Abstract
Seed shattering is an important trait related to the efficiency of harvesting seeds in crops. Although several genes have been identified to control seed shattering in rice (Oryza sativa L.), the genetic architecture underlying extensive variation of seed shattering is yet unclear. ‘Oonari’, a moderate-shattering indica rice cultivar, was developed from a mutant obtained through gamma-ray irradiation of an easy-shattering cultivar ‘Takanari’. The grain pedicel of ‘Oonari’ showed greater resistance to bending than that of ‘Takanari’, while there were no obvious differences in resistance to pulling and abscission zone formation. Investigation of the seed shattering of an F2 population from a cross between ‘Takanari’ and ‘Oonari’ indicated a single semi-dominant locus responsible for seed shattering, which was designated as Sh13. We used bulked DNA of F3 lines with the same shattering degree as that of ‘Oonari’ and DNAs of their parents for whole-genome sequencing, localizing Sh13 at the terminal region on the long arm of chromosome 2. In the candidate genomic region of ‘Oonari’, we identified a tandem duplicated segment containing a microRNA gene, osa-mir172d, whose wheat ortholog is involved in controlling grain threshability. Quantitative RT-PCR analysis indicated that the relative expression of osa-mir172d in ‘Oonari’ was twofold higher than that in ‘Takanari’. We suppose that the duplication of osa-mir172d be involved in reducing seed shattering in ‘Oonari’.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seed shattering is an adaptive trait to facilitate seed dispersal in wild plant species, while reduction of seed shattering is thought to be one of the most important steps in cereal crop domestication, because it drastically enhances the efficiency of seed harvesting. Rice (Oryza sativa L.) is considered to have been domesticated from one or both of two closely related wild species O. nivara and O. rufipogon (Oka 1988; Zong et al. 2007). Although cultivated rice generally has a lower shattering level than the wild species, a wide range of seed-shattering levels, from “very easy” to “very hard” shattering, has been observed in different cultivars (Konishi et al. 2006). A balance between ease of shattering and hard threshing is necessary, and this is determined in part by the harvesting methods employed.
Seed shattering in rice depends on the formation, morphology, composition, and degradation of the abscission zone between a grain and a pedicel (Ebata and Tashiro 1990; Fuller and Allaby 2009; Okubo 2014; Yoon et al. 2014). Quantitative trait loci (QTLs) associated with seed shattering have been identified on nearly every rice chromosome (Xiong et al. 1999; Qin et al. 2010; Htun et al. 2014; Lee et al. 2016), and two genes involved in rice domestication have been cloned (Konishi et al. 2006; Li et al. 2006). The sh4 gene encodes a transcription factor with an Myb3 DNA binding domain, and a nucleotide substitution of G to T in the first exon of sh4 leads to reduced function of sh4 and incomplete development of the abscission zone in cultivated rice (Li et al. 2006). This gene has been considered to be the most important contributor to reduced shattering during rice domestication from its wild ancestor (Li et al. 2006; Purugganan and Fuller 2009). In addition, a one-nucleotide insertion in the second exon of sh4, which can be assumed to be a null-function mutation, disturbs the formation of the abscission zone more seriously and further reduces the shattering degree in an indica rice cultivar (Zhou et al. 2012). The qSH1 gene encodes a BEL1-type homeobox gene, and an SNP (single nucleotide polymorphism) in the 5′ regulatory region of the qSH1 gene causes the absence of abscission zone formation, leading to complete loss of seed shattering in hard-shattering japonica cultivars (Konishi et al. 2006). SH5, a highly homologous gene of qSH1, has been proposed to modulate seed shattering by enhancing abscission zone development and inhibiting lignin biosynthesis (Yoon et al. 2014). Using mutants, more genes involved in seed shattering have been identified (Fukuta and Yagi 1998; Ji et al. 2010; Lin et al. 2012; Zhou et al. 2012). SHAT1, a member of APETALA2 (AP2) (Zhou et al. 2012), and OsSh1, a YABBY-like gene (Lin et al. 2012), also function in specifying abscission zone development. In contrast, another gene Oryza sativa CTD phosphatase-like1 (OsCPL1) has been revealed to repress differentiation of the abscission layer during panicle development (Ji et al. 2006; Ji et al. 2010). More recently, OSH15 (Sato et al. 1999), a KNOX protein, has been revealed to form a dimer with SH5, which enhances seed shattering by directly inhibiting lignin biosynthesis genes (Yoon et al. 2017). Despite such remarkable findings, the genetic architecture responsible for moderate-shattering character is unclear.
‘Takanari’ is a high-yielding indica rice cultivar, which displays an easy-shattering phenotype (Imbe et al. 2004; Kanemura et al. 2007). To reduce its yield loss caused by seed shattering, we recently developed a new cultivar ‘Oonari’ with moderate-shattering character from a mutant line of ‘Takanari’ obtained by gamma-ray irradiation (http://www.naro.affrc.go.jp/english/nics/index.html). Due to the low mutation frequency (one mutation per 6.2 Mb) of gamma-ray-irradiated rice (Sato et al. 2006), the morphological characteristics of both cultivars are almost the same, except that the yield of ‘Oonari’ is higher than that of ‘Takanari’, which is attributed to reduced seed shattering. As a substitute for ‘Takanari’, ‘Oonari’ is expected to be widely used as a high-yielding cultivar. Furthermore, moderate-shattering rice varieties are fitter for harvesting by hands or a large combine harvester. It is therefore of scientific and practical importance to identify the causal genes responsible for reduced seed shattering in ‘Oonari’.
Although all the abovementioned seed shattering–related genes have been isolated by QTL mapping and map-based cloning, this approach is time- and labor-consuming. Alternatively, the combination of bulked segregant analysis and whole-genome sequencing has dramatically accelerated the process of identifying causal genes (Schneeberger and Weigel 2011; Abe et al. 2012; Zou et al. 2016). The underlying principle of this method is that only one or a few mutations are associated with the mutant phenotype in a segregating population which was derived from the cross between a mutant- and wild-type. In the bulked DNA pool from the individuals that exhibit a recessive mutant phenotype, the recessive causal mutation and its closely located mutations are expected to be homozygous for mutant alleles, whereas both mutant alleles and wild-type alleles can be detected in the other mutations.
In this study, we characterized the seed shattering degree of ‘Oonari’ using different methods and identified a mutation associated with reduced seed shattering by combining bulked segregant analysis and whole-genome sequencing.
Materials and methods
Plant materials and growth conditions
Rice cultivar ‘Oonari’ (moderate-shattering phenotype) and its original cultivar ‘Takanari’ (easy-shattering phenotype) were grown in three different cropping seasons, i.e., early season (from seed sowing at the end of March to grain harvesting at the end of September), normal season (from late April to the beginning of October), and late season (from the beginning of June to the end of October), from 2016 to 2018. Seeds were sown in nursery boxes and 4-week-old seedlings were transplanted in a paddy field at the Radiation Breeding Division, Institute of Crop Sciences, NARO (Hitachiomiya, Ibaraki, Japan; 36.52° N 140.40° E). Plants of each cultivar were grown in double rows at a spacing of 20 cm between hills and 14 cm between two paired rows, spacing between the cultivars being 28 cm. To further investigate shattering degrees of ‘Oonari’ and ‘Takanari’, the well-known cultivars ‘Kasalath’ (easy-shattering phenotype) and ‘Nipponbare’ (hard-shattering phenotype) were grown in the normal season for comparison.
