990s that researchers began to take notice of its importance. One of the earliest papers in the 1990s that brought attention to this protein had found that the protein product encoded by the UPF1 gene was necessary for rapid degradation of mRNAs with premature translation codons in yeast, though their understanding of the mechanisms behind this was minimal, if any [41]. Today, the UPF1 gene has been found to be conserved in numerous organisms: chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, fruit fly, mosquito, C. elegans, S. cerevisiae, K. lactis, E. gossypii, S. pombe, M. oryzae, N. crassa, A. thaliana, frog, and others. Aliases for UPF1 include up-frameshift suppressor 1 homolog (hUpf1), nonsense mRNA reducing factor 1 (NORF1), regulator of nonsense transcripts 1 (RENT1), pNORF1, and suppressor with morphogenetic defects in genitalia 2 (SMG2).
Upf1 is part of the SF1 superfamily of RNA helicases based on amino acid sequence similarity and conservation of seven motifs that are common among those in SF1 [42]. The two major domains found in Upf1 are the SQ domain near the C-terminus and the CH domain near the N-terminus. The CH domain is rich in the cysteines and histidines. The CH domain has been found to be involved in inhibition of the ATPase activity of Upf1, and is involved in binding Upf2 [25, 43]. The CH domain has also been found to promote RNA binding by affecting the position of a β-barrel domain also known as domain 1B [26]. When Upf2 binds to Upf1, a conformational change in the CH domain promotes the helicase activity of Upf1 [26].
The SQ motif of Upf1 is rich in amino acids serine and glutamine, and contains critical phosphorylation sites [44]. These phosphorylation sites play an important role in promoting Upf1’s cellular activities and allowing other factors involved in NMD and other processes to bind to it [45]. In comparison to the CH domain, the SQ domain has been found to be less conserved evolutionarily in simpler eukaryotic organisms but more conserved in higher, complex eukaryotic organisms. Evolutionary conservation among different organisms suggests its critical function in the cell since over time, it is likely that those lacking the proper SQ domain have had lower genetic fitness and less reproductive success [46]. The SQ domain has been found to be able to inhibit the helicase activity of Upf1 through direct binding to the helicase domain, thereby having an inhibitory effect on ATP hydrolysis and RNA unwinding [46]. The helicase domain is also essential for Upf1’s function in NMD and other cell processes. The genetic structure of Upf1 protein is showed in Fig. 1a.
The genetic structures of Upf1, 2 and 3 proteins in humans. a Upf1 protein structure in humans. Diagram of the structure of the Upf1 protein with indications of different domains and motifs. Figure not to scale. Numbers positioned above and below indicate the amino acid positions. CH domain is rich in the cysteine and histidine amino acids. SQ motif is rich in the amino acids serine and glutamine and has crucial areas where Upf1 is phosphorylated. The helicase domain is essential for Upf1’s function in NMD and other cell processes. Upf2, eRF3, STAU1, NCBP1 (CBP80), and SMG-1–8–9 complex interaction sites with Upf1 are indicated as well. Important serine and threonine phosphorylation sites are indicated. Asterisk indicates amino acid positions for isoform 1 of Upf1 that contains 1129 amino acids. b Upf2 protein structure in humans. Diagram of the structure of the Upf2 protein with indications of different domains and motifs. Figure not drawn to scale. Numbers positioned above and below indicate the amino acid positions. C-terminus consists of a Upf1 binding domain. SQ motif is rich in the amino acids serine and glutamine and has crucial areas where Upf1 is phosphorylated. The helicase domain is essential for Upf1’s function in NMD and other cell processes. Upf2, eRF3, STAU1, NCBP1 (CBP80), and SMG-1–8–9 complex interaction sites with Upf1 are indicated as well. Important serine and threonine phosphorylation sites are indicated. Asterisk indicates amino acid positions for S. cerevisiae. c Upf3b protein structure in humans. Diagram of the structure of the Upf3b protein with indications of different domains and motifs. Figure not drawn to scale. Numbers positioned above and below indicate the amino acid positions. C-terminus consists primarily of an exon–junction binding domain. Ribonucleoprotein domain (RNP) or RNA recognition motif (RMM) that includes the site of Upf2 interaction is located at the N-terminus, with three proposed amino acid regions labeled #1, #2, and #3. There is a conserved region among several species in the Upf3 protein
There are two isoforms of the human Upf1 protein due to alternative splicing: isoform 1 is 1129 amino acids and isoform 2 is 1118 amino acids due to it missing amino acids 353–363 [47]. Staufen1 (STAU1) is a double stranded RNA-binding protein that interacts with Upf1 between amino acid 1 and 244 [48, 49]. Both Upf2 and eRF3 interact with Upf1 between amino acids 115 and 294 [49]. The CH domain starts at amino acid 115 and ends at amino acid 294 [26]. The helicase domain of Upf1 consists of the amino acids from 295 to 914, though it can interact with other parts of the protein [50]. The SQ domain starts at amino acid 915 and ends at amino acid 1118, though it has been found that the presence of amino acids 967–1019 is sufficient for this domain to exert its inhibitory effects on Upf1 [46]. Furthermore, the amino acids from 419 to 700 in Upf1 have been found to interact with CBP80 on newly synthesized mRNAs [51].
In isoform 2, phosphorylation at threonine residue 28 is required for Smg6 to bind to Upf1 [45]. Binding of the Smg-5:Smg-7 complex requires phosphorylation at serine residue 1096 in isoform 2, which corresponds to serine residue 1107 in isoform 1 [45, 52]. Furthermore, in isoform 2, it was found that the C-terminus region consisting of amino acid 985 to amino acid 1084 is critical for Smg-1:Smg-8:Smg-9 complex binding to Upf1 [52].
Upf2
Studies into the role of the UPF2 gene in cells date back to 1980, but the majority of studies regarding UPF2’s role in NMD began in the 1990’s. Papers from the 1990’s and 2000’s brought attention to the protein encoded by the UPF2 gene and its role in mRNA turnover and mRNA degradation along with the UPF1 protein. The human Upf2 protein (hUpf2 or just Upf2), also known as regulator of nonsense transcripts 2 (Rent2), has been found to be evolutionarily conserved: not only is it conserved in Schizosaccharomyces pombe, but it is also known as Upf2 or Upf2p in S. cerevisiae and known as Smg-3 in C. elegans [29, 53]. The human Upf2 protein is approximately 148 kDa and the gene encoding the protein is found on chromosome 10 [33, 54]. The Upf2 protein has been found to be primarily in the cytoplasm, but it has also been found to be present, though to a much smaller extent, in the nucleoplasm [55].
The structure and functions of the different domains in the Upf2 protein have become well elucidated over time. There are four core regions in its structure that all play important functional roles: three are middle portion of eukaryotic initiation factor 4-gamma (MIF4G) domains and one is the C-terminal region. The C-terminal region’s primary critical involvement in NMD is its role in binding to the CH domain of Upf1. Furthermore, between this C-terminal domain and the MIF4G-3 domain is an acidic region consisting primarily of the amino acids aspartate and glutamate [56]. The MIF4G-3 domain has been found to interact with Upf3b’s RNA recognition motif (RRM) [57]. Furthermore, Smg1 is able to bind non-competitively to the MIF4G-3 domain at the same time as Upf3b [58].
The MIF4G-1 and MIF4G-2 domains play more of a structural or scaffolding role in the Upf2 protein and are necessary for proper NMD to occur. When assembling the Upf–EJC complex in NMD, those two domains play an irreplaceable role in positioning and supporting the EJC, as they are the key linkers between the EJC, CH-domain of Upf1, and the MIF4G-3 domain of Upf2 [58]. MIF4G-1 and MIF4G-2 are not necessarily required for triggering the phosphorylation of Upf1 [58].
