Abstract
Parkinson’s disease (PD) is considered as a high prevalence neurodegenerative disorders worldwide. Pathologically, the demise of dopamine-producing cells, in large part due to an abnormal accumulation of the α-synuclein in the substantia nigra, is one of the main causes of the disease. Up until now, many de novo investigations have been conducted to disclose the mechanisms underlying in PD. Among them, impacts of non-coding RNAs (ncRNAs) on the pathogenesis and/or progression of PD need to be highlighted. microRNAs (miRNAs) and long ncRNAs (lncRNAs) are more noteworthy in this context. miRNAs are small ncRNAs (with 18–25 nucleotide in length) that control the expression of multiple genes at post-transcriptional level, while lncRNAs have longer size (over 200 nucleotides) and are involved in some key biological processes through various mechanisms. Involvement of miRNAs has been well documented in the development of PD, particularly gene expression. Hence, in this current review, we will discuss the impacts of miRNAs in regulation of the expression of PD-related genes and the role of lncRNAs in the pathogenesis of PD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
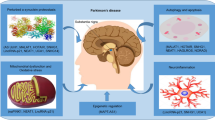
Parkinson’s disease (PD) is one of the high prevalence progressive neurodegenerative disorder just after Alzheimer worldwide, its symptoms can manifest as bradykinesia, rigidity, resting tremor, and posture instability [1]. Mechanistically, degeneration of dopamine-producing cells, because of an abnormal deposition of the α-synuclein which bind to ubiquitin in the cells within substantia nigra, seems to be one of the main causes of the disease [2, 3]. However, as the disease advances, the number of involved brain regions (i.e., cingulate gyrus, amygdala and higher cortical regions) is increased and causes of the emergence of psychosis and dementia [4]. Lewy bodies, as typical characteristic of PD, are cytoplasmic inclusions (as α-synuclein-ubiquitin complex) that cannot be degraded by proteasomes [5]. Newest researches on the pathogenesis of disease have elucidated that the defect in protein trafficking machineries, shuttle proteinaceous compounds between two main cellular compartments—the endoplasmic reticulum and the Golgi apparatus—could be another underlying mechanism for the death of dopaminergic neurons [6]. The α-synuclein, leucine-rich repeat kinase 2 (LRRK-2), parkin (PRKN/PARK2), phosphatase and tensin homologue (PTEN)-induced kinase1 (PINK1/PARK6) and oncogene DJ-1 are suspected to be participated in the initiation/development of PD [7]. Furthermore, some other genes such as synuclein alpha polymorphism, glucoberebrosidase (GBA), microtubule-associated protein, tau/saitohin (MAPT/STH), appear to associate with the risk of PD development/progression [8]. It should be noted that approximately 1.5–2 % of the human genome are protein-coding region, while the remaining is transcribed into transcripts with no protein-coding capacity that is known as non-coding RNAs (ncRNAs) that are believed to have a critical regulatory activity in normal cellular development, function, and pathogenesis of various diseases [9, 10]. As a matter of fact, the importance of ncRNAs in the different basic and translational researches and particularly in the brain function and central nervous system (CNS) disorders become a hot spot for researchers in neuro-regenerative medicine. These RNA molecules are classified as small ncRNAs and long ncRNAs as fewer as and longer than 400 nucleotides in size, respectively. Small ncRNAs per se comprise microRNAs (miRNAs; 19-24 bp), PIWI interacting RNAs (piRNAs; 26–31 bp), transcription initiation RNAs (tiRNAs; 17–18 bp), small nucleolar RNAs (snoRNAs; 60–300 bp), promoter-associated small RNAs (PASRs; 22–200 bp), and TSS-associated RNAs (TSSa-RNAs; 20–90 bp). Long ncRNAs are comprised of transcribed ultraconserved regions (T-UCRs; >200 bp) and large intergenic non-coding RNAs (lincRNAs; >200 bp) [11–14]. Increasing evidence has clearly emphasized the crucial roles of ncRNAs in multiple biological processes (e.g., brain development and differentiation), and various diseases (e.g., neurodegenerative disorders such as Alzheimer’s, Huntington’s, Parkinson’s, and spinocerebellar ataxia diseases) [15]. Here, we aimed to provide a brief introduction about structure and biosynthesis of some important ncRNAs, the molecular mechanisms and main functions of these macromolecules, and finally the major involvements of ncRNAs in neurodegenerative diseases, in particular, Parkinson’s disease.
Non coding RNAs: a brave new RNA world
From their broad biology and function viewpoints, miRNAs are known as the most researched group of ncRNAs [16]. Since their discovery in Caenorhabditis elegans almost 20 years ago by Lee et al. [17], miRNAs are touted as one of the extensively studied classes of small ncRNAs. These macromolecular biomolecules are classified as a conserved class of short and single-stranded RNA molecules with a size of about 22 nucleotides, which has a pivotal function in “fine-tuning” gene expression, mediating through the interaction with DNA, RNA and protein molecules [9, 18]. Calling attention, more miRNA sequences analysis comes to the realization that a wonderful nearly 30 % of them are unique to primates, and even a few, such as miR-941, may be unique to humans [19]. They are thought to underlie phenotypic variation between species, possibly at the foundation of unique human traits [20].
LncRNAs have a unique function in various substantial parts of stem cell biology, epigenetics, cancer, signaling and neurobiology [21–23]. This group of RNAs is comprised of a broad portion of the transcriptome, such that over 18,000 transcripts are presently annotated as lncRNAs and a large number of new lncRNAs are discovered each year [24, 25]. By definition, lncRNAs have over 200 nucleotides in size and do not encode proteins with exceeding lengths of more than 30 amino acids [26, 27]. Generally, this group is less conserved between species and often shows high tissue specificity and low expression levels [25, 28]. Therefore, once discovered, lncRNAs have been referred as “transcriptional noise” [29]. However, increasing studies showed that lncRNAs play significant functions in many aspects of genome function such as, gene transcription, modulating RNA polymerase II function, regulating splicing, and epigenetics [30]. Although a huge amount of the human noncoding transcriptome is occupied by lncRNAs; to illustrate the diversity among the lncRNAs, here we discuss their classification, focusing on four classes, including (a) long intergenic ncRNAs (lincRNA), (b) natural antisense transcripts (NAT), (c) 3′-UTR-associated transcripts (uaRNA), and (d) enhancer RNAs (eRNA).