‘Oonari’ and ‘Takanari’ were crossed to generate three F2 populations, named by F2-a, F2-b, and F2-c. Eighty-eight and 91 individuals in the F2-a and F2-b populations were self-pollinated to generate F3-a and F3-b lines, respectively. Plants of the F3-a and F3-b lines were grown in the late season of 2015 and 2016, respectively. The F3-a population was used for bulked segregant analysis with whole-genome sequencing. The F3-b population was used for validation of bulked segregant analysis. More than eight plants of each F3 line were grown in a single row with a spacing of 15 cm between hills and 30 cm between rows. Seeds of the F2-c population were sown in nursery boxes on March 31, 2016, and a total of 83 4-week-old seedlings were transplanted into 15-cm-diameter pots. After 55 days, about seven tillers per plant were produced. Hence, the tillers of each F2 individual were divided into roughly seven sub-individuals and transplanted into a paddy field in a single row at the same density as that of the F3 populations. Since new tillers can grow from each transplanted sub-individual in the same way as transplanted seedling, the shattering degree of each F2 individual can be evaluated repeatedly. This F2-c population was also used for validation of the mapping results. In each population, ‘Takanari’ and ‘Oonari’ were used as controls and grown in a single row with three replications.
For fine mapping the candidate genes, the recombinant lines together with parent lines were grown in a heated glasshouse (T ≥ 20 °C) from November 2017 to April 2018.
Evaluation of shattering degree
Evaluation methods of pulling strength (PS) and bending strength (BS) were used as described by Ji et al. (2006) and Qin et al. (2010) with minor modifications. Rice panicles from the primary tillers were harvested and put into paper envelopes. The samples were placed in a refrigerator at 4 °C and moisture of about 20% for more than 2 weeks. Breaking tensile strength for detachment of a seed from the pedicel by pulling and bending was measured using a digital force gauge (FGP-0.5, Shimpo, Japan) fitted onto a tension testing machine (AL-107-03A, Alpha Kikai, Japan). For measurement of PS, the panicle axis was clamped by a grip fixed on the tension testing machine and a grain was clamped horizontally by another grip fixed on the force gauge (Supplementary Fig. 1 a and b). Pulling the pedicel by gently rotating the shaft of the tension testing machine, the maximum tensile strength, measured at the moment of pedicel breakage, was recorded in gravitation units of force (gf). For measurement of BS, a grain was clamped vertically and the maximum tensile strength was recorded (Supplementary Fig.1 c and d). To evaluate the changes in PS and BS of grain pedicels during grain development in different cultivars grown in the normal season, we recorded the heading date of the randomly selected primary tillers. One panicle from each of four plants were harvested from each cultivar on the heading date, 10th, 20th, 30th, and 40th day after heading in 2017. PS and BS values of six grains in the upper part of each panicle were respectively measured. So 24 observations for PS and BS were made each time that the trait was measured for a cultivar. In addition, we developed a novel method to evaluate the shattering degree. The grain was cut out with its basal pedicel, the grain being subsequently clamped vertically, following which the lower end of the pedicel was pushed over sideways by a flat strip of metal fixed to the force gauge to test if it would shatter (the force strength was not recorded) (Supplementary Fig. 1 e and f). Twelve grains were tested for each panicle, and the rate of grain shattering by pushing (RSP) was estimated. To confirm the differences in seed shattering among different cultivars, four panicles from each of four plants at the maturity stage were harvested from each cultivar grown in the normal season in 2018. PS, BS, and RSP of 24 grains in the upper part of each plant were respectively measured. Average values of PS, BS, and RSP were calculated for four independent plants in each cultivar.
For rapid selection of F3-a lines with the same shattering degree as that of ‘Oonari’, we adopted a method based on grasping panicles by hand at the maturity stage (Subudhi et al. 2014). A shattering score was given for each line on a scale of 1–9: 1 = no shattering and 9 = heavy shattering. The shattering scores of ‘Oonari’ and ‘Takanari’ were 4 and 6, respectively. Only the lines for which all 12 investigated plants showed shattering scores of 4 were selected for bulked segregant analysis.
The RSP value of each individual in the F2-c population was calculated based on the average value of its four sub-individuals evaluated on the 15–17th day after heading. The shattering degrees of some of the F2-c individuals (n = 53) and F3-b lines (n = 75) were evaluated using the method based on grasping panicles by hand at the maturity stage (Okubo et al. 2012). Three panicles of each plant were grasped each time, and the number of shattered grains was recorded. The rate of grain shattering by grasping (RSG) was calculated by dividing the number of shattered grains by the average number of grains grasped. The average RSG values for five independent plants of an F2 individual or an F3 line were calculated.
Histological analysis of abscission zone formation
The pedicels of plants grown in the early season were collected at the heading stage and those grown in the late season were collected on the 20th day after heading, followed by fixation in FAA solution (5% v/v formaldehyde, 5% v/v acetic acid, and 45% ethanol). After dehydration using a graduated t-butanol/ethanol/water series (0/50/50, 10/50/40, 20/50/30, 35/45/20, 50/40/10, 75/25/0, 100/0/0, in percentage) for 1 h each, the tissues were embedded in Paraplast Plus (McCormick Scientific, Missouri, USA), and cut into 2- or 10-μm-thick sections using a microtome (Leica Biosystems, Nussloch, Germany). The sections were mounted onto Matsunami adhesive silane (MAS)-coated slides (Matsunami Glass, Osaka, Japan), then deparaffinized in xylene and rehydrated through a graded ethanol solution of decreasing concentration (100%, 90%, 70%, 50%, 30%, and 0%) for 3 min each. The sections were then stained with 0.01% toluidine blue and dehydrated through a graded ethanol series and finally mounted in Eukitt (O. Kindler, Freiburg, Germany). The abscission zone was observed through an AX70 or BX53 microscope (OLYMPUS, Tokyo, Japan).
DNA extraction
For whole-genome sequencing, seeds of two plants of each F3-a line with the same shattering degree as that of ‘Oonari’ were harvested and germinated. The leaves of 7-day-old seedlings were harvested and bulked in equal amounts. The leaves of ‘Oonari’ and ‘Takanari’ were harvested from 30-day-old seedlings. DNA was extracted using DNeasy Plant Maxi Kit (Qiagen Inc., Valencia, USA) according to the manufacturer’s protocol. DNA of each line in the F3-b population and that of each individual in the F2-c population were extracted using the simple DNA extraction method (Edwards et al. 1991).
Whole-genome sequencing
The genomic DNAs of F3-a bulk, ‘Takanari’, and ‘Oonari’ were sent to the Beijing Genomics Institute (BGI, Hong Kong, China) for DNA library construction and whole-genome sequencing. Paired-end (2 × 100 bp) sequencing was performed on Illumina HiSeq 4000 (Illumina, Inc. San Diego, CA, USA) to determine genomic sequences to about 30-fold depth for each sample. In addition, the genomic DNA of ‘Oonari’ was sequenced again with paired-end (2 × 150 bp) reads on Illumina HiSeq X Ten.
Bioinformatics analysis
The next-generation sequencing (NGS) data of ‘Takanari’ (Yonemaru et al. 2014) available in the DDBJ Sequenced Read Archive under the accession number DRX016587 was also used in this study. Thus, two sets of NGS data for ‘Takanari’ and ‘Oonari’ and one set for F3 bulk were subjected to whole-genome sequence analysis. After removing adapter sequences, contamination, and low-quality (Q < 20) and unpaired reads from raw reads, genome mapping was conducted using a galaxy workflow (https://galaxy.dna.affrc.go.jp/nias/static/register_en.html). In brief, the clean reads were aligned to the rice reference genome (Nipponbare, IRGSP-1.0, http://rapdb.dna.affrc.go.jp) (Kawahara et al. 2013) using BWA MEM (Galaxy Version 0.7.9) (Li and Durbin 2009), followed by removing the PCR duplicates by the MarkDuplicates tool in Picard-Tools (Galaxy Version 2.7.1.0) (http://broadinstitute.github.io/picard/).
