Abstract
Starch paste viscosity properties are widely used as important indicators for quality estimation in waxy maize. To elucidate the genetic basis of paste viscosity characteristics of waxy maize, seven parameters from the rapid visco analyzer (RVA) profile were analyzed for quantitative trait loci (QTLs) in this study, using a recombinant inbred line population derived from a cross between the inbred lines Tongxi5 and Hengbai522. A high-density linkage map was constructed using 2703 bin markers, covering 1876.20 cM of the whole genome with an average genetic distance of 0.73 cM between adjacent bin markers. Seventy-two QTLs were detected for RVA parameters across 3 years, of which 17 could be identified in 2 years, and 6 identified in all 3 years. Eight QTL clusters were observed to be co-associated with two or more RVA parameters. Three major QTLs, qPV4-1, qTV4-1, and qFV5-2, which explained over 10% of the phenotypic variation, were stably mapped to the chromosomes 4 or 5 in all years. Based on functional annotations, two genes were considered as potential candidate genes for the identified major QTLs. The QTLs and candidate genes identified in this study will be useful for further understanding of the genetic architecture of starch paste viscosity characteristics in waxy maize, and may facilitate molecular breeding for grain quality improvement in breeding programs, and simultaneously provide a basis for cloning of the genes underlying these QTLs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Waxy maize (Zea mays L. var. ceratina) was first discovered in China in 1908 (Collins 1909). It is a special type of maize as it has nearly 100% amylopectin in its endosperm (Perera et al. 2001). For the excellent characteristics in terms of starch composition with low hardness and high digestibility, clarity, and viscosity, waxy maize is mainly consumed as a fresh food in Asia and also used in the food, textile, adhesive, and paper industries (Bao et al. 2012; Fan et al. 2008; Hao et al. 2015b; Lu et al. 2013). In the past several decades, genetic improvements have led to steady increases in waxy maize yield (Ding et al. 2006). Waxy maize breeders are now mandated to develop varieties with good grain quality to meet particular demands of food and other industry target markets.
Starch is the major component in cereal grains, and changes in biochemical characteristics can affect viscosity and gelatinization properties of starch and thus determine the final usage of waxy maize in food, feed, and industry (Cozzolino 2016; Liu et al. 1997; Yang et al. 2014). The starch viscosity profile (RVA profile, as determined by a Rapid Visco Analyzer), referring to the pasting curve generated from cereal flour (Bao et al. 2000; Yang et al. 2014), has been proven to be useful in estimating the quality of cereal crops, including waxy maize (Bao et al. 2000; Ketthaisong et al. 2013; Lu et al. 2013; Yan et al. 2014; Yan et al. 2011). The RVA profile parameters are classic quantitative traits; they are regulated by a complex genetic system involving many starch-related genes and are also influenced by non-genetic and environmental factors (Yan et al. 2014). Due to these quantitatively genetic characteristics, it is difficult for breeders to efficiently accomplish quality improvements by means of conventional approaches (Raihan et al. 2016). Understanding the genetic mechanism of grain quality will allow the genetic manipulation of waxy maize and will be beneficial to breeding programs (Yan et al. 2014).
The advent of maize genomics and development of molecular markers have proved powerful tools for elucidating the genetic basis of waxy maize quality, thus allowing improvements in breeding efficiency. Molecular markers linked to causal genes and/or quantitative trait loci (QTLs) may be useful for marker-assisted selection (MAS) in multi-gene pyramiding (Xu and Crouch 2008). A great number of QTLs for RVA profile parameters have been dissected in diverse crop populations, including rice (Bao et al. 2000; Bao et al. 2002; Hsu et al. 2014; Xu et al. 2015; Yacouba et al. 2013; Yan et al. 2014; Zhang et al. 2013), barley (Wang et al. 2010), and wheat (Liang et al. 2009). In rice, two major QTL clusters, contributing largely to phenotypic variation of several viscosity characteristics, have been consistently mapped to the chromosome 6 region corresponding to the Wx (encoding granule-bound starch synthase-I, GBSSI) and Alk (functions as starch synthase, SS) loci, in different segregating populations of different cross-combinations across different environments (Bao et al. 2000; Hsu et al. 2014; Wang et al. 2007; Xu et al. 2015; Yacouba et al. 2013; Yan et al. 2014). Starch paste viscosity characteristics are also controlled by other genes (Xu et al. 2016), such as pullulanase (PUL) (Kharabian-Masouleh et al. 2012; Yan et al. 2011), soluble starch synthase (SSS) genes (Xu et al. 2016; Xu et al. 2013), starch-branching enzyme (SBE) genes (Han et al. 2004), debranching enzyme (DBE) genes (Yan et al. 2014), and isoamylase (ISA) genes (Hsu et al. 2014; Yang et al. 2014). However, there have been few attempts to address the genetic mechanisms of starch paste viscosity characteristics in maize, including waxy maize (Zhang et al. 2010). QTL analysis of starch paste viscosity characteristics of waxy maize will provide a better understanding of the genetic factors influencing grain quality and also aid discovery of valuable markers for MAS (Wang et al. 2010).