Biosynthesis of ncRNAs
miRNAs
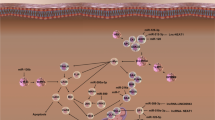
It has been proposed that there are two distinct biosynthesis pathways for small ncRNAs which are divided into multiple steps [31]. At the first step of canonical or Drosha-/Dicer-dependent biosynthesis pathway, RNA polymerase II transcribes primary miRNA (pri-miRNA) from two target genomic loci: miRNA genes or the introns of protein-coding mRNAs [32]. The generated pri-miRNAs, which fold into secondary structures comprised of base-paired stem loops, can subsequently be polyadenylated and regulated by the transcription factors. Next, the pri-miRNAs are cleaved into about 70-nucleotide premature-miRNAs (the so-called pre-miRNAs) containing hairpin structures in the nucleus by Drosha/DGCR8 complex - a RNase III type endonuclease microprocess [33]. The next step is the traverse of pre-miRNAs into the cytoplasmic space across the nuclear membrane governing by Exportin-5 via a Ran-GTP-mediated mechanism. Once inside the cytoplasm, the pre-miRNAs are further cleaved into RNA duplexes of 22 nucleotide by complex of Dicer- a second RNase III-type enzyme- and TAR RNA-binding protein 2. The RNA duplexes bind to a protein of 182 kDa with glycine-tryptophan repeat and argonaute proteins, AGO1-4, resulting in formation of the miRNA-induced silencing complex (RISC) [34, 35]. Notably, the mature guide strand (20–22 nucleotides in length) remains in association with RISC. This strand is also referred as miRNA-5p [36]. The other anti-sense strand, known as passenger miRNA (the so-called miRNA-3p, and is a complementary star-form miRNA, miRNA*) is released from RISC. The first thought was that the antisense strand is degraded in the cytoplasm; however, recently, a number of studies showed that some of them may have biological importance [37].
Subsequently, the mature miRNA exerts its biological function(s) via aligning the RISC to complementary sequences in the 3′ UTR of target mRNA [38, 39]. This association, most commonly, represses the translation of target proteins and recruits some protein complexes contributed in deadenylation and degradation of related target mRNA and finally, as a consequence, down regulation in gene expression [40].
Alternatively, the non-canonical pathway, also termed as Drosha-independent/Dicer-dependent pathway, pre-miRNAs bypass the Drosha/DGCR8 complex and is processed by AGO2 to the mature guide strand. The RNA product of this pathway is very short introns that are often referred as mirtrons. As shown in Fig. 1, following translocation to the cytoplasm from the nucleus, mirtrons act similarly to miRNA produced from the canonical pathway [41].
In the case of lncRNAs, it is suggested that these RNA molecules can be produced by RNA polymerase II, polyadenylated (polyA), even capped, or spliced. lncRNAs can be transcribed from intergenic regions; and in overlapping, antisense, intronic and bidirectional orientations (in comparison with protein-coding genes) from gene regulatory regions as well as specific chromosomal regions [42]. In the following context, we will briefly discuss some structural and biosynthetic features of four distinct lncRNAs.
lincRNAs
Thousands of lincRNAs transcripts have been identified in mammalian genomes, including approximately 3300 ones from six human cell types [24]. Predominately, RNA polymerase II transcribes lincRNAs and generates polyadenylated RNAs ranging in length from 2000 to 20,000 nucleo-tides, and a significant fraction is multiexonic, producing alternatively spliced variants [43]. lncRNA’s genes contain known chromatin signatures, including trimethyation of Lys 36 of histone 3 (H3K36me3) along with their tran-scribed region and trimethyation of Lys 4 of histone 3 (H3K4me3) at promoters. These RNA molecules have some characteristic features [44], such as possessing a lower expression level, being readily detectable, displaying a more tissue-specific pattern, and having a greater conservation rate in sequences together with the patches of higher conservation [45].
NATs
The ncRNA molecules are transcribed from the opposite strand of target genes, originating from protein or non-protein coding genes [46]. In comparison with lincRNAs, NATs are considered as an abundant class of lncRNAs, approximately 70 % of mouse genes, which transcribe in an antisense manner to form sense/antisense pairs of coding and non-coding RNAs [24, 47]. Although, both the 5′ and 3′ regions of protein-coding genes can be a target for antisense transcription [48]. However, it should be noted that, unlike lincRNAs, NATs show very little sequence conservation [49].
uaRNAs
A large number of uaRNAs molecules have been recognized in mouse and humans by the analysis of gene expression and cDNA libraries. Normally, RNA polymerase II–dependent transcriptional start sites have a 5′ cap, nevertheless, uaRNAs do not seem to be independently transcribed [50]. Since, these RNAs lack specific chromatin marks, it is deemed that they are indicative of transcriptional start sites and are not enriched for RNA polymerase II occupancy. Instead, uaRNAs seem to be derived from cleavage of the full-length transcript [51].
eRNAs
Generally, enhancers are regulatory DNA elements which are settled distally from transcription start sites, contain unique histone modifications, and recruiting and depositing transcription factors [52–54]. In fact, eRNAs are lncRNAs with short-life span and approximately 2 kb in length, which are transcribed bi-directionally by RNA polymerase II due to the recruitment of enzyme by enhancer regions in an activity-dependent manner. They have various species, and accordingly about 2000 in mouse and 3000 in humans had so far been identified through multiple studies. Accumulating studies and some special experimental techniques have shown that eRNAs are not polyadenylated and actively involved in promoting mRNA synthesis [55, 56].