SNP and small indel (< 30 bp) calling was performed using the HaplotypeCaller tool in GATK (Version 3.7-0) (McKenna et al. 2010) with the parameter -stand_call_conf = 30. Homozygous variations between ‘Oonari’ and ‘Takanari’ were extracted for further study with three criteria: (1) the variant site had at least five reads across all samples, (2) allele frequencies of both ‘Takanari’ and ‘Oonari’ at the site were higher than 0.9, and (3) genotypes of ‘Takanari’ and ‘Oonari’ at the site were different. For structural variation detection, its calling was performed using Pindel (Ye et al. 2009) and Manta (Chen et al. 2016), and then the structural variations were examined using Integrative Genomics Viewer (IGV) (Robinson et al. 2011). A variation index for F3 bulk, the ratio between the number of ‘Oonari’-type reads and total reads, was calculated in a similar manner to the SNP-index (Abe et al. 2012). Thus, the variation index was assigned to 0 and 1, when all short reads contained alleles from ‘Takanari’ and ‘Oonari’, respectively. Variant annotation was performed using SnpEff v4.2 (Cingolani et al. 2012).
In addition, to reveal whether there is any other candidate gene in the candidate genomic region, we conducted the following additional analyses. (1) We merged the two bam files generated by mapping two sets of NGS data of ‘Takanari’ and ‘Oonari’, respectively, to the genome of ‘Nipponbare’ or an indica rice cultivar ‘Minghui 63’ (http://rice.hzau.edu.cn/rice/) (Zhang et al. 2016), and performed mutation identification. (2) We identified the genomic regions with read depth less than 5 (thereafter called gap regions) using pysamstats software with default parameters (window-size = 300, window-offset = 150) (https://github.com/alimanfoo/pysamstats), followed by annotation analysis with the stand-alone BLAST programs. The genes whose protein coding regions or untranslated regions (UTRs) are contained in the gap regions were extracted.
Validation of the bulked segregant analysis
The mutations in which the variation index was more than 0.85 in the F3-a bulk were selected for further screening of the candidate mutation associated with seed shattering using a small population, which contained 8 easy- and 8 moderate-shattering individuals from the F2-c population. The genotype of each individual was determined by Sanger sequencing. Briefly, primers (Supplementary Table 1) were designed using the Primer3 program (http://bioinfo.ut.ee/primer3-0.4.0/). Polymerase chain reaction (PCR) was performed in a reaction mixture of 10 μl, consisting of 10 ng genomic DNA as a PCR template, 0.5 μM of primers, 0.25 U of PrimeSTAR GXL DNA Polymerase (Takara, Kusatsu, Japan), 1× PrimeSTAR GXL Buffer, and 200 μM of dNTPs. The thermal cycle of PCR was set to be as follows: 1 min pre-incubation at 98 °C, 35 cycles of 10 s denaturation at 98 °C, 15 s annealing at 58 °C, 20 s extension at 68 °C. The amplified fragments were sequenced on a 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) after a sequencing reaction using the BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems).
After the candidate mutation associated with seed shattering was determined, dCAPS markers (Supplementary Table 1) were designed, using dCAPS Finder 2.0 (http://helix.wustl.edu/dcaps/) (Neff et al. 2002) and Primer3 program. Amplified PCR products were digested by the restriction enzyme Bsp1407 I (Takara) at 37 °C, and then analyzed by electrophoresis on 3.0% PrimeGel™ Agarose PCR-Sieve HRS (Takara) gels stained with GelStar® nucleic acid gel stain (Lonza, Rockland, ME, USA).
Validation of structural variations
The identified structural variations were validated by PCR and the Sanger sequencing method using the primers in Supplementary Table 1. Genotyping of duplication based on quantitative real-time PCR (qPCR) was performed using a SYBR Green Supermix Kit (Bio-Rad, Hercules, CA, USA) on a MiniOption Real-time PCR System (Bio-Rad). The PCR reaction mixture (20 μl) consisted of 0.2 μM forward and reverse primers, 1 × SYBR Advantage qPCR Premix, and about 10 ng DNA. The reactions were carried out using the following qPCR protocol: 3 min pre-incubation at 95 °C followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s and finally a dissociation run from 65 to 95 °C. The qSH1 gene was used for normalization of the relative quantities.
Gene expression analyses
Small RNAs were extracted from about 80-mm-long young panicles (about 2 weeks before heading) using a mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, USA). The first-strand cDNAs were synthesized from small RNAs using a Mir-X miRNA first-strand synthesis kit (Clontech, Palo Alto, CA, USA).
The OsmiR172d forward primer was designed based on its full sequences, and the mRQ 3′ primer in the Mir-X miRNA first-strand synthesis kit was used as the reverse primer. U6 snRNA was used as the reference gene for miRNA expression. Primers used for qRT-PCR experiments are listed in Supplementary Table 1.
Statistical analysis
The shattering degrees of different genotypes were compared using pairwise t tests and a Bonferroni correction was used to control for the family-wise type I error rate across the comparisons. Pearson’s correlation coefficient, r, was used to measure the strength of a linear association between the values obtained by different methods for evaluating seed shattering. All statistical analyses were performed in R Statistical Software (version 3.2.2; R Foundation for Statistical Computing, Vienna, Austria).
Accession numbers
All NGS data files are available in the DDBJ Sequenced Read Archive under the accession numbers DRA006863, DRA006864, DRA006865, and DRA006866.
Results
Evaluation of grain shattering
The PS, BS, and RSP values were measured every 10 days from heading to maturity stage. The highest PS values of ‘Nipponbare’ and ‘Kasalath’ appeared on the 10th day after heading, and thereafter, PS values of ‘Nipponbare’ decreased slowly, whereas those of ‘Kasalath’ decreased dramatically (Fig. 1a). In contrast, PS values of ‘Takanari’ and ‘Oonari’ were relatively constant. There was no significant difference (P > 0.05) between ‘Kasalath’, ‘Takanari’, and ‘Oonari’ on the 40th day after heading. The average BS values of these four cultivars were approximately 44.7–52.5 gf on heading date (Fig. 1b). Subsequently, those of ‘Takanari’ and ‘Kasalath’ decreased rapidly and were as low as 18.2 and 7.2 gf on the 40th day after heading, respectively. In contrast, that of ‘Oonari’ was relatively constant and that of ‘Nipponbare’ increased step by step and was as high as 66.1 gf on the 40th day after heading. These results indicated that the difference of shattering degrees among ‘Oonari’, ‘Takanari’, and ‘Kasalath’ was mainly attributed to the difference of their BS values. We noticed that most of the pedicels of ‘Takanari’ were easily separated from the grains and a smooth surface was observed on the abscission zone (Supplementary Fig. 2a) when we pushed the lower end of the pedicel vertically. In contrast, most of the pedicels of ‘Oonari’ could not be separated from the abscission region (Supplementary Fig. 2b). We therefore supposed RSP might also be an appropriate index for discriminating the shattering degree of ‘Oonari’ from that of ‘Takanari’. As a result, the grain pedicel of ‘Nipponbare’ almost never shattered during seed development after the heading stage, and the RSP values remained at less than 10.0% (Fig. 1c). The RSP values of ‘Takanari’, ‘Oonari’, and ‘Kasalath’ were approximately 10.0–20.0% on heading date. Significant difference (P < 0.01) in RSP was observed between ‘Takanari’ and ‘Oonari’ on the 10th day after heading. Furthermore, those of ‘Takanari’ and ‘Kasalath’ increased rapidly to 78.0% and 100.0% on the 20th day after heading, respectively, while those of ‘Oonari’ fluctuated between 10 and 30%.