Development of high-throughput genotyping technologies in maize has enabled development of new single nucleotide polymorphism (SNP) markers, thus leading to an increase in genetic marker density. With the advantage of low time consumption, low cost, and high throughput, SNPs have been widely applied in the construction of high-density genetic linkage maps to significantly increase the detection power, accuracy, and resolution needed to identify QTLs (Wang et al. 2015). Recently, the “skeleton bin map,” which combines co-segregating SNP markers into one bin and separates adjacent bins based on single recombination events, has been widely used to map QTLs for kernel starch content and other important grain-related traits in maize (Raihan et al. 2016; Wang et al. 2015). In this study, a waxy maize recombinant inbred line (RIL) population, derived from two parents of Tongxi5 and Hengbai522 of var. Suyunuo 1, which is currently the most widely grown waxy maize hybrid in China (Xie et al. 1997; Xie et al. 2009), was developed and genotyped using an Affymetrix microarray CGMB50K SNP Array containing 56,000 maize SNPs developed from the B73 reference sequence (Ganal et al. 2011). The starch paste characteristics of the RIL and parental lines were evaluated over 3 years. The objectives were to construct a high-density genetic linkage map of waxy maize using inferred bins from the high-density SNP markers and to dissect the genetic architecture of starch paste viscosity characteristics in waxy maize RIL population. Information on molecular markers tightly linked to QTLs controlling starch paste viscosity characteristics (RVA profiles) identified in this study will facilitate breeding strategies for quality improvements in waxy maize.
Materials and methods
Plant materials and field experiments
A population consisting of 198 waxy maize RILs, produced by single seed descent, was derived from a cross between the inbred lines Tongxi5 and Hengbai522 with similar amylopectin content; these two inbred lines formed the popular commercial hybrid Suyunuo1 (Xie et al. 1997; Xie et al. 2009). The waxy maize inbred lines of Tongxi5 and Hengbai522 differ significantly in starch viscosity, thus providing the means to clarify the genetic basis of the waxy maize grain quality. All 198 RILs and both parents were evaluated in 2013 (designated as environment E1), 2014 (designated as environment E2), and 2015 (designated as environment E3), at the Experimental Farm of Jiangsu Yanjiang Institute of Agricultural Sciences, Nantong, China.
In each environment, field trials were carried out following a randomized complete block design with two replicates (Yan et al. 2014). Each genotype was grown in a single-row plot, 500 cm long and 60 cm apart, with 20 plants per row. Standard agronomic practices were applied to provide adequate nutrition and keep plots disease free. All plants were manually self-pollinated, and mature grains from each RIL were harvested in bulk for phenotypic measurements. Harvested waxy maize grains were ground using a high-speed universal disintegrator (FW-100, Taisite, Tianjin, China) and then sieved through a 100-mesh sieve.
Phenotypic measurements
Pasting properties of the flours (28 g total weight; 10%, w/w, dry basis) were measured using a rapid visco analyzer (RVA, Model 3D, Newport Scientific, Australia) and analyzed using TCW (Thermal Cycle for Windows) software following the method described by Lu and Lu (2012). The sample suspension of each RIL was incubated at 50 °C for 1 min, the temperature increased to 95 °C and maintained for 2.5 min, and finally cooled down to 50 °C and maintained for 1 min. Three primary RVA parameters, peak viscosity (PV), trough viscosity (TV), and final viscosity (FV), were obtained from the pasting curve. Two secondary RVA parameters, breakdown viscosity (BD = PV − TV) and setback viscosity (SB = FV − TV), were calculated from the primary parameters. Pasting time (PT) and pasting temperature (PTP) were also recorded.