Molecular mechanisms and function of ncRNA
miRNAs
Of the ncRNAs, miRNAs play certain roles in the control of multiple processes, including differentiation, proliferation, development and apoptosis [57, 58]. There are several described mechanisms of miRNA action, including (a) mRNA cleavage, (b) cap-40S initiation blockage, (c) 60S ribosomal unit connecting inhibition, (d) elongation inhibition, (e) ribosome premature termination, (f) co-translational protein degradation, (g) decomposition in P-bodies, (h) mRNA destabilization and (j) gene silencing [59, 60].
As main mechanisms, mRNA cleavage or translational repression are two post-transcriptional mechanisms by which miRNAs in complex with RISC can reduce the expression levels of genes. If the miRNA in combination with cytoplasmic RISC has adequate complementarity to the mRNA (usually to 3′ UTR), miRNA will determine the cleavage process [61–63]. If the mRNA does not have enough complimentarily to be degraded, but does have an appropriate constellation of miRNA complementary sites, miRNA will inhibit productive translation [64]. After this process, the miRNA remains unscathed and can guide the recognition and degradation of other targets. Another possible mechanism is that the recent synthesized polypeptide is specifically destructed after translation [65, 66]. In short, despite conduction of several studies on the mode of action of miRNAs, their mediated gene regulation still holds some secrets that need to be disclosed by future biochemical, molecular, and cellular investigation.
LncRNAs
As another class of ncRNA, lncRNAs impose pivotal roles in various aspects of biology [67]. Although the number of well characterized lncRNAs is not too many, until now, they have been confirmed to control the gene expression at various levels [68, 69]. Indeed, they have been implicated in transcriptional and epigenetic mechanisms, gene regulation in post-transcription level, alternative splicing, and in gene silencing [69, 70]. Different RNAs recruit multiple mechanisms that result in different regulatory outcomes. Despite structurally differences found between various classes of lncRNA, there are similarities in their mode of function(s).
LncRNAs have the capability to respond to disparate stimuli by eliciting substantial transcriptional controlling mechanism that regulates their expression. As a result, lncRNAs can act as molecular signals, because particular lncRNAs can be transcribed at a very distinctive time and place to merge developmental cues and explicate responding against several stimuli. It should be pointed out that some lncRNAs encompass regulatory functions, whereas others are exclusively products of transcription [24, 44].
LncRNAs have a central function in positively and negatively regulation of transcription, in large part due to the comprehensive transcription of enhancers and promoters by lncRNAs. Action as a molecular decoy is one of the diverse mechanisms by which lncRNAs may control transcription. After transcription of this type of lncRNA, it binds to a protein target and evaluates it, but does not apply any further functions. Then, the RNAs can exert their function through being as a “molecular sink” for RNA-binding proteins [71]. On the other hand, lncRNAs can bind protein(s), and then direct the ribonucleoprotein complex to particular targets. As discussed above, lncRNAs can exert alternations in gene expression in cis/trans manner that is not readily predicted from lncRNA sequence information. Numerous specific and likely prevalent functions of these RNA molecules in transcriptional regulation dictate somewhat exigency that a given local alteration in chromatin structure has not only local results, and may impose structural counteraction at a distance region [72–74].
In addition, lncRNAs can act as central platforms for assembling of several related molecular components in many various biological signaling events, which is deemed to be the characteristic of exact control by these RNA molecules, and is necessary to the accurate modification of the dynamics and specificity of signaling pathways and interactions between molecules [75, 76].
Roles of ncRNAs in brain health and neurodegenerative diseases
Of the ncRNAs, miRNAs and lncRNAs have rapidly been the center of research interests both in multiple basic fields of biology and translational medicine. These biomolecules have been demonstrated to impose diverse roles in different functions within brain as well as CNS disorders. Despite special attention to this topic, there are surprisingly some inadequacies for the accessible research technique and experimental protocols to disclose these macromolecular biomolecules impacts in brain functions in health and diseased conditions. Accordingly, we will provide brief yet focused insights on the miRNAs and lncRNA functions in modulating the multi-leveled activities within brain and neurodegenerative diseases, in particular, PD.
miRNAs
The expression level of miRNAs is very high within the mature and non-mature brain. They are able to increase the cellular homeostasis, moderate stress responses, and to modify numerous parameters related to the synaptic plasticity [77–81]. Actually, a range of miRNAs are only limited to human and have a brain-specific expression pattern. These RNA molecules can promote the integrity of specific differentiated neural phenotypes by inhibiting the expression of genes affecting the maintenance of the undifferentiated neural cell state. Subsequently, some changes in the expression mode of miRNAs may initiate the dedifferentiation and cellular transformation [82, 83]. However, it suggested that human brain-specific miRNA genes are not necessarily related to human special cognitive function, since these miRNA molecules have low expression levels and few conserved targets [84]. A recent study found a human brain-specific miRNA, miR-941, whose target genes are contributed in neurotransmitter signaling [19]. Another human brain-enriched and primate-specific miRNA termed miR-1202, which has been reported to be associated with the pathophysiology of depression [85].
One of the well- studied subgroups of miRNA targets comprises transcripts encoding synapse-related proteins such as synaptotagmin, synapsin 1, and the fragile-X mental retardation protein (FMRP) [86]. Myriad studies showed that during the crucial phases of memory formation, miRNAs positively exert their functions in synaptic tagging to confirm synaptic input specificity [87–89]. In the dendritic spines, miRNAs can interact with the cellular machinery in a reversible manner to generate long-term alternations in synaptic function. In response to synaptic activity, alternation in the RISC or conformational changes in dendritic mRNAs implicating availability to their 3′ UTRs induced miRNA-mediated silencing [90, 91] [92].
LncRNAs
The expression of LncRNAs is also high in various parts of the CNS, so that in a study of 1328 lncRNAs examined 849 cases were found to be expressed within the mouse brain [28]. In addition, it has been confirmed that lncRNAs are expressed across various regions within the brain [93]. Recently, huge numbers of studies have reported that lncRNAs possess main function in spatial–temporal modulation of gene expression involved in the brain development. Given that the CNS is one of the most intricate biological systems, it encompasses an enormous array of neuronal.