In humans, the MIF4G-1 domain extends from amino acids 121–429; however, only amino acids 168–364 are part of the typical ten helices that all MIF4G domains are supposed to have [13, 59]. Similarly, the MIF4G-2 domain extends from amino acid 457 to 757, but amino acids 569–758 are part of the typical ten helices [60]. Likewise, the MIF4G-3 domain starts at amino acid 768 and ends at 1015, but only the amino acids from 773 to amino acid 986 are part of the ten helices [54]. At the C terminal, amino acids 1105–1207 are involved in Upf1 binding [58]. The region between MIFG-3 and the C-terminal Upf1 binding domain is an acidic domain, ranging from amino acid 1016–1104 [61]. Several important phosphorylation sites for NMD near the N-terminal on the Upf2 protein in Saccharomyces cerevisiae include aspartic acid-31, serine-32, lysine-35, and arginine-36 [62]. The genetic structure of Upf2 protein is showed in Fig. 1b.
Upf2 plays a role not only in NMD, but also possibly in several other processes in the cell. Impairments in its function in NMD and other cellular functions can lead to a variety of phenotypic effects. One such crucial process involves fetal liver development, which is impaired upon deletion of Upf2 most likely due to its role in NMD. Furthermore, though the exact role in adult livers is not known, loss of Upf2 results in its functional deterioration and hepatic steatosis, indicating Upf2’s involvement in lipid metabolism with other NMD machinery. Loss of Upf2 by hematopoietic-specific deletion results in extinction of all hematopoietic stem cells, though this effect does not occur to the same extent in already differentiated cells, indicating Upf2’s essential role in early proliferating cells through its involvement in NMD [63]. Additionally, Sertoli cell-specific knockout of UPF2 indicated Upf2’s significant role in male fertility and testicular development, as sterility and impairment in the development of the testes were observed due to Upf2’s absence from NMD [64]. Moreover, in regards to male germ cells, Upf2 has been found to be critical for enriching mRNA transcripts with short 3′ UTRs and degrading transcripts with long 3′ UTRs, a process that is essential for proper spermatogenesis and male fertility. Upf2’s role in this enrichment of short 3′ UTRs involves other NMD machinery but is separate from its typical role of degrading transcripts with PTCs [65]. Upf2, in addition to Upf1, is necessary for proper development and viability through its role in NMD, as seen in Drosophila melanogaster. Moreover, copy number variants in the UPF2 gene have been linked to several disorders involving neurodevelopment, such as thrombocytopenia with absent radius (TAR) syndrome [34].
Upf3
Likewise to Upf1 and Upf2, Upf3 was discovered and investigated starting in a similar era, with publications on Upf3 in Saccharomyces cerevisiae starting to arise in the 1990s [41]. Today, the UPF3 gene is most noted for its role in nonsense mediated decay, and it has been found in many organisms including, but not limited to, humans, mice, Arabidopsis, zebrafish, and Drosophila. The Upf3 protein is also known as RENT3 and SMG-4 and is functionally homologous to the yeast Upf3p protein [66]. Moreover, in human cells, there are two different isoforms of the Upf3 protein: Upf3a and Upf3b [67]. The Upf3a protein is 452 amino acids long and Upf3b is 470 amino acids long. Both of these isoforms have a ribonucleoprotein (RNP) domain at the N-terminal that is also known as the RNA recognition motif (RRM); however, neither isoform has been found to directly bind to RNA. In Upf3b, three highly similar amino acid regions have been found to be involved in binding to Upf2, which is critical in NMD for Upf2 recruitment to an EJC: residues 42–143, 49–143, and 52–137 [28, 43, 58, 67,68,69].Furthermore, the amino acids from 421 to 434, known as the EJC binding motif (EBM), in Upf3b have been found to be critical for Upf3b’s binding to Y14, an essential component of EJCs, and for activating NMD [28]. The amino acid region from residue 49 to residue 279 is highly conserved among the Upf3 proteins in various species [13, 28]. Upf3a has been found to not be as critical as Upf3b in NMD; however, Upf3a’s expression is increased when Upf3b levels are low, and it is able to mimic, to a certain extent, Upf3b’s role in NMD. UPF3’s existence as a paralog pair allows for a division of labor and for both proteins to serve specialized functions for modulating gene expression, with Upf3a having been shown to inhibit NMD through isolating Upf2 from other NMD substrates and Upf3b stimulating NMD [70].
The Upf3 protein has shown to be involved in lots of critical processes related to NMD in humans and other organisms. In Arabidopsis, Upf3’s homolog was found to be involved in the plant’s biological response to salt stress [71]. More importantly, mutations in the UPF3 gene have been found to be in a multitude of neurodevelopmental disorders, including schizophrenia, autism spectrum disorders, mental retardation, renal dysplasia, and more [35,36,37,38]. These findings further support Upf3’s critical role in the differentiation of neural stem cells [69]. Furthermore, the Upf3 homolog in S. cerevisiae was found to play a role in proper respiratory function [72]. The normal and balanced expression of the NMD factor Upf3 is necessary for the NMD homeostasis and normal physiological processes. The genetic structure of Upf3 protein is showed in Fig. 1c.
Interaction between Upf1 and other NMD factors
There are a multitude of other NMD factors that the Upf proteins interact with, including Smg1, Smg5–9, eRF1, eRF3, and the exon junction complex components.
The primary release factors that interact with Upf1 are eRF1 and eRF3. In yeast, the homolog of eRF1 is Sup45 and the homolog of eRF3 is Sup35 [73]. In the process of NMD, these release factors interact with all three of the Upf proteins—Upf1, Upf2, and Upf3. The Upf1, eRF1, and eRF3 interactions are important, as the binding of eRF1 and eRF3 to Upf1 has been found to inhibit Upf1’s ability to hydrolyze ATP RNA-dependent [74]. Furthermore, eRF1 and eRF3 are part of the SURF (Smg-1–Upf1-release factor) complex that contains SMG-1, which phosphorylates Upf1 after the SURF complex interacts with a exon junction complex that has Upf2 bound to it [75]. Upf2 has been found to be able to interact with eRF3 directly, though Upf3’s binding to Upf2 could negatively affect this ability [54].
In regards to the Smg proteins, Smg1, Smg5, Smg6, Smg7, Smg8, and Smg9 have all been found to interact with the Upf1 protein as well. As mentioned earlier, Smg1 phosphorylates Upf1 after the SURF complex binds with a Upf2–EJC complex, thereby forming a decay inducing complex (DECID) complex [75]. Though it has been found that Smg5, Smg6, and Smg7 are involved in the dephosphorylation of through Upf1 through interactions with phosphatase PP2A, the exact mechanistic details are still unclear and need to be further researched [76,77,78]. Smg8 and Smg9 interact with Smg1, have been found to play a role in regulating the interactions between SURF complexes and downstream EJCs, and they have been found to be involved in dephosphorylation of Upf1, possibly through activating or recruiting other proteins and factors needed for dephosphorylation [79]. Smg8 and Smg9 have also been found to increase Upf1’s affinity for Smg1 when both of those Smg proteins form a complex with Smg1 [52]. Furthermore, Upf2 has been found to promote the release of phosphorylated Upf1 from this complex when Upf2 binds to the Upf1–Smg8–Smg9–Smg1 complex, thereby possibly initiating the start of degradation of the mRNA [52]. Moreover, mutations in SMG9 have been found to lead to irregular embryogenesis and linked to a multiple congenital anomaly syndrome involving congenital heart disease and the Dandy–Walker malformation [80].