Statistical analysis
Statistical analyses of all phenotypic data of the pasting properties across 3 years were performed using SAS 9.13 software (SAS Institute, Cary, NC, USA). Analysis of variance (ANOVA) was performed using the PROC GLM procedure. The broad-sense heritability (H 2) of each RVA parameter was estimated according to the following formula: H 2 = σ 2 g/(σ 2 g + σ 2 e), where σ 2 g denotes genotypic variance components and σ 2 e denotes error variance components, which were evaluated using PROC VARCOMP. The best linear unbiased predictors (BLUPs) for each line in the tested RILs were evaluated to account for the effects of environmental factors using PROC MIXED. PROC CORR was used to estimate the correlations between the RVA parameters based on the BLUPs.
Genotyping and construction of genetic linkage maps
All RILs and parental inbred lines were genotyped using an Affymetrix microarray CGMB50K SNP Array containing 56,000 maize SNPs developed from the B73 reference sequence (Ganal et al. 2011), at China Golden Marker (Beijing) Biotech, China. The harvested genotypic matrix was corrected using a hidden Markov model approach from the R package MPR50 with some modifications, and the co-segregated SNP markers were grouped in bins using a home-made Perl script (Zhang et al. 2016). Chi-squared tests were conducted to test segregation distortion of the bins at the significance level of P > 0.05. All called bins were used to construct the genetic linkage map using JoinMap version 4.0 software (Van Ooijen 2006), with the Kosambi mapping function used to calculate the marker genetic distance.
QTL analysis
QTL mapping and estimation of QTL effects on each RVA parameter were performed using QTL IciMapping v4.1 (Lei et al. 2015). The method of inclusive composite interval mapping (ICIM) was applied to detect additive QTLs. For ICIM, the scanning step size was set to 1 cM, and the largest P value for entering variables in stepwise regression of phenotype on marker variables (PIN) was set at 0.001. QTLs were considered significant at a LOD threshold of 2.5 as this is commonly used for QTL mapping in maize (Song et al. 2016). QTLs detected in different environments and explaining more than 10% of phenotypic variance were considered as major QTLs (Raihan et al. 2016).
Results
Phenotypic variations and correlations
In the 3 years investigated, the variation of pasting properties (RVA profiles) revealed a continuous distribution among the 198 RILs, and significant differences were observed between the two parents for most pasting properties (Table 1). The RVA parameters of PV, TV, BD, FV, and SB for Tongxi5 were significantly higher than those for Hengbai522 in all years, except for PT and PTP. Transgressive segregation was detected in both directions for all traits in the RIL population. Analysis of variance for all tested RVA parameters revealed that the variances of genotypes (G) and the interactions between genotype and all environmental factors (G × E) were highly significant at the 0.001 probability level (Table 1), which indicated that all these pasting properties were quantitative traits affected by the environment. The estimated broad-sense heritability (H 2) of RVA parameters in the RIL population ranged from 68.9% (PTP) to 80.6% (FV).
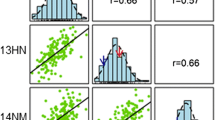
To explore the relationships among pasting properties, pairwise correlation analysis was carried out based on the BLUPs in the RIL population. The RVA parameters were significantly correlated with each other, except for BD and PTP, and SB and PT (Table 2). Significant positive correlations were observed between any two parameters among PV, TV, BD, FV, and SB. Nevertheless, PT was negatively correlated with PV, TV, BD, and FV, whereas PTP was negatively correlated with PV, TV, and FV, and positively correlated with SB and PT.
Construction of a high-density bin map
The Tongxi5/Hengbai522 RIL population and both parental inbred lines were genotyped using 56,000 SNPs. Following quality control, 12,268 SNPs with precise physical positions based on the B73 reference sequence showed polymorphism between the two parental lines. Based on these polymorphic SNPs, a skeleton bin map consisting of 2703 recombinant bins was constructed for all 198 RILs. The resulting high-density bin map spanned a total length of 1876.20 cM of the whole genome, with an average genetic distance of 0.73 cM between adjacent bin markers (Table 3, Supplementary Fig. S1). The number of bin markers in each chromosome ranged from 189 (chromosome 2) to 352 (chromosome 1), with an average of 270 bin markers per chromosome. The genetic length of the 10 chromosomes ranged from 129.15 cM (chromosome 6) to 273.47 cM (chromosome 2), with an average size of 187.62 cM. There were 16 gaps larger than 5 cM in the bin map, six of which were located on chromosome 2.