Comparison of shattering degrees between different cultivars at different developmental stages in 2017. a Comparison of pulling strength (PS). b Comparison of bending strength (BS). c Comparison of rate of grain shattering by pushing (RSP). d Correlation of RSP and BS from 10th to 40th day after heading (DAH). Bars indicate mean values ± standard deviation (n = 4)
We calculated the correlations between RSP and BS values in four cultivars from the 10th to 40th day after heading (Fig. 1d). No significant correlation was observed on the 10th day after heading (r = − 0.29, P = 0.28). However, RSP was significantly negatively correlated with BS on the 20th day after heading (r = − 0.78, P < 0.001), and the correlation coefficient reached to − 0.96 (P < 0.001) on the 40th day after heading. Since the RSP method does not require the recording of force strength using the digital force gauge, it is relatively more convenient and labor-saving compared with that of BS.
We re-evaluated the seed shattering of different cultivars grown in 2018 at maturity stage using fourfold amount of grains used in 2017. In the same manner, there were no significant differences in PS among ‘Kasalath’, ‘Takanari’, and ‘Oonari’ (P > 0.05) (Supplementary Fig. 3). For the BS, ‘Kasalath’, ‘Takanari’, ‘Oonari’, and ‘Nipponbare’ were showed in ascending order, and in decreasing order for RSP.
Histological analysis of abscission zone formation
A total of 14 pedicels at heading stage were observed in ‘Takanari’ and ‘Oonari’ grown in the early season of 2016, and no obvious differences were observed (Supplementary Fig. 4a, b). The abscission zone cells, which are flattened and densely aligned transversely to the organ axis, were observed in both cultivars. Both had an incomplete abscission layer, i.e., absence of the abscission zone cells near the vascular bundle, which is a common characteristic in most of the indica cultivars (Li et al. 2006; Ishikawa et al. 2017). Five pedicels on the 20th day after heading in ‘Takanari’ and ‘Oonari’, which were grown in the late season of 2017, were also observed, no obvious differences being observed between them (Supplementary Fig. 5a, b).
Segregation of seed shattering in F2 population
The frequency of the shattering degrees of F2-c individuals (n = 83) that were evaluated by RSP on the 15–17th day after heading showed a bimodal distribution (Supplementary Fig. 6). Twenty-five individuals shown RSP value less than 20.0% as ‘Oonari’, 20 individuals fell within or higher than the range of ‘Takanari’, and 38 individuals between the parents. This segregation ratio fits a 1:2:1 ratio as determined by a chi-square test (χ2 = 1.19, P = 0.55), revealing evidence of a major locus with a semi-dominant inheritance mode responsible for seed shattering.
Bulked segregant analysis based on whole-genome sequencing
Among F3-a lines, 11 lines showing shattering scores comparable to that of ‘Oonari’ were selected for bulked segregant analysis. To identify highly reliable mutations in ‘Oonari’, two sets of NGS data were obtained for ‘Oonari’ and ‘Takanari’ (Supplementary Table 2). After removing duplicated reads, 68.4–96.6% of the clean reads were mapped to the ‘Nipponbare’ reference genome. The average coverages were 15.8–26.2 times and more than 85.7% of the genome had at least 5× coverage.
SNPs and small indels were first called between parental lines and the reference genome using GATK, and then homozygous variations unique to a single parent were identified. As a result, 62 SNPs, 20 small deletions (1~20 bp), and 4 small insertions (1~8 bp) were identified between ‘Takanari’ and ‘Oonari’ (Supplementary Table 3). Furthermore, using Pindel, we identified a tandem duplicated segment of 6.2 kb on chromosome 2 (Supplementary Fig. 7a) and an inversion of 881.6 kb on chromosome 6 (Supplementary Fig. 8a). These structural variations were validated by PCR and Sanger sequencing (Supplementary Fig. 7b and Supplementary Fig. 8b, c). To identify mutations associated with reduced seed shattering, the variation index of each mutation except of the tandem duplication mutation TO21 in the F3-a bulk was calculated (Supplementary Table 3) and plotted on all the 12 chromosomes of the rice genome (Fig. 2). Since only the paired reads crossing the rejoined site of the duplication are regarded as mutant alleles, and the paired reads completely aligned in duplicated region are all considered as wild alleles (https://software.broadinstitute.org/software/igv/interpreting_pair_orientations), therefore, the variation index of the duplication mutation TO21 cannot be correctly calculated. As a result, one SNP named TO20 and two deletions named TO42 and TO75, of which variation indices were more than 0.85, were identified on chromosome 2, chromosome 4, and chromosome 9, respectively.
Identification of candidate mutations associated with reduced seed shattering using the method combining bulked segregant analysis with whole-genome sequencing. The X-axis represents the position of 12 chromosomes and the Y-axis represents the variation index of bulked F3 individuals with moderate-shattering trait. The horizontal dashed green line represents the threshold of the variation index, being 0.85. The candidate mutations were labeled by their IDs as in Supplementary Table 2
Validation of the bulked segregant analysis
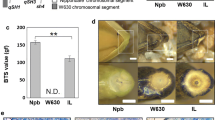
Eight easy- and 8 moderate-shattering individuals from the F2-c population were first used for screening of a candidate mutation associated with RSP on the 15–17th day after heading. The RSP values of individuals with homozygous ‘Takanari’ and ‘Oonari’ alleles at the TO20 site were 53.0 ± 7.9 and 8.0 ± 4.8, respectively, which showed significant difference (P < 0.001) from each other (Supplementary Fig. 9). No significant differences (P = 1.0) were observed between different homozygous alleles at the TO42 and TO75 sites (Supplementary Fig. 9). Hence, association of the TO20 genotype and the RSP value in the whole F2-c population (n = 83) was further investigated. The RSP values of individuals with homozygous ‘Takanari’ and ‘Oonari’ alleles at the TO20 site were close to those of ‘Takanari’ and ‘Oonari’, respectively (P = 1.0), and those with heterozygous were intermediate between the parents (Fig. 3). This result confirms the findings that the mutation at TO20 is associated with the reduced shattering trait and that the causal mutation has a semi-dominant effect. At the maturity stage of rice, RSG was investigated in some of the F2-c individuals (n = 53) and F3-b lines (n = 75). The Pearson’s correlation coefficient between RSG and RSP was 0.83 (Supplementary Fig. 10), indicating that RSG can also be used as an index for evaluating seed shattering. In both the F2-c and F3-b populations, the RSG values of individuals or lines with homozygous ‘Takanari’ allele at TO20 were notably higher than those with ‘Oonari’ allele (P < 0.01) (Supplementary Fig. 11a and b). Subudhi et al. (2014) and Lee et al. (2016) have localized minor QTLs for seed shattering at marker interval RM13910-RM13938 (corresponding to 30.0–30.7 Mb) and RM525-RM450 (corresponding to 28.2–28.6 Mb) on chromosome 2, both of which are distant from TO20 at 34.8 Mb of chromosome 2. Since the names of sh/SH/qSH/Sh/SS 1 to 12 have been used as seed shattering-related loci, we designated it as Sh13.