Upf1 in nonsense-mediated mRNA decay
Within NMD, though there are numerous proteins involved, Upf1 is considered the primary protein through which NMD operates. During normal translation termination, due to the proper length of the 3′-UTR and stimulatory activity of PABPC1 (poly(A)-binding protein), the release factors eRF1 and eRF3 are efficiently recruited to the A site of terminating ribosome. This prevents Upf2/3 from associating with the termination complex and Upf1 from associating with the 40S ribosomal subunit. The coordinated interactions of release factors and ABCE1, including GTP hydrolysis by eRF3, the conformational change of eRF1, release of eRF3–GDP and recruitment of ABCE1, all together lead to peptide release. After ATP hydrolysis by ABCE1 and actions of eIF1, eIF1A and eLF3j, the ribosomal subunits (including 60S and 40S subunits) dissociate and recycle, and the mRNA decays. Sometimes the mRNAs can also be ‘recycled’, which means they can be translated multiple times before they are degraded.
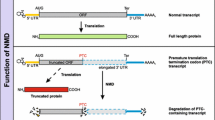
On the other hand, during premature translation termination, lack of proper 3′-UTR-related stimulatory activity leads to inefficient recruitment of eRF1 and eRF3 to the A site of the terminating ribosome. Upf2 and Upf3 interactions with Upf1 promotes Upf1’s binding to ATP and then subsequent association with the terminating ribosome [81]. The three Upf factors have critical interactions and functions in GTP hydrolysis by eRF3, the conformational change of eRF1, dissociation of eRF3–GDP, and release of the peptide. At the next stage, ATP hydrolysis by Upf1 promotes Upf2 and Upf3 release, and dissociation and recycling of 60S subunits and tRNA [30]. During the process of normal termination and NMD, the ABCE and Upf factors play similar roles, but the detailed mechanisms are different. Next we will focus on the specific processes and mechanisms of NMD. The whole process of NMD is shown in Fig. 2.
An overview of premature translation termination. During premature translation termination, because of the lack of a 3′-UTR-based stimulatory activity, eRF1 and eRF3 don’t efficiently bind to the A site of the 40S ribosome. After Upf1 binding to release factors and Smg-1, these factors form SURF complex. The joining of Upf2 and Upf3 stabilizes Upf1’s binding to the ribosome and promotes the formation of DECID. Binding of SURF to the EJC induces Upf1 phosphorylation, stimulating eRF1 and eRF3 activities and promoting peptide releasing. Later, the complex induces dissociation and recycling of ribosomal subunits and tRNA. At the end of the premature termination process, phosphorylated Upf1 binds to 40S subunit which is still attached to mRNA. Phosphorylated Upf1 is implicated in later decay events, including endonucleolytic cleavage and deadenylation pathways. In the endonucleolytic cleavage pathway, the cytoplasmic exosome participates in the digestion of 5′ cleavage product, and the 3′ cleavage product is digested by Xrn1. In the deadenylation pathway, due to the interplay between Smg7, Pop2 and the Ccr4-Not deadenylase, Ccr4-Not deadenylase complex (Ccr4–Not1–Not2–Not3) is recruited to the mRNA and activates deadenylation. After that the mRNA is digested by Xrn1 and decapped by Dcp1–Dcp2–Edc4 complex
Release factor recruitment
NMD is physically and functionally related to the eukaryotic release factors eRF1 and eRF3. In yeast, the two release factors are Sup45 and Sup35, respectively [30]. The eRF1 protein, connecting the genetic code to translational output, is structurally and functionally similar to a tRNA. eRF1 is able to bind to the A site, decipher the A site codon, and subsequently induce peptide release [82, 83]. eRF3 serves as an accessory factor by increasing the rate of peptide release in a GTP hydrolysis-dependent manner [84, 85]. Another critical factor is ATPase Rli1/ABCE1, which also increases the peptide release rate in an ATP hydrolysis-dependent way [86]. The yeast and human forms of Upf1 interact with both eRF1 and eRF3, and the Upf1–eRF1 complex is more sensitive to increasing salt concentrations than the Upf1–eRF3 complex [74]. As a result, it’s reasonable to assume that the interaction between Upf1 and eRF1 may involve ionic bonding. In addition, eRF3 and RNA compete for binding to Upf1, and ATP binding to Upf1 functionally increases the interplay between Upf1 and eRF3 but reduces Upf1’s affinity for RNA [74, 87]. As a result, the ATP-bound form of Upf1 prefers interacting with eRF3 when eRF3 and RNA are competing for binding to Upf1.
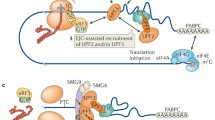
Upf1 binding to eRF1 and eRF3 inhibits Upf1’s ATPase and helicase activities [74], while Upf2 and Upf3 link Upf1 to the EJC and activate its RNA helicase and ATPase activity [25]. These results support the conclusion that Upf1 is initially recruited to a premature terminating ribosome and remains in an inactive form because of the release factors, but it is subsequently activated through bridging by Upf2 and Upf3 to the EJC. Moreover, Upf1 also interacts with other NMD factors and induces the formation of SURF (Smg-1–Upf1-release factor) complex in a Upf2- and Y14-independent manner [75, 79]. In this complex, Upf1 interacts with both Smg-1–ND and Smg-1–CD, whereas Upf2 and Y14, a part of EJC, can only bind Smg-1–CD, indicating that Smg-1 interplays with Upf1 and Upf2-Y14 through different structural domains. The formation of the SURF complex occurs just after recognition of the termination codon on a post-spliced mRNA. When SURF binds to Upf2 and EJC, these factors function together and form the DECID (decay-inducing complex) to induce Upf1 phosphorylation and dissociation of eRF3 from Upf1 [75].
Another highly debated part of NMD involves the first release factor that is released. Early on, Wang et al. postulated that eRF1 was released first. In yeast, Upf2 and Upf3 interact with eRF3 and compete for binding to specific eRF3 domain with eRF1 [88]. The three factors, Upf2, Upf3, and eRF1, interact with similar sites on eRF3, while Upf1 binds to a more N-terminal site on eRF3. As a result, the authors put forth a model to explain this. First, Upf1 is associated with the eRF1–eRF3 complex during the termination process. After hydrolysis of the peptidyl-tRNA bond, eRF1 is released from the Upf1–eRF3 complex and Upf2 or Upf3 binds to the complex. The complex then rearranges and Upf3 or Upf2 joins it and displaces eRF3, forming a final surveillance complex [88]. However, other investigators believe that eRF3 is released first based on two phenomena: eRF3 is located in lateral and eRF1 still functions after peptide release. Daniel et al. revealed that after peptide and ribosomal subunit release, eRF1 remained bound to the ribosomal complexes, and released eRF3 assisted the dissociation of eRF1 from the post-termination complex [89]. Although there is still no definite conclusion about which release factor is dissociated first, increasing number of people believe after GTP hydrolysis by eRF3, eRF3–GDP is released first and that eRF1 changes its conformation and is released after the peptide releases. More research still needs to be done to reach an accurate conclusion.
Upf1 recruitment and activation models
The core of the latest scientific research on NMD is focused on the mechanisms behind the initiation of NMD, and the complexities that underlie this process helps make this area of investigation intriguing. There are three controversial models for how our cells are able to initiate NMD: Upf1 3′-UTR sensing and potentiation model, exon–junction complex (EJC) model, and the faux 3′-UTR model.