Identification of QTLs for pasting properties
Based on the ICIM mapping, a total of 72 QTLs were identified for all seven RVA parameters involved in pasting properties; these were distributed on all 10 chromosomes (Supplementary Table S1). Of these QTLs, 23 were stably identified in at least 2 years (Table 4), but the remaining 49 were only detected in a specific year, indicating that most QTLs identified in the RIL population were affected by environment.
Peak viscosity
Ten QTLs were detected for PV across the 3 years; these were located on chromosomes 1, 2, 3, 4, 5, and 9 (Supplementary Table S1). Of these QTLs, qPV4-1, qPV5-1, and qPV5-2 were identified in 2 or more years (Table 4). A major QTL of qPV4-1, flanked by PZE-104141018 and PZE-104141424 on chromosome 4, was stably detected in all 3 years and accounted 15.51, 14.43, and 11.82% of phenotypic variation, respectively. The allele from Tongxi5 increased the trait value. The other two QTLs (qPV5-1 and qPV5-2) were detected in 2 years (2013 and 2014); both were located on chromosome 5.
Trough viscosity
For TV, 12 QTLs were identified across 3 years, which were located on chromosomes 2, 3, 4, 5, 6, 7, and 9 (Supplementary Table S1). Among them, three QTLs, qTV4-1, qTV5-2, and qTV5-3, were identified in at least two of the years (Table 4). A major QTL of qTV4-1 was stably identified in all years, which was located at the interval of PZE-104141018_PZE-104141424 on chromosome 4 and explained 11.87 to 13.33% of phenotypic variation in the 3 years. The favorable allele was from Tongxi5, which could increase the TV value by 192.8 cP on average.
Breakdown viscosity
In total, nine QTLs were detected for BD in the 3 years investigated; these were located on chromosomes 1, 3, 5, 7, 8, and 10 (Supplementary Table S1). Of these QTLs, qBD1-1, qBD3-3, and qBD8-1 were detected in 2 or more years (Table 4). Only one stable QTL of qBD1-1 was identified for BD in all years, which was flanked by PZE-101125948 and SYN25740 on chromosome 1 and accounted for 2.29 to 4.19% of phenotypic variation in different years. The allele from Tongxi5 showed an increasing effect.
Final viscosity
Twelve QTLs were identified for FV across the 3 years; these were located on chromosomes 2, 5, 6, 7, and 9 (Supplementary Table S1). Four of these QTLs for FV (qFV2-1, qFV5-1, qFV5-2, and qFV5-3) were identified in 2 or more years (Table 4). Among which, two stable QTLs, qFV2-1 (in the marker interval of SYN5616_PZE-102161143 on chromosome 2) and qFV5-2 (a major QTL flanked by PZE-105159228 and PZE-105158503 on chromosome 5), were consistently detected in all years, accounting for 4.41 and 13.95% of phenotypic variation on average, respectively. Furthermore, qFV2-1 and qFV5-2 showed oppositely additive effects: the Tongxi5 allele of qFV2-1 increased FV by a mean value of 139.56 cP, whereas that of qFV5-2 decreased it by 254.77 cP on average.
Setback viscosity
Seven QTLs were identified for SB across the 3 years, which were located on chromosomes 2, 3, 4, 5, and 7 (Supplementary Table S1). Among them, two QTLs, qSB2-1 (flanked by SYN5616 and PZE-102161143 on chromosome 2) and qSB5-1 (flanked by PZE-105041558 and PZE-105041198 on chromosome 5), were persistently detected in two of the years and explained 5.63 and 7.42% of the average phenotypic variation, respectively (Table 4). All favorable alleles of qSB2-1 and qSB5-1 were from Tongxi5, which could increase SB by a mean value of 42.69 and 44.19 cP, respectively. No major QTL was identified in all years.
Pasting time
For PT, a total of 14 QTLs were identified across 3 years, distributing on eight chromosomes, the exceptions being chromosomes 2 and 6 (Supplementary Table S1). Of these QTLs, eight were detected in two of the years and explained 0.92 to 2.70% of total phenotypic variance (Table 4). In all years, only one QTL of qPT8-1 was stably identified and explained only a small percentage of the phenotypic variance, with an average of 0.92%, and was flanked by PZE-108100090 and PZE-108100367 on chromosome 8. The allele from Tongxi5 could increase the trait value by 0.12 min.