Parkinson’s disease: molecular mechanisms and etiology
Parkinson’s disease (PD) is defined with some pathological hallmarks such as the progressive death of dopaminergic neurons, which contains apparent amounts of neuromelanin in a midbrain structure, so-called the substantia nigra pars compacta (SNpc), and the presence of intraneuronal cytoplasmic inclusions, Lewy bodies [94]. These inclusions are made from neurofilaments and ubiquitin. Lewy bodies are also found in the peripheral nervous system of PD patients [95]. The motor symptoms, particularly akinesia, are caused by intense dopamine depletion in the striatum. Despite the increasing number of studies about the etiology of parkinsonian degeneration, the exact phenomena underlying such a degeneration process is yet to be fully defined [95, 96]. It should be stated that the dopaminergic neurons in the substantia nigra are more susceptible to degeneration. Some data supporting such findings are based on the high levels of reactive oxygen species, non-enzymatic auto-oxidation of dopamine which generates neuromelanin, and decrease in cells synthesizing glutathion peroxidase [97, 98]. Apparently, oxidative stress is one of the key causes of the nigral degeneration. The increase in ROS production during the degeneration of substantia nigra is caused by two major biochemical phenomena, (a) elevation in iron level possibly because of reduction of cellular ferritin and an increase in expression of lactoferrin receptor. This situation can result in an increased generation of ROS due to the auto-oxidation of neuromelanin [99, 100], and (b) reduction in antioxidant defense level. An increase in the dopamine turnover rates can cause a rise in basal generation of hydrogen peroxide, which in turn depletes the glutathione reservoirs [101].
Mitochondrial impairment, especially defect in complex I (nicotinamide adenine dinucleotide coenzyme Q reductase) of the respiratory chain appears to be another possible mechanism for Parkinsonian degeneration. No matter what the nature of the mitochondrial impairment is, it may eventually cause a marked drop in ATP levels and a defect in proton pumping. As a result, the mitochondrial membrane potential can be decreased, and cell can undergo the apoptosis [102].
Parkinson’s diseases related genes
Although there are 28 discrete chromosomal loci shown to be related to PD, only six of them have been illustrated to cause heritable monogenic PD, including α-synuclein (PARK1/PARK4), Parkin (PARK2), PINK1 (PARK6), DJ-1 (PARK7), LRRK2 (PARK8), and ATP13A2 (PARK9) [103]. Of these, α-synuclein is one of the most important genes with a definitive role in the pathogenesis of PD. Some significant genetic alternations in this gene are included point mutations, duplications, and triplications result in the death of dopamine neurons due to α-synuclein aggregation and accumulation in fibrillar form and neurotoxicity. α-synuclein is a prominent component of Lewy bodies. A dose–response relevance of α-synuclein gene has been reported to associate with the development of PD at an earlier age and its elevation is directly related to dementia severity [104, 105]. Moreover, mutation in leucine-rich repeat kinase 2 (LRRK2) has been known as the most prevalent cause of dominantly inherited PD and is a risk factor for sporadic PD. Recent accumulating studies suggested that LRRK2 may be involved in membrane trafficking and cytoskeletal dynamics, but its certain normal function has not been clearly described. LRRK2 has been suggested to deteriorate in the PD pathogenesis via a gain-of-function mechanism. Actually, a glycine to serine substitution, at position 2019 (G2019S) of LRRK2, results in an elevated activity of the activation loop of the kinase domain. The finding that LRRK2 inhibition suppresses neurotoxicity provides further support for the gain-of-function mechanism [106–108]. Another important PD- related gene, DJ-1, is a multifunctional protein that is associated with different cellular processes including the transcriptional regulation, cellular transformation, antioxidative stress reaction, chaperone, protease, and mitochondrial regulation. Further, oxidation of DJ-1, which causes inactivation of DJ-1, has been seen in patients with sporadic PD, revealing that DJ-1 also takes part in the onset and pathogenesis of the familial and sporadic PD [109]. Another causative gene for an autosomal recessive form of PD is Paskin, an ubiquitin-protein ligase (E3). Parkin is a component of the ubiquitin system and major adenosine triphosphate-dependent protein degradation machinery. It encodes a protein in an incomparable structure with a ubiquitin-like domain in the N-terminal and a RING finger motif in the C-terminal. Furthermore, it has been shown that CDCrel-1, a synaptic vesicle associated protein is a substrate for Parkin. In fact, several deletion and point mutations have been detected in patients with autosomal recessive PD. The substantia nigra and the locus coeruleus optionally undergo neurodegeneration without producing Lewy bodies [110, 111]. Mutations of the mitochondrial PINK1 appear to be another reason for the recessive PD. Studies on loss of function and overexpression revealed that PINK1 plays a significant role in apoptosis, abnormal mitochondrial morphology, impaired dopamine release and motor deficits. However, the basic mechanism underlying these different phenotypes remains to be elucidated. It has been shown that PINK1 deficiency or clinical mutations may affect the function of Complex I of the mitochondrial respiratory chain, resulting in mitochondrial depolarization and an elevated sensitivity to apoptosis. Therefore, multiple studies have suggested that Complex I deficiency underlies, at least partially, in the pathogenesis of hereditary form of PD [112, 113]. Finally, Human ATP13A2, a lysosomal P-type ATPase, has been shown to be related with autosomal recessive PD. The protein coded by ATP13A2 has a highly neurons-specific expression pattern and acts as a cation pump, but the substrate specificity is uncertain. Among the several genes involved in familial forms of PD, the ATP13A2 gene may display the first genetic link between PD pathogenesis and lysosomal pathways. Impairment in the lysosomal targeting of ATP13A2 is caused by missense or truncation mutations in the related gene. As a consequence, lysosomes encounter with a ATP13A2 deficiency because of the retention of mutant ATP13A2 in the endoplasmic reticulum cisterna [114, 115].