In the first model involving sensing of the 3′-UTR, Upf1 is believed to associate with transcripts that contain NMD-inducing long 3′-UTRs at the UTR itself, and then decay begins after association with another unidentified mRNP structure that plays a role in activating Upf1 [90]. They found that this Upf1 binding to long 3′-UTRs is dependent on length but not sequence [90]. However, this raises some questions as several researchers have found that Upf1 can also bind to non-NMD inducing long 3′-UTRs, so there must be some other factors that play a role [91, 92]. Furthermore, some human mRNAs with long 3′-UTRs have been shown to evade NMD through use of cis acting elements located near the PTC or through other independent mechanisms [93]. More specifically, those with long 3′ UTRs that can avoid NMD have been found to usually have AU-rich sequences within the first 200 nucleotides of the 3′ UTR, which needs to be investigated to determine whether those AU sequences play an important role in evading NMD [93]. It is hypothesized that there is a trans factor that has a strong affinity for those AU sequences and that mediates a cis element’s inhibition of NMD [93]. Interestingly, new research has found that many Upf1 targets have GC rich nucleotide sequences in the 3′ UTR [49]. Moreover, it has been found that Upf1 in its steady state binds indiscriminately to mRNAs, regardless of NMD fate [94]. Therefore, NMD targets can be detected based on binding to phosphorylated Upf1 but not steady state Upf1 [95].
The second model involves a faux-3′ UTR and is extremely similar to the model described previously except that it also places emphasis on the concept that termination at premature translation codons is significantly less efficient than termination at normal translation termination codons [96]. Possible reasons for this inefficient termination include slower ribosome dissociation from premature termination codons and lack of a critical factor, a termination-promoting signal [96, 97]. Moreover, in this model, extension of the 3′-UTR can lead to initiation of NMD, and reduction of the length of the 3′-UTR through tethering of PABPC1 between the 3′ poly(A) tail and PTC leads to inhibition of NMD [98, 99,100,]. The termination factor eRF3 is normally able to bind non-competitively to PABPC1; however, when the distance between a PTC and PABPC1 is shortened, EJCs and other NMD-promoting factors are able to compete with PABPC1 for binding to eRF3, leading to NMD initiation [100,101,102]. In such situations, it is proposed that Upf1 competitively binds to eRF3 and then is subsequently phosphorylated by SMG1, leading to NMD [103].
The third model significantly involves exon junction complexes and is the model that is more generally accepted by most researchers working on NMD. EJCs have four core components: eIF4A3, CASC3 (MLN51), RBM8A (Y14), and either MAGOH or MAGOHB [2, 104, 105]. In the EJC model, EJCs play a critical role in recruiting Upf1 to the mRNA that is to undergo NMD. During the first round of translation, ribosomes remove EJCs that are present on the mRNAs; however, when there is a PTC in the A site of the ribosome, EJCs downstream of the PTC are not removed since the ribosome is halted [106, 107]. These downstream EJCs that are not removed are now part of the 3′ untranslated region.
In the EJC model, a SURF complex is formed near the PTC. This SURF complex is formed when the release factors eRF1 and eRF3 recruit SMG1 and Upf1 and associate with each other. An EJC that is downstream to this SURF complex has Upf2 and Upf3 associated with it, which then activate Upf1 in the SURF complex, forming a DECID complex, thereby leading to the initiation of NMD [108]. Once activated, Upf1 then recruits directly or indirectly any protein factors needed for degradation of the transcript, such as a SMG5–SMG7 heterodimer, CCR4–NOT deadenylase complex, and more [109,110,]. As with the other proposed models, evidence against this EJC model has been proposed. Previous research has found NMD to occur on mRNAs without introns before splicing, mRNAs that have not been spliced, and mRNAs without the appropriate EJC spacing or components [16, 110,111,112].
Release of peptide and ribosomal subunits
The Upf1 protein binds to ATP and RNA, and works as a RNA-dependent ATPase and exhibits 5′-to-3′ RNA helicase activities [113]. Activation of the Upf1 ATPase and helicase activities are implicated in assisting the mRNAs for decay by dissociating the peptide and 60S ribosomal subunit from the termination site [97]. In yeast extract expressing ATPase- or helicase-deficient Upf1, mRNAs fail to reinitiate translation after encountering a PTC, and the efficiency of the 60S joining step at initiation also decreases [24]. The decreased efficiency is thought to be the result of incomplete dissociation and recycling of ribosomal subunits at a premature termination event. As a result, Upf1 plays a critical role in the mechanisms involving termination at a PTC, subsequent ribosome release, and reinitiation of another round of translation [24]. Similarly, in human cells lacking Upf1 ATPase activity, partially degraded mRNA intermediates and NMD-related factors accumulate in processing bodies [23]. Therefore, the disassembly of messenger ribonucleoprotein (mRNPs) and release of their protein components require ATPase activity of Upf1. A model has been put forward to illustrate the roles of Upf1. Upf1 functions at premature terminating ribosomes, and the ATP hydrolysis by Upf1 induces conformational alterations and compositional transitions of mRNPs, including release of Upf2 and Upf3, along with dissociation and recycling of 60S subunit and tRNA. These alterations are necessary for the initiation and completion of NMD. ATPase-deficient Upf1 results in mRNPs stalling at an intermediate step and impairment of NMD. In addition, Upf1 is also required for rapid degradation of truncated polypeptides derived from PTC-containing mRNAs by proteasomes [114]. This Upf1-dependent proteasome-mediated degradation is also activated by mRNAs containing a faux 3′-UTR [114]. Upf1’s function in polypeptide degradation is related to its role as an E3 ubiquitin ligase. The CH domain of Upf1 binds to Upf2, and this binding is involved in inhibition of Upf1’s ATPase activity. Takahashi et al. showed that the CH domain was also related to classical E3-RING finger domains and exhibited ligase activity [115].
RNA degradation
Upf1 (123–124 kDA) has been found to be the primary factor involved in determining whether a certain mRNA transcript is to undergo NMD and also its subsequent degradation [95]. Upf1 is able to bind to RNA, hydrolyze ATP dependent on RNA, and unwind RNA using ATP [25, 26, 46]. Upf1 is initially phosphorylated by SMG1 at several motifs at the C-terminus; however, SMG1 is inhibited by SMG8 and SMG9 [116, 117]. It is only after interacting with Upf2 and DHX34, which facilitate the release of SMG8 and SMG9 and a conformational change in SMG1, that SMG1 is able to phosphorylate Upf1 [26, 79, 118]. Once phosphorylated, Upf1 can recruit an SMG-5/SMG-7 heterodimer through interactions with their 14-3-3-like domains [119]. SMG5 recruits mRNA-decapping protein DCP2 and DCP1a, while SMG7 recruits CCR4–NOT deadenylase complex [78, 120, 121]. CCR4–NOT deadenylase complex is dependent on decapping, and subsequently, the RNA is degraded by XRN1 (exoribonuclease 1) [122, 123].
Another more frequent method of degradation is through SMG6, which has endonuclease activity that allows it to cleave NMD substrates [124, 125]. Moreover, SMG6 has been shown to be recruited to NMD substrates through its interactions with phosphorylated Upf1 [45]. There is also evidence indicating that SMG6’s proper function is dependent on its interactions with the helicase domain and SQ domain of Upf1 [47]. The 5′ end of the RNA fragment is digested by the exosome and the 3′ RNA fragment is degraded by XRN1 [126]. The decay pathway through the SMG5–SMG7 heterodimer is less frequent and is primarily a backup for the SMG6 pathway [127]. These subsequent steps occur after the cell has determined that an mRNA is to undergo NMD.