Pasting temperature
Eight QTLs were detected for PTP in all years, which were located on chromosomes 1, 2, 5, 6, and 7 (Supplementary Table S1). Among these QTLs, three were identified in 2013, four in 2014, and one in 2015, collectively explaining 16.78, 8.49, and 8.94% of phenotypic variation, respectively. Nevertheless, none was detected in 2 or more years, so these QTLs may have been the result of environmental effects.
Co-localization of QTLs underlying pasting properties
Eight QTLs for RVA parameters were identified as being co-located on the same genome regions of waxy maize (Supplementary Table S1, Table 4). The qBD3-3 for BD and qSB3-1 for SB were co-located in the marker interval of PZE-103066568_PZE-103123538 on chromosome 3, and the QTL for BD (qBD3-2) on chromosome 3 coincided with the QTL for PT (qPT3-1). Three QTLs for PV, qPV4-1 (flanked by PZE-104141018 and PZE-104141424 on chromosome 4), qPV5-2 (flanked by PZE-105028457 and PZE-105027956 on chromosome 5), and qPV9-1 (flanked by PZE-109094529 and PZE-109096246 on chromosome 9), were also identified for TV (qTV4-1, qTV5-3, and qTV9-1), and the QTL for PV (qPV5-1) in the interval of PZE-105159228_PZE-105158503 on chromosome 5 was simultaneously found for TV (qTV5-2) and FV (qFV5-2). The QTLs for FV (qFV2-1) and SB (qSB2-1) on chromosome 2 were co-located in the interval of SYN5616_PZE-102161143. A QTL for TV on chromosome 7, qTV7-1 (flanked by SYN12703 and SYN10108), was also found for FV (qFV7-3).
Candidate gene prediction
Candidate genes in the physical intervals of PZE-104141018_PZE-104141424 on chromosome 4 and PZE-105159228_PZE-105158503 on chromosome 5, where the major QTLs of qPV4-1 and qTV4-1, and qFV5-2 were located, were predicted based on B73 reference genome sequence Version 5b.60 (http://www.maizegdb.org). These two genomic intervals of PZE-104141018_PZE-104141424 and PZE-105159228_PZE-105158503 were approximately 475 and 181 kb, encompassed 28 and 11 protein-coding genes, respectively (Supplementary Table S2). According to the maize gene annotation database at MaizeGDB (http://www.maizegdb.org), the putative genes within these two major QTL intervals indicated that genes of GRMZM2G146028 on chromosome 4 and GRMZM2G142709 on chromosome 5 were the most likely candidate genes for qPV4-1 and qTV4-1, and qFV5-2, respectively. The candidate gene of GRMZM2G146028 encodes an AP2/EREBP family transcription factor (Supplementary Table S2). In rice, an AP2/EREBP family transcription factor of RSR1 has been identified as a starch biosynthesis regulator underlying the amylopectin structure and consequently determining the physicochemical properties of starch (Fu and Xue 2010). Another candidate gene of GRMZM2G142709 encodes a glucosyltransferase (Supplementary Table S2), which was involved in starch pasting properties in cassava (Thanyasiriwat et al. 2014).
Discussion
Waxy maize is generally reserved for fresh food in Asia, but is also widely used as industry resource for the textile, adhesive, and paper industries. The quality of food and industrial applications represents an important index reflecting the waxy maize grain quality (Yang et al. 2014), and high-quality grain is becoming an increasingly important objective in waxy maize breeding. Starch paste viscosity properties (RVA profile) are widely used as indicators of cereal kernel quality (Bao et al. 2000; Ketthaisong et al. 2013; Lu et al. 2013; Yan et al. 2014; Yan et al. 2011; Yang et al. 2014). A few studies have focused on the relationship between starch biosynthesis-related genes and starch paste viscosity properties in maize (non-waxy maize) (Wilson et al. 2004; Yang et al. 2014), but little effort has been made to elucidate the genetic basis of starch paste viscosity in waxy maize. Waxy maize kernel contains nearly 100% amylopectin. Unfortunately, the genetic information gained from non-waxy maize cannot be applied directly for waxy maize analysis and quality improvement, similarly to glutinous rice vs. non-glutinous rice (Yan et al. 2011). In this study, the genetic basis of starch paste viscosity properties in waxy maize was dissected using high-density QTL mapping. This provided an overview of the genetic mechanism of the starch viscosity profile in waxy maize and may facilitate improvements in the grain quality of waxy maize.