The impacts of miRNAs on Parkinson’ s disease
Because of the highlighted importance of miRNAs in pathogenesis of brain diseases, various studies have been conducted to clarify either direct or indirect roles of miRNAs in PD. Evidence for a direct or indirect function(s) of miRNAs in the pathophysiology of PD is now accumulating. One of the first investigations of a role for miRNAs in the maintenance of midbrain dopaminergic neurons was reported by Kim et al. [116]. Deletion of Dicer in embryonic stem cells when post-mitotic Dopaminergic neurons first arise resulted in complete loss of the dopaminergic neurons, while the generation of other mature neuronal classes was less affected. More important, the phenotype was rescued by transfection of miRNA. It has been demonstrated that several genes related to PD can be affected by miRNAs. Of these genes, the functional expression of α-synuclein is modulated by miR-153 and miR-7 which bind directly to the 3´UTR of α-synuclein. Further investigations reported other miRNAs can be involved in initiation and/or development of PD [117, 118]. For example, increased levels of fibroblast growth factor 20 (FGF20) gene also resulted in overexpression of α-synuclein and hence loss of dopaminergic neurons. Binding of miR-433 to a single nucleotide polymorphism (SNP) in the promoter region of FGF20 was shown to interrupt the target site for miR-433, causing the overexpression of α-synuclein [119]. In one of the first works investigating miRNAs involved in the pathology of PD, about six miRNAs were identified, including miR-1, miR-22*, and miR-29a (decreased levels in untreated patients in comparison with healthy controls), and miR-16-2*, miR-26a-2*, and miR-30a (increased levels in treated patients in comparison with untreated ones). It is worthy of mentioning that all of these miRNA molecules can be indirectly associated with the acting of α-synuclein. Of note, miR-30a and miR-1 were implicated in regulation of dopamine transport [120]. Valadi et al. investigated the miRNA expression in blood leukocytes of PD patients pre- and post-deep brain stimulation (DBS) by using the combination of exon microarray technologies and small RNA-seq. They compared the pattern of obtained miRNA with parallel healthy controls [121]. A set of five miRNAs (miR-4293, miR-378c, miR-18b*, miR-20a, and miR-1249) that are under-expressed in PD versus DBS has inversed/higher expression in controls when compared with PD, suggesting both treatment and disease-related changes in miRNA expression. Interestingly, down-regulation of miR-29a, miR-29c, miR-30c, and miR-19b can also be observed in white blood cells of PD DBS-off versus DBs-on patients and/or in the serum of PD versus control.
miRNA profiles in PD patients plasma samples have also been reported. In a study by Cardo et al. qRT-PCR analysis was performed in 25 controls and 31 patients after the onset of symptoms; they found only one significantly up-regulated miRNA, miR-331-5p. Another study identified miR-450b-3p, miR-1826, miR-505, and miR-626 in PD patients [122]. Soreq et al. identified 16 miRNAs altered in PD patients in comparison with healthy controls. They revealed that miR-20a, miR-16, and miR-320 presented in blood leukocytes [123]. As the development of circulating biomarkers for PD detection has great importance, hence, finding measurable molecular biomarkers can be potential clinical tools to facilitate early PD diagnosis. In the case of using of different protein biomarkers, there are some problems such as invasiveness and conflicting results among CSF proteins due to assay differences and/or blood contamination. Therefore, it is essential for detecting alternative biomarkers with high rate of efficacy and safety [124]. As above-mentioned, miRNAs open new opportunities for prognostic, diagnostic, and therapeutic interventions during PD onset.
LRRK2 as the most fundamental gene in PD pathophysiology negatively controls Let-7 and miR-184 in the dopamine generating cells. It initiates the overexpression of E2F1 and DP, resulting in inevitable defects in cell proliferation capacity and promoting cell death [125]. The miR-133b, a dopaminergic neuron-specific miRNA was reported to generate a negative feedback loop with the transcription factor PITX3 in midbrain dopamine producing neurons. Downregulation of miR-133b plays an important neuroprotective role [116]. miR-205, as another miRNA that regulates LRRK2, has significantly lower levels in the frontal cortex and striatum of PD patients. In murine models, the inhibition of miR-205 was reported to impose upregulation of LRRK2 protein expression [126]. Further investigations have been centralized on the other PD-related genes, when meaningful relationships have been found that exist with miRNAs. In one of these studies, it was revealed that DJ-1 and Parkin are respectively controlled by miR-34b and miR-34c, respectively. These miRNAs were downregulated at advanced stages of PD.
Furthermore, decrease in dopamine signaling in the striatum may involve to up-regulate acetylcholine esterase (AChE), because an acetylcholine imbalance promotes the death of dopaminergic neurons [127]. miR-132 is an important molecule that negatively regulates dopamine neuron differentiation. Yang et al. [128] previously reported that the inhibition of miR-132 significantly increases differentiation of dopamine neurons, whereas prolific expression of miR-132 in the embryonic stem cells dramatically represses dopamine neuron differentiation. It also has been found that miR-132 inhibits AChE, demonstrating a neuro-protective role in dopaminergic neurons [129].
A brain-enriched miRNA, miR-124, was reported to play a critical role in the neuronal differentiation during the CNS development. Kanagaraj et al. [130] showed a reduction in the expression miR-124 in the substantia nigra of the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD mouse model. Further, in vitro study revealed a decrease in the expression levels of miR-124 in MN9D dopaminergic neurons treated with MPP iodide. Gong et al. [131] reported that miR-124 suppression increased neuronal autophagy and apoptosis by regulating the AMPK/mTOR signaling pathway in PD. In addition, miR-124 is down-regulated in MPTP-treated PD cells, indicating its role on the induction of neurons apoptosis and autophagy. In addition to its possible role in PD, the integrated interplay between miR-124 and glucocorticoids seems to be important [132]. In response to the induction of miR-124 by glucocorticoids, miRNA can directly interact with the 3′-UTR of glucocorticoids receptor-α mRNA to inhibit the expression of glucocorticoids receptor α and decrease the anti-inflammatory effects of glucocorticoids [133].
On the other hand, previous studies unveiled that the deregulation of glucocorticoid receptor and function are possibly substantial in the degeneration of dopamine neurons through the establishment of chronic inflammation [134].