Comparison of rate of grain shattering by pushing (RSP) between different genotypes at the TO20 mutation site. T/T_TO20 (n = 18) represents a homozygous allele for ‘Takanari’, O/O_TO20 (n = 24) for ‘Oonari’, and T/O_TO20 (n = 41) for a heterozygous allele at the TO20 mutation site. Different lowercase and uppercase letters over the bars show significance levels of 0.05 and 0.01, respectively. Bars indicate mean values ± standard deviation
Identification of candidate genes
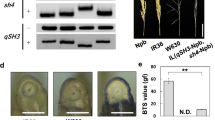
Annotation analysis revealed that TO20 was an intronic SNP (Supplementary Table 3). Furthermore, we found that one F2 individual, TS64, which had heterozygous ‘Oonari’ allele at TO20, had an RSP value as low as that of ‘Oonari’ (Supplementary Fig. 12), and two F3 lines, TS185 and TS198 with homozygous ‘Oonari’ allele, had RSP values as high as or higher than that of ‘Takanari’. These results indicate that although TO20 linked to Sh13 but it may not be the causal mutation. Hence, the duplication mutation TO21 at 35.1 Mb of chromosome 2 was subjected to further analysis. Although there was no protein coding gene in the duplicated 6.2-kb region, we identified one small RNA sequence that corresponds to osa-miR172d, whose wheat ortholog has recently been revealed to play a crucial role in grain threshability (Debernardi et al. 2017). For genotype analysis of TO21, we designed a pair of primers, one of which covers the junction of the duplicated regions (Fig. 4a). Using this primer set, the same PCR products were yielded in both homozygous ‘Oonari’ allele and heterozygous allele, while not in the homozygous ‘Takanari’ allele (Fig. 4b). As a result, TS185 and TS198 were identified to have homozygous ‘Takanari’ alleles at TO21. qPCR analysis revealed that TS64 had a homozygous ‘Oonari’ allele at TO21(Fig. 4c). From the descendants of TS64, we isolated the plants with homozygous ‘Takanari’ allele (TS64-2-T) and the plants with homozygous ‘Oonari’ allele (TS64-2-O) (Supplementary Fig. 12a) at the TO20 site and found that there was no significant difference in RSP values between them (Fig. 4d), indicating that the duplication of osa-miR172d is more closely associated with the moderate-shattering trait than TO20 (Supplementary Fig. 13). RT-qPCR analysis showed that the gene expression of osa-miR172d in young panicles of ‘Oonari’ was about three times as high as that of ‘Takanari’ (Fig. 5) (P < 0.01).
Identification of candidate gene associated with reduced seed shattering. a Schematic representation of the primer positions for genotype analysis of the duplication mutation TO21. The gray arrow-shaped boxes indicate the positions of osa-miR172d. Arrowheads indicate the primer positions, and the predicted PCR product sizes are shown at the lower part. DUP_F3 covers the junction of the duplicated regions. Note that the diagram is not drawn to scale. b Agarose gel electrophoresis of the amplification products obtained by PCR using the primer set of DUP in a, and the primer set of qSH1 was used a positive control in PCR. No PCR product of DUP was observed in the lines with homozygous ‘Takanari’ allele at the TO21 site. c Genotyping analysis of the duplication using qPCR. The Y-axis represents the fold change between the duplication and qSH1 amplicons. ‘Oonari’ was normalized to 1 and all other lines were compared to this. Value more than 0.8 was regarded as a homozygous allele for ‘Oonari’, otherwise a heterozygous allele. Bars indicate mean values ± standard deviation (n = 3). d Comparison of rate of grain shattering by pushing (RSP) between the plants of TS64-2-T and TS64-2-O grown in a heated greenhouse from Nov. 2017 to Apr. 2018. Different lowercase and uppercase letters over the bars show significance levels of 0.05 and 0.01, respectively. Bars indicate mean values ± standard deviation (n = 4)
We merged the bam files of ‘Oonari’ and ‘Takanari’, respectively, to reduce the ratio of gap regions to less than 9.3% of the reference genomes (Supplementary Table 4), followed by mutation detection. However, we yet did not identify more mutations at the candidate genomic region from TO19 (at 32.0 Mb) to the end of the long arm of chromosome 2. We identified 21 genes in or containing gap regions when the ‘Nipponbare’ genome was used as a reference genome, of which 11 genes were assumed to be lost in the ‘Minghui 63’ and ‘Takanari’ genomes (Supplementary Table 5). Furthermore, the coverages of the remaining nine genes were more than 5× when the ‘Minghui 63’ genome was used as a reference genome. On the other hand, we identified three genes in the gap regions in this analysis (Supplementary Table 6), of which one was similar to the Pib gene and the other two were retrotransposon genes. These results suggested that there was no other candidate gene when the ‘Nipponbare’ or ‘Minghui 63’ genome was used as a reference genome.
Discussion
Seed shattering can be evaluated by different methods. Hand grasping followed by recording on a scale of 1–9 and RSG at the seed maturity stage are simple traditional methods (Lee et al. 2016). However, these methods are affected by the water content of the panicle and human errors. Use of a digital force gauge to evaluate the shattering degree by pulling or bending grain pedicels which have the same water content can reduce the abovementioned effects and provide more precise data. It has been revealed that PS is positively correlated with BS (Qin et al. 2010; Lee et al. 2016), and that the degree of abscission zone formation is significantly negatively correlated with PS and BS (Lee et al. 2016). However, there was obvious difference between ‘Oonari’ and ‘Takanari’ only in BS but not in either PS (Fig. 1 and Supplementary Fig. 3) or abscission zone formation (Supplementary Figs. 4 and 5), indicating that there is a specific unknown factor involved in control of BS. In fact, a previous study has shown that some different cultivars have the same PS values but different BS values (Jin and Inouye 1981), and several specific loci for BS have been identified (Qin et al. 2010; Lee et al. 2016). However, related genes have not been isolated. A more intensive study on the morphological features of the abscission zone using scanning electron microscope (Ebata and Tashiro 1990; Okubo 2014) might be useful to better understand the varietal differences between ‘Oonari’ and ‘Takanari’.
RSP used in this study was significantly correlated with BS and RSG (negatively and positively, respectively) (Fig. 1 and Supplementary Fig. 10). The RSP evaluation method can save labor compared with the BS method and is considered to be more precise than the RSG method. Furthermore, since the RSP values from the 10th day after heading forward showed significant difference between the two parents (Fig. 1), evaluation of seed shattering of their segregation population and genetic analysis could be conducted before the maturity stage.
Although some major genes, such as qSH1 (Konishi et al. 2006), SH4 (Li et al. 2006), SHAT1 (Zhou et al. 2012), OsSh1 (Lin et al. 2012), etc., have been identified as controlling seed shattering, none of the minor genes have been cloned (Htun et al. 2014; Inoue et al. 2015; Kwon et al. 2015; Yao et al. 2015; Lee et al. 2016; Ishikawa et al. 2017). The mechanism for the moderate-shattering trait is still unclear. The moderate-shattering mutant cultivar ‘Oonari’ is useful for uncovering this mechanism. Whole-genome sequencing analysis of ‘Oonari’ and its original cultivar ‘Takanari’ identified 88 mutations in ‘Oonari’, 33.3% of which were indels and structural variations (Supplementary Table 3). This mutation characterization is similar to those of mutants obtained by C-ion beams (30.0 keV μm−1) (Kazama et al. 2017) and fast-neutron irradiations (Li et al. 2017). The high proportion of indels and structural variations, including duplication, transversion, and translocation, suggests that the MutMap developed for identifying the causal SNP mutation in EMS mutants (Abe et al. 2012) is not enough for identifying the causal mutation for a specific mutant phenotype induced by radiation. As with the calculation method of SNP-index in MutMap, we calculated the variation index for each mutation (except for the duplication mutation). Although the bulked DNA of only 11 mutant-type F3 lines was used for bulked segregant analysis, only three mutations with high variation index were identified due to the low mutation frequency in the gamma-ray mutant (Fig. 2). If the bulk of wild-type F3 (or F2) progenies were used for this analysis as with the MutMap+ approach (Fekih et al. 2013), the candidate genomic region might have been directly identified. Alternatively, after a simple screening using a small-size population followed by a further confirmation with about 160 F2 individuals and F3 lines, we identified that TO20 is associated with moderate shattering (Supplementary Figs. 9 and 11). Hence, fine mapping using recombinant lines between TO20 and its near duplication mutation TO21 revealed that TO21 is more closely associated with the reduced seed shattering (Supplementary Fig. 13). Since there are no other mutations near TO21, the osa-miR172d gene in duplicated region of TO21 might be considered as a candidate gene for seed shattering. In a previous study, a segmental duplication encompassing S-haplotype induced by gamma irradiation has been suggested to trigger pollen-part self-compatibility in Japanese pear (Pyrus pyrifolia) (Mase et al. 2014). According to our knowledge, this is the first report on identification of a candidate gene for irradiation-induced mutation using the NGS-based bulked segregant analysis approach. We anticipate that this approach will facilitate gene isolation and utilization of irradiation-induced mutation for gene identification.