Upf1 processing-body (P-body) localization
Recent research has shown that in many eukaryotic cells, lots of key NMD factors including decapping enzyme Dcp1p/Dcp2p (for the removal of 5′-cap), activators of decapping (Dhh1p, Pat1p, Lsm1–7p), and XRN1 (for the 5′–3′ degradation) localize in large granules called P-bodies [128, 129]. Moreover, the proteins involved in mRNA degradation, mRNA surveillance, and RNA-mediated gene silencing also accumulate in the P-bodies [130, 131]. In mammalian and Drosophila S2 cells, the P-bodies are also called Gawky (GW) bodies, because other than decay factors, they contain the GW182 protein involved in the regulation of mRNA stability, normal translation, and RNA interference pathway [132, 133]. As a result, some people have put forward a view that mRNA decay factors are active when assembled in P-bodies, or that at least mRNA decay occurs preferentially in P-bodies [134].
Several observations raise the possibility that NMD might take place in P-bodies. First, in P-bodies, Dcp1p, Dcp2p, and Xrn1p are all involved in catalyzing the degradative process of NMD [131]. Secondly, Smg5, Smg7 and Upf1 are localized to P-bodies in cells under conditions where either DCP1, DCP2, or XRN1 lack, or Upf2 and Upf3 are depleted [78, 135, 136]. Third, the PTC-containing mRNA accumulates in P-bodies and the level of mRNA reduces when Upf1 is deleted [136]. One controversial problem is that Upf1 proteins don’t diffusely localize in P-bodies in wild-type cells, but some people argue that this is because of NMD substrates transporting through P-bodies only transiently. Although neither NMD factors nor substrates are normally examined, they accumulate in P-bodies when NMD is impaired and NMD-related factors deletion [134]. Therefore it is proposed that Upf1 leads PTC-containing mRNAs to P-bodies and elicits mRNA decay.
Other functions of Upf1
The Upf1 protein, apart from playing a critical role in NMD, might take part in other nuclear-related functions unrelated to NMD. Here we review the evidence connecting Upf1 to non-NMD functions. Abundant research shows that Upf1 mutants or RNA interference cells display deficiencies in cell cycle (especially S phase) progression, DNA replication, and telomere homeostasis [137]. Upf1 is recruited by mammalian Staufen1, an RNA binding protein functioning in mRNA transportation and translational regulation, to the specific mRNA 3′-UTRs regions to induce mRNA decay [138]. Upf1 is also recruited by SLBP (stem-loop binding protein) to elicit replication-dependent histone mRNA degradation through phosphorylation [139]. Upf1 is one of the components of HIV-1 RNP, implicated in HIV-1 genomic RNA stability and involved in the major structural protein pr55Gag synthesis [140]. Moreover, Upf1 regulates the expression of genes that mediate inflammation and myeloid cell differentiation via hnRNP E2 [141].
S phase progression and DNA replication
Previous studies demonstrated that the depletion of Upf1 damaged normal nuclear functions and DNA replication. Drosophila cells depleted of Upf1 are arrested at the S phase, display misexpression of a cohort of genes functioning in cell cycle progression, and impair cell proliferation [142]. In murine embryonic stem cells, disruption of Rent1 gene, which encodes a mammalian ortholog of Upf1 protein, inhibits cell cycle and leads to apoptosis [143]. In addition, the depletion of Upf1 in HeLa cells also changed the transcript levels of 25 genes related to the cell cycle [144].
Azzalin and Lingner revealed the direct roles of Upf1 during DNA replication. The shRNA-mediated depletion of Upf1 led to Hela cells arresting early in S phase, which was in accordance with previous discoveries. These Hela cells were able to initiate DNA replication, but not progress or complete it, which induced an ATR (ataxia telangiectasia mutated and Rad3-related)-dependent DNA injuries response. However, the cells with depletion of Upf2 were able to properly complete cell cycle, but NMD was damaged. These results indicated that the reason of defective cell-cycle progression is the deficiency of Upf1, but not the loss of proper NMD [145]. In addition, the reciprocal coimmunoprecipitation of Upf1 and p125, the catalytic subunit of DNA polymerase δ was observed, and this interaction was promoted in S phase. This indicated a direct physical association between Upf1 and polymerase δ. However, the physical interaction between Upf1 and Upf2 also existed, and no interaction was detected between p125 and Upf2. Thus, it is reasonable to speculate that Upf1 assembles into two distinguishable parts: one part interacts with Upf2 and performs functions in NMD, and another part physically interplays with of polymerase δ to facilitate fork progression and perform DNA synthesis [145].
Telomere homeostasis
Telomeres are chromosomal heterochromatic structures located at the terminal regions. Their functions include compensating for incomplete DNA semi-conservative replication and protecting the structure of chromosomes from nuclease trimming [146]. Lots of proteins associated with telomeric DNA are involved in telomere length maintenance. Mutations of Upf proteins shorten telomere length and impair telomeric homeostasis.
The first indication that Upf1-related regulations might participate in telomere homeostasis was the observation that deletion of Upf1, Upf3 and RLF4 (RLF4 is the allelic gene of Upf2 and required for NMD) in yeast caused telomere-associated defects, including reduced telomeric silencing and shortened telomere DNA tracts [147]. Furthermore, in Saccharomyces cerevisiae, Northern blots and high-density oligonucleotide microarrays demonstrated that Upf mutations could increase mRNA levels of the genes regulating telomere activities, including those encoding telomerase catalytic subunit (Est2p), telomeric structure regulators (Sas2p and Orc5p), and telomerase mediators (Est1p, Est3p, Stn1p and Ten1p) [148].
The detailed mechanism of how NMD factors affect telomere homeostasis is not yet understood. However, the key proteins could be Upf1 and its functional modulators, such as SMG1 and SMG6 [137]. SMG 6 (also called Est1a) is a homolog of the yeast protein EVER SHORTER TELOMERES 1 (Est1p), which was first described as a protein involved in telomere biology [149]. A recent research speculated a reasonable model explaining Upf1 association and function at telomeres [150]. The association of Upf1 with telomeres is stimulated by the phosphorylation by ATR (ataxia telangiectasia mutated and Rad3-related), which is the PI3K (phosphoinositide 3-kinase)-related protein kinase [151]. Moreover, Upf1 localizes at telomeres and physically interplays with telomerase and telomeric factor TPP1, a mammalian telomeric shelterin complex [152]. This interplay is also regulated by ATR. The Upf1’s ATPase activity is responsible for inhibiting telomeric damage, as the depletion of Upf1 causes inefficient telomere leading-strand replication and assembly of DNA damage repair factors in telomere regions [150].
Staufen1-mediated mRNA decay (SMD)
Mammalian Staufen1 is a double-stranded RNA binding protein that binds to extensive RNA secondary structures and functions in mRNA transport, localization, and translation [153]. Staufen1 localizes to the somatodendritic domain of hippocampal neurons [154] and exists in RNA granules, which migrate within the dendrites in a kinase-driven manner [155]. Furthermore, Staufen1 interacts with telomerase RNA, indicating that it plays a critical role in DNA replication and cell division [156, 157].