In this study, a RIL population, comprising 198 families derived from a cross between two waxy maize parents with similar amylopectin content, was used to map QTLs for starch viscosity properties using the ICIM method across 3 years. The estimates of broad-sense heritability revealed moderate heritabilities for the tested RVA parameters, with the H 2 value ranging from 68.9% for PTP to 80.6% for FV, which was consistent with the results from previous studies in rice (Wang et al. 2007). The results of linkage analysis indicated that these grain quality traits were genetically complex and significantly influenced by the environment. Seventy-two QTLs were detected for seven RVA parameters, each accounting for 0.55–18.46% of phenotypic variation. Among these QTLs, 17 were identified in two of the years, six were identified in all 3 years. These results indicated that most QTLs for starch viscosity properties in waxy maize might be affected by environmental factors with QTL-by-environment interaction (G × E, QEI) and under the control of multiple genes with minor effects (Hao et al. 2015a; Raihan et al. 2016; Zhou et al. 2016). The QEI may be due to the specific expression of certain genes during the process of adaptation to different ecological environments (Zhou et al. 2016). Due to the QEI, the specific QTL might be strongly expressed in a particular environment, while not or weakly expressed in another environment; or the specific QTL could have opposite effects on the phenotypic variation in different environments (Li et al. 2003). The stable and highly heritable QTLs, which could be expressed in different environments, might be useful for the future MAS for the improvement of grain quality in a wide range of environments in waxy maize breeding programs (Zhou et al. 2016); while particular environmental conditions were important in the expression of grain quality, environment-favorable QTL could be used for MAS within specific target environments (Li et al. 2015; Shang et al. 2016). In maize, MAS has been successfully used for the improvement of multiple grain quality-related traits, such as provitamin A content (Azmach et al. 2013) and kernel oil content (Hao et al. 2014). In this study, the stable QTLs identified for the RVA parameters with relatively high heritability, including qPV4-1, qTV4-1, qBD1-1, qFV2-1, qFV5-2, and qPT8-1, are likely important QTLs for starch viscosity properties in waxy maize and should be considered priority candidates for MAS in wide range environments in modern waxy maize breeding programs.
In this study, eight QTL clusters, mainly distributed on chromosomes 2, 3, 4, 5, 7, and 9, were observed to be co-associated with two or more RVA parameters, which was coincided with significant correlations among the studied parameters. For example, a QTL cluster in the interval of PZE-104141018_PZE-104141424 on chromosome 4 was responsible for two major QTLs of qPV4–1 and qTV4–1, detected in all years; a major QTL for FV (qFV5-2) was in the interval of PZE-105159228_PZE-105158503 on chromosome 5, and the other two QTLs of qPV5-1 for PV and qTV5-2 for TV were mapped to the same region; significant positive correlations were observed among these RVA parameters. The co-locations of QTLs for different RVA parameters in the studied population suggest pleiotropy of a single causal gene or close linkage of multiple causal genes (Yan et al. 2014; Zhang et al. 2013). In modern waxy maize breeding schemes for grain quality improvement, MAS of these co-associated genetic loci could simultaneously improve multi-target traits of waxy maize grain quality.
In previous studies, a few gene loci involved in starch biosynthesis were revealed to be related to starch paste viscosity properties in rice, including Wx, Alk, PUL, ISA, SBE, and SS (Han et al. 2004; Xu et al. 2013; Yan et al. 2014; Yan et al. 2011; Zhang et al. 2013). Based on candidate gene association analysis, genes of isa2, ae1, and sh2 were identified to be significantly associated with starch viscosity properties in maize (Wilson et al. 2004; Yang et al. 2014). To further address the natural variation of known starch metabolism-related genes and possible molecular mechanisms underlying the detected QTLs for starch paste viscosity properties in this study, their physical positions were compared based on B73 reference genome sequence Version 5b.60. Unexpectedly, no QTL was found in the wx locus on chromosome 9, and no other known starch metabolism-related genes co-localized within the QTLs detected in this study. These results suggest that both parents with similar amylopectin content might have the same wx allele, or that some of the genes involved in starch viscosity properties did not show allelic variation between the two parents (Yan et al. 2014; Zhao et al. 2009); therefore, novel molecular mechanisms might account for the QTLs identified in the studied population (Wang et al. 2015) or there may be genotype-by-environment interactions in waxy maize (Liu et al. 2016). The other explanation for this phenomenon might be that other components of waxy maize kernels also influence starch paste viscosity properties, such as protein (the second most abundant component following starch), which significantly affects the physicochemical properties of starch because that gelatinized starch granules are either coated on the peripheral surface of the protein network or trapped in the protein matrix (Lu and Lu 2012).