Although there is no available reference genome sequence of ‘Takanari’, and the genome of ‘Nipponbare’ or ‘Minghui63’ was used as a reference genome, analysis of the genes in or containing gaps suggested that there was no other candidate gene (Supplementary Tables 5 and 6). We could not exclude the possibility that there might be a small fraction of genomic regions specific to ‘Takanari’. However, since the mutation frequency induced by gamma radiation is very low, for example, one mutation per 4.2 Mb in this study, and one mutation per 6.2 Mb in a previous study (Sato et al. 2006), the possibility is very low that the causative mutation might reside in these specific regions.
In recent years, microRNAs (miRNAs) have been shown to play important regulative roles in developmental processes and stress responses by regulating target mRNA expression post-transcriptionally in a wide variety of organisms (Bartel 2004; Kidner and Martienssen 2005). In plants, miRNA172 is involved in the regulation of phase transition and in determination of floral organ identity through targeting AP2 and AP2-like genes (Park et al. 2002; Aukerman and Sakai 2003; Dai et al. 2016). Q in wheat is an AP2-like gene involved in controlling seed shattering (Simons et al. 2006). During wheat domestication, a nucleotide change at the miR172 binding site of the Q gene reduced their interaction, which contributed to subcompact spikes and freethreshing grains (i.e., grains readily separated from glumes) (Debernardi et al. 2017). Overexpressing the miR172d gene under the control of the maize UBIQUITIN promoter in wheat made more than sevenfold increase of the expression of miR172d, leading to reduced threshability (Debernardi et al. 2017). However, overexpressing miR172d under the control of the UBIQUITIN or 35S promoter in wheat and rice cause pleiotropic effects including spikelet and inflorescence developmental defects (Dai et al. 2016; Debernardi et al. 2017), so that these materials cannot be used in breeding. The AP2-like gene SHAT1 (Zhou et al. 2012) is regarded as one of the target genes of osa-miR172d (Liu et al. 2016). Null mutation in SHAT1 causes hard-shattering and developmental defects in the spikelet and inflorescence (Zhou et al. 2012). In the SHAT1-RNAi transgenic plants, the pedicel breaking tensile strength correlated well with the magnitude of the reduction in SHAT1 mRNA (Zhou et al. 2012). Since at least 13 putative targets of osa-miR172d were identified in rice (Liu et al. 2016), we suppose that the twofold increase of miR172d expression in ‘Oonari’ (Fig. 4) causes only slight repression of the expression of each target gene, thus causing only moderate reduction of shattering degree without obvious morphological difference between ‘Oonari’ and ‘Takanari’.
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30(2):174–178. https://doi.org/10.1038/nbt.2095
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15(11):2730–2741. https://doi.org/10.1105/tpc.016238
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, Cox AJ, Kruglyak S, Saunders CT (2016) Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32(8):1220–1222. https://doi.org/10.1093/bioinformatics/btv710
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu XY, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6(2):80–92. https://doi.org/10.4161/fly.19695
Dai ZY, Wang J, Zhu ML, Miao XX, Shi ZY (2016) OsMADS1 represses microRNA172 in elongation of palea/lemma development in rice. Front Plant Sci 7:1891. https://doi.org/10.3389/fpls.2016.01891
Debernardi JM, Lin HQ, Chuck G, Faris JD, Dubcovsky J (2017) microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144(11):1966–1975
Ebata M, Tashiro T (1990) Studies on shedding habit of rice plant .1. Morphology of separation zone. Jpn J Crop Sci 59(1):63–71
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19(6):1349–1349
Fekih R, Takagi H, Tamiru M, Abe A, Natsume S, Yaegashi H, Sharma S, Sharma S, Kanzaki H, Matsumura H, Saitoh H, Mitsuoka C, Utsushi H, Uemura A, Kanzaki E, Kosugi S, Yoshida K, Cano L, Kamoun S, Terauchi R (2013) MutMap+: genetic mapping and mutant identification without crossing in rice. PLoS One 8(7):e68529. https://doi.org/10.1371/journal.pone.0068529
Fukuta Y, Yagi T (1998) Mapping of a shattering resistance gene in a mutant line SR-5 induced from an indica rice variety, Nan-jing11. Breed Sci 48(4):345–348. https://doi.org/10.1270/jsbbs1951.48.345
Fuller DQ, Allaby R (2009) Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation. In: Ostergaard L (ed) Annual plant reviews, vol Fruit Development and Seed Dispersal. Wiley-Blackwell, Oxford, pp 238–295
Htun TM, Inoue C, Chhourn O, Ishii T, Ishikawa R (2014) Effect of quantitative trait loci for seed shattering on abscission layer formation in Asian wild rice Oryza rufipogon. Breed Sci 64(3):199–205. https://doi.org/10.1270/jsbbs.64.199
Imbe T, Akama Y, Nakane A, Hata T, Ise K, Ando I, Uchiyamada H, Nakagawa N, Furutachi H, Horisue N, Noto M, Fujita Y, Kimura K, Mori K, akayanagi K, Uehara Y, Ishizaka S, Nakagahra M, Yamada T, Koga Y (2004) Development of a multipurpose high-yielding rice variety ‘Takanari’. Bull Natl Inst Crop Sci 5:35–51
Inoue C, Htun TM, Inoue K, Ikeda K, Ishii T, Ishikawa R (2015) Inhibition of abscission layer formation by an interaction of two seed-shattering loci, sh4 and qSH3, in rice. Genes Genet Syst 90(1):1–9
Ishikawa R, Nishimura A, Htun TM, Nishioka R, Oka Y, Tsujimura Y, Inoue C, Ishii T (2017) Estimation of loci involved in non-shattering of seeds in early rice domestication. Genetica 145(2):201–207. https://doi.org/10.1007/s10709-017-9958-x
Ji H, Kim SR, Kim YH, Kim H, Eun MY, Jin ID, Cha YS, Yun DW, Ahn BO, Lee MC, Lee GS, Yoon UH, Lee JS, Lee YH, Suh SC, Jiang WZ, Yang JI, Jin P, McCouch SR, An G, Koh HJ (2010) Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J 61(1):96–106. https://doi.org/10.1111/j.1365-313X.2009.04039.x
Ji HS, Chu SH, Jiang WZ, Cho YI, Hahn JH, Eun MY, McCouch SR, Koh HJ (2006) Characterization and mapping of a shattering mutant in rice that corresponds to a block of domestication genes. Genetics 173(2):995–1005. https://doi.org/10.1534/genetics.105.054031
Jin ID, Inouye J (1981) On the degree of grain shedding of japonica–indica hybrid rice bred in Korea. Jpn J Crop Sci 50:181–185. https://doi.org/10.1626/jcs.50.181
Kanemura T, Homma K, Ohsumi A, Shiraiwa T, Horie T (2007) Evaluation of genotypic variation in leaf photosynthetic rate and its associated factors by using rice diversity research set of germplasm. Photosynth Res 94(1):23–30. https://doi.org/10.1007/s11120-007-9208-7
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG, Childs KL, Davidson RM, Lin HN, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. https://doi.org/10.1186/1939-8433-6-4