Many of these studies, which have been conducted recently, provide important evidence that dysregulated miRNA acts as an essential molecular trigger in the pathogenesis of PD [135]. The list of miRNAs involved in PD become longer every year, and discussion about all these miRNAs is out of the scope of this review.
LncRNAs: new target for PD research
In 2014, for the first time, Soreq et al. [136] utilized a whole-transcriptome RNA sequencing, consisting general RNA-Seq analysis techniques, to determine all the transcripts that code proteins in leukocyte and lncRNAs in the control group as well as PD patients. They recognized a decreased lncRNA expression and selective PD-induced alternation in 13 of over 6000 detected leukocyte lncRNAs. These researchers also found five lncRNAs that their expression levels were increased in the disease and inversely reduce following DBS. The candidate lncRNAs include the spliceosome component U1, which support the idea that splicing modulations is involved in diseases. Additionally, elevated levels of the muscular dystrophy-associated RP11-462G22.1 (lnc-FRG1-3) may be related to the muscle rigidity in PD, since routinely is screened as one of the six disease indications of motor symptoms. Another disease-altered lncRNAs, post-DBS (RP11-79P5.3), was also detected as differentially expressed by analysis of an additional external, independent PD brain RNA-Seq data-set. Based upon their findings, the authors concluded that lncRNAs may be the exquisite biomarkers for PD and other neurodegenerative diseases and a considerable mean in future personalized neurology. In 2015, in a new study was conducted by Carrieri et al. [137] to study the involvement of antisense lncRNAs in PD based on assumption that antisense transcription can regulate sense gene expression functioning at distinct regulatory levels. These researchers, in a previous study, identified AS Uchl1 as an antisense to the mouse Ubiquitin carboxy-terminal hydrolase L1 (Uchl1) gene (AS Uchl1), the synthetic locus of UCHL1/PARK5. It should be highlighted that UCHL1/PARK5 one of the 28 genes involved in PD- is mutated in rare cases of early-onset familial PD. More important, manipulation of UchL1 expression has been proposed as a tool for therapeutic intervention. AS Uchl1 enhances UchL1 translation and expression and is the indicative member of SINEUPs (SINEB2 sequence to upregulate translation) from a new functional group of NATs. It was shown that Nurr1, a key transcription factor implicated in dopaminergic cells’ differentiation and maintenance, controls the functional expression of AS Uchl1. Moreover, AS Uch1 RNA levels are significantly down regulated in neurochemical models of PD in vitro and in vivo. Therefore, AS Uchl1 RNA is a part of Nurr1-dependent gene network and a target of cellular stress, which can extend our understanding of the role of AS transcription in the brain. Taken together, since elevating evidence indicated that increase expression levels of Uchl1 could be advantageous in neurodegenerative diseases, the application of AS Uchl1 as RNA-based drugs may display a novel therapeutic strategy.
Conclusion
Detection and analysis of ncRNAs, which appear to control the gene expression, will provide a better understanding of the molecular mechanisms, particularly in the nervous system. However, the pathogenesis of various neurodegenerative diseases is yet to be fully understood. Such information may favor the development of diagnostics and treatment of severe human disorders including, PD. Therefore, it is suggested that the general synthetic pathways and classification of ncRNAs and the possible mechanism of ncRNAs involved in PD need to be clarified towards the roles of two classes of ncRNAs, i.e., miRNA and lncRNA. These ncRNAs seem to play pivotal roles can be implicated in the unusual deposition of α-synuclein and affect other PD-related genes. Further, ncRNAs are particularly important for their roles in normal development and function of the CNS and the CNS-related disorders. They are implicated in targeted gene expression and function as major modulators in various neuroprotective pathways. Changes in the patterns of miRNA and presumably lncRNA expression will probably act as diagnostic indication of brain function, and the pathology of neurological disorders and neurodegenerative diseases. Further research seems to be necessary in ncRNA expression patterns and profiling, which may result in discovery of many different modern biomarkers. Profound studies need to be capitalized on identifying the function of lncRNAs in RNA-mediated gene control is likely to remain an area of extreme research interest. However, it opens a new horizon in the human genomic studies towards clarification of the transcriptional complexity of the human brain and potentially novel ways in the regulatory network governing PD pathogenesis.
Future perspectives
A pivotal challenge in targeting lncRNAs in different milieu is that they have potency to elicit off-target interactions and recognize unexpected targets when added to different cells or tissues [138]. However, it is recently acclaimed that any manipulation or chemical modification can tailor their affinity to intended effects, reduce toxicity rate and contribute to improved pharmacokinetic properties [139]. In addition, the determination of alternative hypotheses to pinpoint interesting results, assessment of several negative controls, calculating the copy number of the RNA target per cell, designing independent experimental approaches and establishment of independent empirical approaches could ameliorate their side effects [138].
Commensurate with these findings, emerging computational models and reliable instruments can help us to decrease the time and overall cost of biological trials by calculating the inherent association probability of candidate lncRNA-disease pair and confirming most favorable lncRNA-disease pairs with high scores related to ongoing biological experimental validation. Further investigation in the development of computational models by combining multiple biological datasets have no doubt to increase our understanding of the disease-related lncRNAs in health and disease. It also will remarkably benefit the introduction of lncRNA therapeutic strategies aimed at the modulation of uncontrolled aberrancies [140].