Kim et al. found that Staufen1 could bind the Upf1 protein and 3′-UTR of the mRNA encoding Arf1 (ADP-ribosylation factor). Staufen1 elicited Arf1 mRNA decay via a distinctive mechanism that occurred independently of splicing and independently of when Upf2 or Upf3b was downregulated. Downregulating either Stau1 or Upf1 enhanced Arf1 mRNA abundance and stability. Similarly, tethering Staufen1 downstream of a normal termination codon also decreased mRNA abundance by the same mechanism as that of Arf1. Additionally, microarray analyses displayed that lots of transcripts other than Arf1 mRNA also binded Staufen1 and were regulated by SMD [138]. SMD works on the process where Staufen1 binds the Arf1 mRNA 3′-UTR and recruits Upf1 independently of an EJC. Similar to EJC-dependent NMD, the Staufen1 binding site resides more than 20 to 25 nt downstream of the normal termination codon to induce mRNA decay [138].
Replication-dependent histone mRNA decay
In mammalian cells, the regulation of histone synthesis mostly belongs to post-transcriptional modifications involving the abundance of histone mRNA. Histone mRNA levels increase as the cells enter S phase and decrease at the end of S phase [158]. In metazoan mRNAs, the only histone mRNAs that are not polyadenylated are replication-dependent histone mRNAs [159]. These mRNAs contain special 3′UTRs which harbor a conserved stem-loop structure. This structure, as the only cis-acting element functioning in coupling regulation of mRNA half-life with DNA synthesis, interacts with stem-loop binding proteins (SLBP), also called hairpin-binding protein (HBP) [159]. At the end of S phase, Upf1 interplays with SLBP and elicits histone mRNA degradation by its phosphorylation [139].
HIV-1 genomic RNA stability
The stability and proper abundance of viral genomic RNA are necessary to a successful viral infection and RNA replication. As a result, viral RNAs have a distinctive ability to avoid RNA damage by the host machinery. The HIV-1 RNP (ribonucleoprotein) consists of viral genomic RNA, the major structural protein pr55Gag, and the host protein Staufen1. Ajamian et al. found that Upf1 was also one of HIV-1 RNP components and was implicated in HIV-1 genomic RNA stability. siRNA knockdown of Upf1 reduced HIV-1 RNA and pr55Gag synthesis. In addition, overexpression of Upf1 resulted in increased regulation of steady-state HIV-1 RNA and protein synthesis levels. The effects of Upf1 on HIV-1 genomic stability were dependent on Upf1 ATPase activity, but were independent of NMD and interactions with Upf2 [140].
Mediation of inflammation and myeloid cell differentiation
In addition to the functions in NMD, Upf1 is also involved in other posttranscriptional gene regulation. A recent paper suggested that Upf1 regulates the expression of genes that mediate inflammation and myeloid cell differentiation via heterogeneous nuclear ribonucleoprotein (hnRNP) E2 [141]. hnRNP E2 is a member of minor hnRNP proteins, and it is involved in splicing [160, 161] and regulating translational repression [162]. Saul et al. presented that the balance between hnRNP E2 and microRNA miR-328 controlled the expression of genes involved in inflammation myeloid cell differentiation. On the other hand, the genes which were downregulated by Upf1 knockdown had binding sites for hnRNP E2 at their 5′-UTR. Upf1 is responsible for downregulating hnRNP E2 and regulating the balance between hnRNP E2, thus further affecting myeloid cell maturation and functions [141].
Conclusion and perspectives
The Upf1 protein plays a critical role in primarily non-sense mediated decay. Upf1 binds to ATP and RNA, manifesting RNA-dependent ATPase and 5′-to-3′ RNA helicase activities. The ATP-binding and hydrolysis activities of Upf1 activate NMD [30]. Upf1 interacts with eRF1, eRF3 and Smg-1, and forms SURF complex. Binding of SURF to the EJC induces Upf1 phosphorylation and NMD [75]. The association of Upf2 and Upf3 stabilizes Upf1’s binding to the premature terminating ribosome, and they function together in stimulating eRF1 and eRF3 activities and promoting peptide releases. Later, the complex induces dissociation and recycling of ribosomal subunits and tRNA. At the end of the premature termination process, phosphorylated Upf1 binds to the 40S subunit which is still attached to mRNA. Phosphorylated Upf1 is implicated in later decay events, including endonucleolytic cleavage and deadenylation pathways [30]. The interactions between Upf1 and other factors are shown in Fig. 3. Furthermore, Upf1 is also involved in a variety of other physiological processes, including Staufen1-mediated mRNA decay (SMD), Replication-dependent histone mRNA decay, S-phase progression and DNA replication, telomere homeostasis, and HIV-1 genomic RNA stability. Thus, any errors during the transcription and translation process of the protein can lead to a multitude of diseases and illnesses, such as Marfan syndrome, β-thalassemias, and several neurodegenerative diseases [40]. Nonetheless, the Upf2 and Upf3 proteins play crucial roles in NMD and any anomalies in their functions can lead to many clinical phenotypes as well.
The interactions between Upf1 and other NMD-related factors. Upf1, also called hUpf1, NORF1, RENT1, pNORF1, and Smg2. The two major domains in Upf1 are the SQ domain located near the C-terminus and the CH domain located near the N-terminus. The CH domain is involved in inhibition of the ATPase activity of Upf1, and it’s the domain involved in binding Upf2 (RENT2, Smg3). The SQ domain is implicated in inhibition of the helicase activity of Upf1 through direct binding to the helicase domain, thereby having an inhibitory effect on ATP hydrolysis and RNA unwinding. There are four core regions in Upf2 playing important functional roles: three MIF4G domains and one C-terminal region for binding to the CH domain of Upf1. The MIF4G-1 and MIF4G-2 domains work as the key linkers between the EJC, CH-domain of Upf1, and the MIF4G-3 domain of Upf2. The MIF4G-3 domain has been found to interact with Upf3b (RENT3, Smg4)’s RNA recognition motif (RRM), and Smg1 is able to bind non-competitively to the MIF4G-3 domain. The primary release factors interacting with Upf1 are eRF1 (Sup45 in yeast) and eRF3 (Sup35 in yeast). In the process of NMD, release factors interact with Upf1, Upf2, and Upf3. The binding of eRF1 and eRF3 to Upf1 inhibits Upf1’s ability to hydrolyze ATP in RNA-dependent way. Furthermore, eRF1 and eRF3 are part of the SURF (Smg-1–Upf1-release factor) complex that contains Smg 1, which phosphorylates Upf1 after the SURF complex interacts with a exon junction complex (including Y14, BTZ, MAGOH and eIF4AIII) that has Upf2 bound to it. In regards to the Smg proteins, Smg1, Smg5, Smg6, Smg7, Smg8, and Smg9 have all been found to interact with the Upf1 protein as well. Smg5, Smg6, and Smg7 are involved in the dephosphorylation of through Upf1 through interactions with phosphatase PP2A, but the exact mechanistic details are still unclear and need to be further researched. Smg8 and Smg9 interact with Smg1, have been found to play a role in regulating the interactions between SURF complexes and downstream EJCs, and they have been found to be involved in dephosphorylation of Upf1, possibly through activating or recruiting other proteins and factors needed for dephosphorylation. In addition, Once Upf1 is phosphorylated, it can recruit an Smg5/Smg7 heterodimer. Smg5 recruits mRNA-decapping protein DCP2 and DCP1a, while Smg7 recruits ccr4–NOT deadenylase complex. ccr4–NOT deadenylase complex is dependent on decapping, and then subsequently, the RNA is degraded by Xrn1
Though there has been significant progress on the understanding of Upf1 in nonsense mediated decay, the exact model through which NMD is initiated is not yet understood. Experts have yet to come to a consensus on the entire mechanistic process of aberrant mRNA degradation. EJC model is a good predictor of whether a given mRNA is subjected to NMD, which consists of a SURF complex with Upf1 forming near a PTC and then subsequent activation by factors from a downstream EJC. For mRNA transcripts without EJCs, the distance between PABPC1 and the PTC is critical factor in initiating NMD, since if the distance is too large, Upf1 is able to out-compete PABPC1 for binding to eRF3. Nonetheless, as degradation of mRNAs without EJCs has been found previously, our team emphasizes that investigators continue to look at the importance of nucleotide sequences located near the PTC and in the 3′ UTR, especially G and C nucleotides [101]. Though there are several primary models proposed for NMD initiation, none of them are able to explain the reasoning for all mRNAs that undergo NMD. A thorough understanding and common consensus among leading researchers in the field is still lacking regarding the complete mechanisms of NMD. In order to grasp a better understanding of NMD, more research on factors other than Upf1 need to be initiated, such as on Upf2 and Upf3. Though these factors seem minute in importance, it is possible that their involvement in NMD can lead to critical findings. Collaboration among investigators on Upf1 and other related proteins is necessary for advancements to proceed rapidly. Thoroughly understanding this one pathway for mRNA degradation can lead to breakthrough therapies for a multitude of diseases.