Kazama Y, Ishii K, Hirano T, Wakana T, Yamada M, Ohbu S, Abe T (2017) Different mutational function of low- and high-linear energy transfer heavy-ion irradiation demonstrated by whole-genome resequencing of Arabidopsis mutants. Plant J 92(6):1020–1030. https://doi.org/10.1111/tpj.13738
Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8(1):38–44
Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312(5778):1392–1396. https://doi.org/10.1126/science.1126410
Kwon SJ, Yu J, Park YJ, Son JH, Kim NS, Lee JK (2015) Genetic analysis of seed-shattering genes in rice using an F3:4 population derived from an Oryza sativa x Oryza rufipogon cross. Genet Mol Res 14(1):1347–1361. https://doi.org/10.4238/2015.February.13.14
Lee GH, Kang IK, Kim KM (2016) Mapping of novel QTL regulating grain shattering using doubled haploid population in rice (Oryza sativa L.). Int J Genomics:1–8. https://doi.org/10.1155/2016/2128010
Li CB, Zhou AL, Sang T (2006) Rice domestication by reducing shattering. Science 311(5769):1936–1939. https://doi.org/10.1126/science.1123604
Li G, Jain R, Chern M, Pham NT, Martin JA, Wei T, Schackwitz WS, Lipzen AM, Duong PQ, Jones KC, Jiang L, Ruan D, Bauer D, Peng Y, Barry KW, Schmutz J, Ronald PC (2017) The sequences of 1504 mutants in the model rice variety Kitaake facilitate rapid functional genomic studies. Plant Cell 29(6):1218–1231. https://doi.org/10.1105/tpc.17.00154
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Lin ZW, Li XR, Shannon LM, Yeh CT, Wang ML, Bai GH, Peng Z, Li JR, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu JM (2012) Parallel domestication of the Shattering1 genes in cereals. Nat Genet 44(6):720–724
Liu WT, Yang CC, Chen RK, Jwo WS, Wu CW, Ting WY, Shung DP, Liu CC, Chen JJ (2016) RiceATM: a platform for identifying the association between rice agronomic traits and miRNA expression. Database (Oxford) 2016. https://doi.org/10.1093/database/baw151
Mase N, Sawamura Y, Yamamoto T, Takada N, Nishio S, Saito T, Iketani H (2014) A segmental duplication encompassing S-haplotype triggers pollen-part self-compatibility in Japanese pear (Pyrus pyrifolia). Mol Breed 33:117–128. https://doi.org/10.1007/s11032-013-9938-5
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. https://doi.org/10.1101/gr.107524.110
Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18(12):613–615
Oka HI (1988) Origin of cultivated rice. Japan Scientific Society Press, Tokyo
Okubo K (2014) Morphological evaluation of the trace of grain detachment in japonica rice cultivars with different shattering habits. Plant Prod Sci 17(4):291–297
Okubo K, Watanabe T, Miyatake N, Maeda S, Inoue T (2012) Evaluation method for threshability of rice varieties by grasping the panicle with hand. Jpn J Crop Sci 81(2):201–206. https://doi.org/10.1626/jcs.81.201
Park W, Li JJ, Song RT, Messing J, Chen XM (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12(17):1484–1495
Purugganan MD, Fuller DQ (2009) The nature of selection during plant domestication. Nature 457(7231):843–848. https://doi.org/10.1038/nature07895
Qin Y, Kim SM, Zhao XH, Jia BY, Lee HS, Kim KM, Eun MY, Jin ID, Sohn JK (2010) Identification for quantitative trait loci controlling grain shattering in rice. Genes Genom 32(2):173–180. https://doi.org/10.1007/s13258-009-0879-7
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26. https://doi.org/10.1038/nbt.1754
Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M (1999) Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J 18(4):992–1002. https://doi.org/10.1093/emboj/18.4.992
Sato Y, Shirasawa K, Takahashi Y, Nishimura M, Nishio T (2006) Mutant selection from progeny of gamma-ray-irradiated rice by DNA heteroduplex cleavage using Brassica petiole extract. Breed Sci 56(2):179–183. https://doi.org/10.1270/jsbbs.56.179
Schneeberger K, Weigel D (2011) Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci 16(5):282–288. https://doi.org/10.1016/j.tplants.2011.02.006
Simons KJ, Fellers JP, Trick HN, Zhang ZC, Tai YS, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172(1):547–555. https://doi.org/10.1534/genetics.105.044727
Subudhi PK, Singh PK, DeLeon T, Parco A, Karan R, Biradar H, Cohn MA, Sasaki T (2014) Mapping of seed shattering loci provides insights into origin of weedy rice and rice domestication. J Hered 105(2):276–287. https://doi.org/10.1093/jhered/est089
Xiong LX, Liu KD, Dai XK, Xu CG, Zhang QF (1999) Identification of genetic factors controlling domestication-related traits of rice using an F-2 population of a cross between Oryza sativa and O. rufipogon. Theor Appl Genet 98(2):243–251
Yao N, Wang L, Yan HX, Liu YS, Lu BR (2015) Mapping quantitative trait loci (QTL) determining seed-shattering in weedy rice: evolution of seed shattering in weedy rice through de-domestication. Euphytica 204(3):513–522. https://doi.org/10.1007/s10681-014-1331-x
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z (2009) Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25(21):2865–2871. https://doi.org/10.1093/bioinformatics/btp394
Yonemaru J, Mizobuchi R, Kato H, Yamamoto T, Yamamoto E, Matsubara K, Hirabayashi H, Takeuchi Y, Tsunematsu H, Ishii T, Ohta H, Maeda H, Ebana K, Yano M (2014) Genomic regions involved in yield potential detected by genome-wide association analysis in Japanese high-yielding rice cultivars. BMC Genomics 15:346. https://doi.org/10.1186/1471-2164-15-346
Yoon J, Cho LH, Antt HW, Koh HJ, An G (2017) KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol 174(1):312–325. https://doi.org/10.1104/pp.17.00298
Yoon J, Cho LH, Kim SL, Choi H, Koh HJ, An G (2014) The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J 79(5):717–728. https://doi.org/10.1111/tpj.12581
Zhang JW, Chen LL, Xing F, Kudrna DA, Yao W, Copetti D, Mu T, Li WM, Song JM, Xie WB, Lee S, Talag J, Shao L, An Y, Zhang CL, Ouyang YD, Sun S, Jiao WB, Lv F, Du BG, Luo MZ, Maldonado CE, Goicoechea JL, Xiong LZ, Wu CY, Xing YZ, Zhou DX, Yu SB, Zhao Y, Wang GW, Yu YS, Luo YJ, Zhou ZW, Hurtado BEP, Danowitz A, Wing RA, Zhang QF (2016) Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. P Natl Acad Sci USA 113(35):E5163–E5171. https://doi.org/10.1073/pnas.1611012113
Zhou Y, Lu DF, Li CY, Luo JH, Zhu BF, Zhu JJ, Shangguan YY, Wang ZX, Sang T, Zhou B, Han B (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24(3):1034–1048. https://doi.org/10.1105/tpc.111.094383
Zong Y, Chen Z, Innes JB, Chen C, Wang Z, Wang H (2007) Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449(7161):459–463. https://doi.org/10.1038/nature06135
Zou C, Wang P, Xu Y (2016) Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol J 14(10):1941–1955. https://doi.org/10.1111/pbi.12559
Acknowledgments
We gratefully acknowledge the Advanced Analysis Center of NARO for use of high-speed processor system, Prof. Takeshi Nishio for valuable revision and comments, Mrs. Sayaka Niwa and Mrs. Kanako Matsumoto for experiment support, and Mr. Masayoshi Tobita for field support.