References
Wu Y, Le W, Jankovic J (2011) Preclinical biomarkers of Parkinson disease. Arch Neurol 68(1):22–30
Zheng B et al (2010) PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med . doi:10.1126/scitranslmed.3001059
Papapetropoulos S, McCorquodale D (2007) Gene-expression profiling in Parkinson’s disease: discovery of valid biomarkers, molecular targets and biochemical pathways. Future Neurol 2:29–38
Hamza TH et al (2010) Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet 42(9):781–785
Ibanez P et al (2004) Causal relation between α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364(9440):1169–1171
Chiba-Falek O, Lopez GJ, Nussbaum RL (2006) Levels of α-synuclein mRNA in sporadic Parkinson disease patients. Mov Disord 21(10):1703–1708
Mandel S et al (2007) Applying transcriptomic and proteomic knowledge to Parkinson’s disease drug discovery. Exp Opin Drug Disc 2(9):1225–1240
Grünblatt E (2012) Parkinson’s disease: molecular risk factors. Parkinsonism Relat Disord 18:S45–S48
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6(4):259–269
Hammond SM (2005) MicroRNAs as tumor suppressors. Nat Genet 39(5):582–583
Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10(10):704–714
Nicoloso MS et al (2009) MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 9(4):293–302
Salta E, De Strooper B (2012) Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol 11(2):189–200
Ambros V (2001) microRNAs: tiny regulators with great potential. Cell 107(7):823–826
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318(5858):1931–1934
Hu HY et al (2012) Evolution of the human-specific microRNA miR-941. Nature Commun 3:1145
Robles AI, Harris CC (2013) A primate-specific microRNA enters the lung cancer landscape. Proc Natl Acad Sci 110(47):18748–18749
Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12(2):136–149
Wang X et al (2011) The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol 3:a003756
Gupta RA et al (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291):1071–1076
Guttman M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227
Pauli A et al (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22(3):577–591
Li X et al (2013) Long noncoding RNAs: insights from biological features and functions to diseases. Med Res Rev 33(3):517–553
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10(3):155–159
Mercer TR et al (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105(2):716–721
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136(4):629–641
Managadze D et al (2011) Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol Evol 3:1390–1404
Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463(7281):621–626
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Ameres SL, Zamore PD (2013) Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14(8):475–488
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11(9):597–610
Rüegger S, Großhans H (2012) MicroRNA turnover: when, how, and why. Trends Biochem Sci 37(10):436–446
Westholm JO, Lai EC (2011) Mirtrons: microRNA biogenesis via splicing. Biochimie 93(11):1897–1904
Fabian MR, Sonenberg N (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19(6):586–593
Gregory RI et al (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123(4):631–640
Schwarz DS et al (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2):199–208
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acids Res 32(suppl 1):D109–D111
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9(2):102–114
Chen G et al (2013) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41(D1):D983–D986
Khalil AM et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106(28):11667–11672
Guttman M et al (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28(5):503–510
Dinger ME et al (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18(9):1433–1445
Katayama S et al (2005) Antisense transcription in the mammalian transcriptome. Science 309(5740):1564–1566
Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10(9):637–643
He Y et al (2008) The antisense transcriptomes of human cells. Science 322(5909):1855–1857
Werner A, Sayer JA (2009) Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens 18(4):343–349
Rastinejad F, Blau HM (1993) Genetic complementation reveals a novel regulatory role for 3′ untranslated regions in growth and differentiation. Cell 72(6):903–917
Sanchez-Herrero E, Akam M (1989) Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107(2):321–329
Heintzman ND et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39(3):311–318
Visel A et al (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457(7231):854–858
Kim T-K et al (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465(7295):182–187
Tisseur M, Kwapisz M, Morillon A (2011) Pervasive transcription–lessons from yeast. Biochimie 93(11):1889–1896
Mattick JS (2010) Linc-ing long noncoding RNAs and enhancer function. Dev Cell 19(4):485–486
Jalali S et al (2013) Systematic transcriptome wide analysis of lncRNA–miRNA interactions. PLoS One 8(2):e53823
Ouellet DL et al. (2006) MicroRNAs in gene regulation: when the smallest governs it all. BioMed Research International
Friedman RC et al (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell 120(1):15–20
Hutvágner G, Zamore PD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297(5589):2056–2060
Zeng Y, Cullen BR (2003) Sequence requirements for micro RNA processing and function in human cells. RNA 9(1):112–123
Zeng Y, Wagner EJ, Cullen BR (2002) Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 9(6):1327–1333
Doench JG, Petersen CP, Sharp PA (2003) siRNAs can function as miRNAs. Genes Dev 17(4):438–442
Seggerson K, Tang L, Moss EG (2002) Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Developmental biology 243(2):215–225
Brennecke J et al (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1):25–36
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21(6):354–361
Bernstein E, Allis CD (2005) RNA meets chromatin. Genes Dev 19(14):1635–1655
Whitehead J, Pandey GK, Kanduri C (2009) Regulation of the mammalian epigenome by long noncoding RNAs. Biochimica et Biophysica Acta 1790(9):936–947
Bracken AP, Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9(11):773–784
Guenther MG et al (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130(1):77–88
Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330(6004):612–616
Hung T, Chang HY (2010) Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 7(5):582–585
Lee JT (2009) Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 23(16):1831–1842
Spitale RC, Tsai M-C, Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6(5):539–543
Good MC, Zalatan JG, Lim WA (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332(6030):680–686
Cheng L-C, Tavazoie M, Doetsch F (2005) Stem cells: from epigeneticsto microRNAs. Neuron 46(3):363–367
Jin P, Alisch RS, Warren ST (2004) RNA and microRNAs in fragile X mental retardation. Nat Cell Biol 6(11):1048–1053
Schratt GM et al (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439(7074):283–289
Sempere LF et al (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5(3):R13