References
Frazer KA (2012) Decoding the human genome. Genome Res 22:1599–1601
Kurosaki T, Maquat LE (2016) Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129:461–467
Gandhi R, Manzoor M, Hudak KA (2008) Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J Biol Chem 283:32218–32228
Simms CL, Thomas EN, Zaher HS (2017) Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev RNA 8:e1366
Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258–2261
Stalder L, Mühlemann O (2008) The meaning of nonsense. Trends Cell Biol 18:315–321
He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12:1439–1452
McGlincy N, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci 33:385–393
Ni J, Grate L, Donohue J, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21:708–718
Thompson DM, Parker R (2007) Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27:92–101
Tani H, Torimura M, Akimitsu N (2013) The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE 8:e55684
Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Seki M (2009) Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA 106:2453–2458
Karousis ED, Nasif S, Mühlemann O (2016) Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA 7:661–682
Nagy E, Maquat LE (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23:198–199
Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17:3484–3494
Zhang J, Sun X, Qian Y, Maquat LE (1998) Intron function in the nonsense-mediated decay of beta- globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4:801–815
Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE (2010) NMD: RNA biology meets human genetic medicine. Biochem J 430:365–377
Culbertson MR, Underbrink KM, Fink GR (1980) Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95:833–853
Page MF, Carr B, Anders KR, Grimson A, Anderson P (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19:5943–5951
Chiu SY, Serin G, Ohara O, Maquat LE (2003) Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9:77–87
Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226–1235
Min EE, Roy B, Amrani N, He F, Jacobson A (2013) Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA 19:1105–1115
Franks TM, Singh G, Lykke-Andersen J (2010) Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 143:938–950
Ghosh S, Ganesan R, Amrani N, Jacobson A (2010) Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA 16:1832–1847
Chamieh H, Ballut L, Bonneau F, Le Hir H (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15:85–93
Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E (2011) Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41:693–703
Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U et al (2010) Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. PNAS 107:10050–10055
Gehring NH, Neu-Yilik G, Schel T, Hentze MW, Kulozik AE (2003) Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11:939–949
Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21:209–223
He F, Jacobson A (2015) Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet 49:339–366
Liu C, Karam R, Zhou Y, Su F, Ji Y, Li G, Xu G, Lu L, Wang C, Song M, Zhu J, Wang Y, Zhao Y, Foo WC, Zuo M, Valasek MA, Javle M, Wilkinson MF, Lu Y (2014) The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med 20:596–598
Jolly LA, Homan CC, Jacob R, Barry S, Gecz J (2013) The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet 22:4673–4687
Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L et al (2012) Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry 17:1103–1115
Nguyen LS, Kim H-G, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gécz J (2013) Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet 22:1816–1825
Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C (2007) Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39:1127–1133
Lynch SA, Nguyen LS, Ng LY, Waldron M, McDonald D, Gecz J (2012) Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet 55:476–479
Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Inoff-Germain G et al (2011) A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry 16:238–239
Xu X, Zhang L, Tong P, Xun G, Su W (2013) Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet 83:560–564
Daar IO, Maquat LE (1988) Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol 8:802–813
Caputi M, Kendzior RJ, Beemon KL (2002) A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev 16:1754–1759
Leeds P, Peltz SW, Jacobson A, Culbertson MR (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5:2303–2314
Applequist SE, Selg M, Raman C, Jäck HM (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25:814–821
Kadlec J, Guilligay D, Ravelli RB, Cusack S (2006) Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12:1817–1824
Fiorini F, Bagchi D, Le Hir H, Croquette V (2015) Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat Commun 6:7581
Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S (2012) N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40:1251–1266
Fiorini F, Boudvillain M, Le Hir H (2013) Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res 41:2404–2415
Nicholson P, Josi C, Kurosawa H, Yamashita A, Mühlemann O (2014) A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res 42:9217–9235
Park E, Maquat LE (2013) Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 4:423–435
Imamachi N, Tani H, Akimitsu N (2012) Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. Drug Discov Ther 6:55–61
Cheng Z, Muhlrad D, Lim MK, Parker R, Song H (2007) Structural and functional insights into the human Upf1 helicase core. EMBO J 26:253–264
Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE (2010) UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39:396–409
Deniaud A, Karuppasamy M, Bock T, Masiulis S, Huard K, Garzoni F et al (2015) A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res 43:7600–7611
Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol 20:8944–8957
López-Perrote A, Castaño R, Melero R, Zamarro T, Kurosawa H, Ohnishi T et al (2016) Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res 44:1909–1923
Tatsuno T, Nakamura Y, Ma S, Tomosugi N, Ishigaki Y (2016) Nonsense-mediated mRNA decay factor Upf2 exists in both the nucleoplasm and the cytoplasm. Mol Med Rep 14:655–660
Clerici M, Mourão A, Gutsche I, Gehring NH, Hentze MW, Kulozik A et al (2009) Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28:2293–2306
Kadlec J, Izaurralde E, Cusack S (2004) The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11:330–337
Clerici M, Deniaud A, Boehm V, Gehring NH, Schaffitzel C, Cusack S (2014) Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res 42:2673–2686
LaRonde-LeBlanc N, Santhanam AN, Baker AR, Wlodawer A, Colburn NH (2007) Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol Cell Biol 27:147–156
Melero R, Uchiyama A, Castaño R, Kataoka N, Kurosawa H, Ohno S, Yamashita A, Llorca O (2014) Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 22:1105–1119
Fourati Z, Roy B, Millan C, Coureux PD, Kervestin S, van Tilbeurgh H et al (2014) A highly conserved region essential for NMD in the Upf2 N-terminal domain. J Mol Biol 426:3689–3702
Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI (2006) Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol 26:3390–3400
Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT (2008) NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22:1381–1396
Bao J, Tang C, Yuan S, Porse BT, Yan W (2015) UPF2, a nonsense-mediated mRNA decay factor, is required for prepubertal Sertoli cell development and male fertility by ensuring fidelity of the transcriptome. Development 142:352–362
Bao J, Vitting-Seerup K, Waage J, Tang C, Ge Y, Porse BT, Yan W (2016) UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′UTR transcripts. PLoS Genet 12:e1005863
Thapar R, Denmon AP (2013) Signaling pathways that control mRNA turnover. Cell Signal 25:1699–1710
Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121–1131
Melero R, Buchwald G, Castaño R, Raabe M, Gil D, Lázaro M, Urlaub H, Conti E, Llorca O (2012) The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol 19:498–505, S1–S2
Alrahbeni T, Sartor F, Anderson J, Miedzybrodzka Z, McCaig C, Müller B (2015) Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol Brain 8:33
Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK, Wilkinson MF (2016) The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 165:382–395
Vexler K, Cymerman MA, Berezin I, Fridman A, Golani L, Lasnoy M, Saul H, Shaul O (2016) The Arabidopsis NMD factor UPF3 is feedback-regulated at multiple levels and plays a role in plant response to salt stress. Front Plant Sci 1376: eCollection
de Pinto B, Lippolis R, Castaldo R, Altamura N (2004) Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol Microbiol 51:1129–1142
Ito K, Ebihara K, Nakamura Y (1998) The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA 4:958–972
Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Haley A, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12:1665–1677
Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R et al (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20:355–367
Anders KR, Grimson A, Anderson P (2003) SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22:641–650
Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12:1187–1200
Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16:587–596
Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B (2009) SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23:1091–1105