Funding
This work was supported by Cabinet Office, Government of Japan, Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Fig. 1
Evaluation of shattering degree using a tension testing machine equipped with a digital force gauge. (a) Measurement method and the direction of pulling strength (PS). (b) Close-up view corresponding to the red box in (a). (c) Measurement method and the direction of bending strength (BS). (d) Close-up view corresponding to the red box in (c). (e) Measurement method and the direction of press stress for the rate of grain shattering by pushing (RSP). (f) Close-up view corresponding to the red box in (e). (PPTX 2088 kb)
Supplementary Fig. 2
Representative images of pedicels of ‘Takanari’ (a) and ‘Oonari’ (b) after pushing the lower end of a pedicel vertically. (PPTX 5755 kb)
Supplementary Fig. 3
Comparison of shattering degrees between different cultivars at maturity stage in 2018. (a) Comparison of pulling strength (PS). (b) Comparison of bending strength (BS). (c) Comparison of rate of grain shattering by pushing (RSP). Bars indicate mean values ± standard deviation (n = 4). (PPTX 53 kb)
Supplementary Fig. 4
Longitudinal sections (10-μm thick) across the abscission zone of ‘Takanari’ (a) and ‘Oonari’ (b) grown in the early season. The sections were stained by toluidine blue. The read arrows indicate the abscission zones. (PPTX 1295 kb)
Supplementary Fig. 5
Longitudinal sections (2-μm thick) across the abscission zone of ‘Takanari’ (a) and ‘Oonari’ (b) grown in the late season. The sections were stained by toluidine blue. The read arrows indicate the abscission zones. (PPTX 1002 kb)
Supplementary Fig. 6
Frequency distribution of the rate of grain shattering by pushing (RSP) in F2_c population. Horizontal bar and solid dot indicate the RSP range and mean value of ‘Oonari’ or ‘Takanari’ (n = 3 independent replicates with 4 plants grown in a row analyzed in each assay). (PPTX 43 kb)
Supplementary Fig. 7
Validation of the tandem duplication mutation on chromosome 2 of ‘Oonari’. (a) Schematic representation of the duplication mutation of the genomic region from ‘Takanari’ to ‘Oonari’. Directions of arrow-shaped boxes indicate 5’ to 3’ directions based on the ‘Nipponbare’ genome sequence. Arrowheads indicate the primer positions, and the predicted PCR product sizes are shown at the lower part. The positions of break points and rejoined sites of the duplication are shown at the upper part. Note that the diagram is not drawn to scale. (b) Agarose gel electrophoresis of the amplification products obtained by PCR. The primer sets are shown at the lower part, and sample names are shown at the upper part. T and O represent ‘Takanari’ and ‘Oonari’, respectively. (PPTX 576 kb)
Supplementary Fig. 8
Validation of the inversion on chromosome 6 of ‘Oonari’. (a) Schematic representation of inversion of the genomic region in ‘Takanari’ to that of ‘Oonari’. Directions of arrow-shaped boxes indicate 5’ to 3’ directions based on the ‘Nipponbare’ genome sequence. Arrowheads indicate the primer positions, and the predicted PCR product sizes are shown at the lower part. The positions of break points and rejoined sites of the inversion are shown at the upper part. Note that the diagram is not drawn to scale. (b) Agarose gel electrophoresis of the amplification products obtained after PCR. The primer sets are shown at the lower part, and sample names are shown at the upper part. T and O represent ‘Takanari’ and ‘Oonari’, respectively. (c) The nucleotide sequences of break points and rejoined sites of the inversion were determined by sanger sequencing. (PPTX 2155 kb)
Supplementary Fig. 9
Variance of rate of grain shattering by pushing (RSP) at the three mutation sites with different parental genotypes in partial F2-c population (n = 16). Bars indicate mean values ± standard deviation. T/T and O/O represent homozygous ‘Takanari’ and ‘Oonari’ alleles, respectively. Double asterisks (**) represent significant difference in RSP between the different genotypes at P < 0.01 level. (PPTX 43 kb)
Supplementary Fig. 10
Correlation between rate of grain shattering by grasping (RSG) and rate of grain shattering by pushing (RSP) (n = 53). (PPTX 43 kb)
Supplementary Fig. 11
Variance of rate of grain shattering by grasping (RSG) between different genotypes at the TO20 mutation site in F2-c population (n = 53) (a) and F3-b population (n = 75) (b). T/T_TO20 represents a homozygous allele for ‘Takanari’, O/O_TO20 for ‘Oonari’, and T/O_TO20 for a heterozygous allele at the TO20 mutation site. Different small and capital letters over the bars show significance levels at 0.05 and 0.01, respectively. Bars indicate mean values ± standard deviation. (PPTX 47 kb)
Supplementary Fig. 12
Identification of individuals and lines in which TO20 genotypes were not associated with seed shattering. (a) Detection of TO20 genotype with the dCAPS marker. F1: an F1 from the crossing of ‘Takanari’ and ‘Oonari’, T: ‘Takanari’, O: ‘Oonari’, TS185 and TS198: two F3 lines in the F3_c population, TS64: an individual in the F2-b population, TS64-2-T and TS64-2-O: F4 lines with homozygous ‘Takanari’ and ‘Ooanri’ alleles at TO20 site, respectively, derived from TS64. (b) Rate of grain shattering by pushing (RSP) of exceptional lines on 18th day after heading in the late season of 2016. T and O were used for controls. Different lowercase and uppercase letters over the bars show significance levels at 0.05 and 0.01, respectively. Bars indicate mean values ± standard deviation (n = 4). (PPTX 170 kb)
Supplementary Fig. 13
Genotypes at TO20 and TO21 sites in different cultivars or lines and their shattering degrees. There are an SNP (A/G) and a tandem duplication at TO20 and TO21 sites, respectively. The red arrows indicate the positions of the osmir172d genes. Note that the diagram is not drawn to scale. The cultivars or lines with duplication showed moderate shattering, while those without duplication showed easy shattering. (PPTX 52 kb)
Supplementary Table 1
Primers used in this study. (XLSX 9 kb)
Supplementary Table 2
Summary of Illumina sequencing data. (XLSX 10 kb)
Supplementary Table 3
Mutations identified in ‘Oonari’, ‘Tankari’, and F3 bulk (XLSX 35 kb)
Supplementary Table 4
Summary of the alignment of all of the NGS data of ‘Takanari’ and ‘Oonari’, respectively, to the ‘Nipponbare’ or ‘Minghui 63’ genome. (XLSX 31 kb)
Supplementary Table 5
Genes in gap regions when the ‘Nipponbare’ genome was used as a reference genome (XLSX 51 kb)
Supplementary Table 6
Genes in gap regions when ‘Minghui 63’ genome was used as a reference genome (XLSX 27 kb)
Rights and permissions
About this article
Cite this article
Li, F., Numa, H., Hara, N. et al. Identification of a locus for seed shattering in rice (Oryza sativa L.) by combining bulked segregant analysis with whole-genome sequencing. Mol Breeding 39, 36 (2019). https://doi.org/10.1007/s11032-019-0941-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0941-3