Smirnova L et al (2005) Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21(6):1469–1477
Kosik KS, Krichevsky AM (2005) The elegance of the microRNAs: a neuronal perspective. Neuron 47(6):779–782
Krichevsky AM et al (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9(10):1274–1281
Chen W, Qin C (2015) General hallmarks of microRNAs in brain evolution and development. RNA Biol 12(7):701–708
Lopez JP et al (2014) miR-1202: a primate specific and brain enriched mirna involved in major depression and antidepressant treatment. Nat Med 20(7):764
John B et al (2004) Human microRNA targets. PLoS Biol 2(11):e363
Kim J et al (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci 101(1):360–365
Martin KC, Kosik KS (2002) Synaptic tagging—who’s it? Nat Rev Neurosci 3(10):813–820
Schaeffer C et al (2003) The RNA binding protein FMRP: new connections and missing links. Biol Cell 95(3–4):221–228
Lugli G et al (2005) Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94(4):896–905
Ashraf SI et al (2006) Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124(1):191–205
Mehler MF, Mattick JS (2007) Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 87(3):799–823
Qureshi IA, Mattick JS, Mehler MF (2010) Long non-coding RNAs in nervous system function and disease. Brain Res 1338:20–35
Dauer W, Przedborski S (2003) Parkinson’s disease: mechanisms and models. Neuron 39(6):889–909
Marsden C (1982) Neuromelanin and Parkinson’s disease. J Neural Transm Suppl 19:121–141
Wu D-C et al (2003) NADPH oxidase mediates oxidative stress in the 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine model of Parkinson’s disease. Proc Natl Acad Sci 100(10):6145–6150
Uhl GR (1998) Hypothesis: the role of dopaminergic transporters in selective vulnerability of cells in Parkinson’s disease. Ann Neurol 43(5):555–560
Damier P et al (1993) Glutathione peroxidase, glial cells and Parkinson’s disease. Neuroscience 52(1):1–6
Dexter D et al (1989) Increased nigral iron content and alterations in other metal ions occurring in brain in Parkinson’s disease. J Neurochem 52(6):1830–1836
Riederer P et al (1989) Transition metals, ferritin, glutathione, and ascorbic acid in parkinsonian brains. J Neurochem 52(2):515–520
Sofic E et al (1992) Reduced and oxidized glutathione in the substantia nigra of patients with Parkinson’s disease. Neurosci Lett 142(2):128–130
Blum D et al (2001) Molecular pathways involved in the neurotoxicity of 6-OHDA, dopamine and MPTP: contribution to the apoptotic theory in Parkinson’s disease. Prog Neurobiol 65(2):135–172
Klein C, Westenberger A (2012) Genetics of Parkinson’s disease. Cold Spring Harb Perspect Medicin 2(1):a008888
Farrer M et al (2004) Comparison of kindreds with parkinsonism and α-synuclein genomic multiplications. Ann Neurol 55(2):174–179
Singleton A et al (2003) α-Synuclein locus triplication causes Parkinson’s disease. Science 302(5646):841
West AB et al (2005) Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA 102(46):16842–16847
Brice A (2005) Genetics of Parkinson’s disease: lRRK2 on the rise. Brain 128(12):2760–2762
Heman-Ackah SM et al (2013) RISC in PD: the impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Frontiers in molecular neuroscience. doi:10.3389/fnmol.2013.00040
Lev N et al (2006) Role of DJ-1 in Parkinson’s disease. J Mol Neurosci 29(3):215–225
Mizuno Y et al (2001) Parkin and Parkinson’s disease. Curr Opin Neurol 14(4):477–482
Dawson TM, Dawson VL (2010) The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord 25(S1):S32–S39
Jones R (2010) The roles of PINK1 and Parkin in Parkinson’s disease. PLoS Biol 8(1):e1000299
Gandhi S et al (2006) PINK1 protein in normal human brain and Parkinson’s disease. Brain 129(7):1720–1731
Dehay B et al (2012) Lysosomal dysfunction in Parkinson disease: aTP13A2 gets into the groove. Autophagy 8(9):1389–1391
Park JS. et al (2014) Parkinson’s disease-associated human ATP13A2 (PARK9) deficiency causes zinc dyshomeostasis and mitochondrial dysfunction. Human mole genet p ddt623
Kim J et al (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317(5842):1220–1224
Doxakis E (2010) Post-transcriptional regulation of α-synuclein expression by mir-7 and mir-153. J Biol Chem 285(17):12726–12734
Junn E et al (2009) Repression of α-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci 106(31):13052–13057
Wang G et al (2008) Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-synuclein. Am J Hum Genet 82(2):283–289
Margis R, Margis R, Rieder CR (2011) Identification of blood microRNAs associated to Parkinsońs disease. J Biotechnol 152(3):96–101
Valadi H et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659
Cardo LF et al (2013) Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J Neurol 260(5):1420
Soreq L et al (2013) Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front Mol Neurosci 6:10
Khoo SK et al (2012) Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. J Parkinson’s Dis 2(4):321–331
Gehrke S et al (2010) Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466(7306):637–641
Cho HJ et al (2013) MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet 22(3):608–620
Shaked I et al (2009) MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31(6):965–973
Yang D et al (2012) miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J Cell Sci 125(7):1673–1682
Soreq H (2015) MicroRNA-target interactions in neurodegenerative diseases. SpringerPlus 4(Suppl 1):L1
Kanagaraj N et al (2014) Downregulation of miR-124 in MPTP-treated mouse model of Parkinson’s disease and MPP iodide-treated MN9D cells modulates the expression of the calpain/cdk5 pathway proteins. Neuroscience 272:167–179
Gong X et al (2016) miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am J Transl Res 8(5):2127–2137
Kim J et al (2015) MicroRNA-124 regulates glucocorticoid sensitivity by targeting phosphodiesterase 4B in diffuse large B cell lymphoma. Gene 558(1):173–180
Ledderose C et al (2012) Corticosteroid resistance in sepsis is influenced by microRNA-124–induced downregulation of glucocorticoid receptor-α*. Crit Care Med 40(10):2745–2753
Herrero, M.-T., et al., Inflammation in Parkinson’s disease: role of glucocorticoids. Front Neuroanat 2015. 9
Miñones-Moyano E et al (2011) MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet 20(15):3067–3078
Soreq L et al (2014) Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput Biol 10(3):e1003517
Carrieri C et al (2015) Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cellular Neurosci 9:114
Matsui M, Corey DR (2016) Non-coding RNAs as drug targets. Nat Rev Drug Discov. doi:10.1038/nrd.2016.117
Bennett CF, Swayze EE (2010) RNA targeting therapeutics: molecular mechanisms ofAntisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50:259–293
Chen X (2015) Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep 17(5):13186
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors have declared no conflicts of interest.
Rights and permissions
About this article
Cite this article
Majidinia, M., Mihanfar, A., Rahbarghazi, R. et al. The roles of non-coding RNAs in Parkinson’s disease. Mol Biol Rep 43, 1193–1204 (2016). https://doi.org/10.1007/s11033-016-4054-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4054-3