Shaheen R, Anazi S, Ben-Omran T, Seidahmed MZ, Caddle LB, Palmer K et al (2016) Mutations in SMG9, encoding an essential component of nonsense-mediated decay machinery, cause a multiple congenital anomaly syndrome in humans and mice. Am J Hum Genet 98:643–652
Serdar LD, Whiteside DL, Baker KE (2016) ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat Commun 7:14021
Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5:1014–1020
Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100:311–321
Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV (2006) In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125:1125–1136
Eyler DE, Green R (2011) Distinct response of yeast ribosomes to a miscoding event during translation. RNA 17:925–932
Shoemaker CJ, Green R (2011) Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA 108:1392–1398
Weng Y, Czaplinski K, Peltz SW (1996) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16:5477–5490
Wang W, Czaplinski K, Rao Y, Peltz SW (2001) The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20:880–890
Daniel EE, Karen AW, Rachel G (2013) Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem 288:29530–29538
Hogg JR, Goff SP (2010) Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143:379–389
Hurt JA, Robertson AD, Burge CB (2013) Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23:1636–1650
Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Akimitsu N (2012) Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9:1370–1379
Toma KG, Rebbapragada I, Durand S, Lykke-Andersen J (2015) Identification of elements in human long 3′ UTRs that inhibit nonsense-mediated decay. RNA 21:887–897
Lee SR, Pratt G, Martinez F, Yeo GW, Lykke-Andersen J (2015) Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell 59:413–425
Kurosaki T, Li W, Hoque M, Popp MWL, Ermolenko DN, Tian B, Maquat LE (2014) A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 28:1900–1916
Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112–118
Kervestin S, Jacobson A (2012) NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13:700–712
Muhlrad D, Parker R (1999) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299–1307
Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O (2008) Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6:e92
Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L (2008) Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14:563–576
Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26:1591–1601
Zahdeh F, Carmel L (2016) The role of nucleotide composition in premature termination codon recognition. BMC Bioinform 17:519
Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27:736–747
Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE (2009) The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol 7:e1000120
Bono F, Gehring NH (2011) Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol 8:24–29
Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607–617
Lejeune F, Ishigaki Y, Li X, Maquat LE (2002) The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J 21:3536–3545
Hug N, Longman D, Cáceres JF (2016) Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res 44:1483–1495
Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O (2013) Nonsense-mediated mRNA decay: mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829:612–623
LeBlanc JJ, Beemon KL (2004) Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J Virol 78:5139–5146
Wen J, Brogna S (2010) Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J 29:1537–1551
Wang J, Gudikote JP, Olivas OR, Wilkinson MF (2002) Boundary-independent polar nonsense-mediated decay. EMBO Rep 3:274–279
Czaplinski K, Weng Y, Hagan KW, Peltz SW (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1:610–623
Kuroha K, Tatematsu T, Inada T (2009) Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep 10:1265–1271
Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino S, Kontani K, Nishina H, Katada T (2008) Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14:1950–1958
Arias-Palomo E, Yamashita A, Fernández IS, Núñez-Ramírez R, Bamba Y, Izumi N et al (2011) The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev 25:153–164
Yamashita A (2013) Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 18:161–175
Hug N, Cáceres JF (2014) The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep 8:1845–1856
Jonas S, Weichenrieder O, Izaurralde E (2013) An unusual arrangement of two 14–3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 27:211–225
Cho H, Kim KM, Kim YK (2009) Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33:75–86
Loh B, Jonas S, Izaurralde E (2013) The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27:2125–2138
Lejeune F, Li X, Maquat LE (2003) Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12:675–687
Mühlemann O, Lykke-Andersen J (2010) How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 7:28–32
Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14:2609–2617
Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen T (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16:49–55
Gatfield D, Izaurralde E (2004) Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429:575–578
Lykke-Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, Sandelin A, Jensen TH (2014) Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev 28:2498–2517
van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21:6915–6924
Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T (2002) The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489–1501
Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22
Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808
Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ (2006) Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J Cell Biol 174:349–358
Ding L, Han M (2007) GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol 17:411–416
Brogna S, Ramanathan P, Wen J (2008) UPF1 P-body localization. Biochem Soc Trans 36:698–700
Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17:537–547
Sheth U, Parker R (2006) Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095–1109
Varsally W, Brogna S (2012) UPF1 involvement in nuclear functions. Biochem Soc Trans 40:778–784
Kim YK, Furic L, Desgroseillers L, Maquat LE (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195–208
Kaygun H, Marzluff WF (2005) Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12:794–800
Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE et al (2008) Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14:914–927
Saul MJ, Stein S, Grez M, Jakobsson PJ, Steinhilber D, Suess B (2016) UPF1 regulates myeloid cell functions and S100A9 expression by the hnRNP E2/miRNA-328 balance. Sci Rep 6:31995
Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11:1530–1544
Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10:99–105
Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36:1073–1078
Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16:433–439
Zakian VA (1995) Telomeres: beginning to understand the end. Science 270:1601–1607
Lew JE, Enomoto S, Berman J (1998) Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol 18:6121–6130
Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J (2003) mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryot Cell 2:134–142
Reichenbach P, Höss M, Azzalin CM, Nabholz M, Bucher P, Lingner J (2003) A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13:568–574
Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM (2011) Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J 30:4047–4058
Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9:616–627
Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42:301–334
Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L (1999) Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19:2220–2230
Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG (1999) The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci 19:288–297
Kanai Y, Dohmae N, Hirokawa N (2004) Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43:513–525
Bachand F, Triki I, Autexier C (2001) Human telomerase RNA-protein interactions. Nucleic Acids Res 29:3385–3393
Le S, Sternglanz R. Greider CW (2000) Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell 11:999–1010
Baumbach LL, Stein GS, Stein JL (1987) Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry 26:6178–6187
Jaeger S, Barends S, Giegé R, Eriani G, Martin F (2005) Expression of metazoan replication-dependent histone genes. Biochimie 87:827–834
Broderick J, Wang J, Andreadis A (2004) Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene 331:107–114
Motta-Mena LB, Smith SA, Mallory MJ, Jackson J, Wang J, Lynch KW (2011) A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs). J Biol Chem 286:20043–20053
Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, Iervolino A, Condorelli F, Gambacorti-Passerini C, Caligiuri MA, Calabretta B (2002) BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet 30:48–58
Acknowledgements
The authors express gratitude to Skanda Setty, Magid Abdo, Karthik Dhanireddy, Zhihua Li and Mingye Yan in their support of this project. Moreover, we would like to thank the members of BRAX for their encouragement as well.
Author information
Authors and Affiliations
Contributions
Y-RL and PG conceived and authored the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
Rights and permissions
About this article
Cite this article
Gupta, P., Li, YR. Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay. Mol Biol Rep 45, 39–55 (2018). https://doi.org/10.1007/s11033-017-4139-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-017-4139-7