The quality of genetic maps significantly influences accuracy of QTL mapping. The use of high-density genetic maps enables capture of more recombination events and makes fuller use of the linkage information, thus improving the resolution and accuracy of QTL mappings performed (Chen et al. 2016). In the present study, a high-density genetic map was constructed using 56,000 SNPs, with an average genetic distance of 0.73 cM between adjacent bin markers. Due to the reduction of QTL intervals, candidate genes underlying the mapped locus could be predicted; this will provide valuable information for positional cloning of these QTLs (Zhou et al. 2016). Among three major QTLs detected in this study, the physical distances for qPV4-1 and qTV4-1 in the interval of PZE-104141018_PZE-104141424 on chromosome 4 and for qFV5-2 in the interval of PZE-105159228_PZE-105158503 on chromosome 5 were approximately 475 and 181 kb, respectively. These two genomic intervals encompass 28 and 11 protein-coding genes, respectively. According to the B73 reference genome sequence Version 5b.60 and the gene annotation data available at MaizeGDB, two genes, GRMZM2G146028 (encoding an AP2/EREBP family transcription factor) within the qPV4-1 and qTV4-1 interval on chromosome 4 and GRMZM2G142709 (encoding a glucosyltransferase) within the qFV5-2 interval on chromosome 5, were the most likely candidate genes that might regulate starch pasting properties, as previously reported in rice (Fu and Xue 2010) and in cassava (Thanyasiriwat et al. 2014). These results not only promote further research into the genetic mechanisms of starch viscosity profile in waxy maize but also provide a basis for MAS of starch pasting properties in waxy maize quality improvement programs. However, their association with starch pasting properties in waxy maize requires further study, and this would involve strategies such as fine mapping of identified QTLs by backcrosses or knock-outs and over-expression of candidate genes in the QTL intervals (Wang et al. 2015).
References
Azmach G, Gedil M, Menkir A, Spillane C (2013) Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol 13:277
Bao JD, Yao JQ, Zhu JQ, Hu WM, Cai DG, Li Y, Shu QY, Fan LJ (2012) Identification of glutinous maize landraces and inbred lines with altered transcription of waxy gene. Mol Breeding 30:1707–1714
Bao J, Zheng X, Xia Y, He P, Shu Q, Lu X, Chen Y, Zhu L (2000) QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.) Theor Appl Genet 100:280–284
Bao JS, Wu YR, Hu B, Wu P, Cui HR, Shu QY (2002) QTL for rice grain quality based on a DH population derived from parents with similar apparent amylose content. Euphytica 128:317–324
Chen L, Li C, Li Y, Song Y, Zhang D, Wang T, Li Y, Shi Y (2016) Quantitative trait loci mapping of yield and related traits using a high-density genetic map of maize. Mol Breeding 36:134. doi:10.1007/s11032-016-0545-0
Collins GN (1909) A new type of Indian corn from China. Bur of Plant Ind (Bulletin) 161:1–30
Cozzolino D (2016) The use of the rapid visco analyser (RVA) in breeding and selection of cereals. J Cereal Sci 70:282–290
Ding ZH, Meng ZD, Zhang FJ, Wang LM, Sun Q, Zhang QW (2006) General situation and development priority of waxy corn breeding in China. J Maize Sci 14:46–48
Fan L, Quan L, Leng X, Guo X, Hu W, Ruan S, Ma H, Zeng M (2008) Molecular evidence for post-domestication selection in the waxy gene of Chinese waxy maize. Mol Breeding 22:329–338
Fu FF, Xue HW (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154:927–938
Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Clarke JD, Graner EM, Hansen M, Joets J (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One 6:e28334
Han Y, Xu M, Liu X, Yan C, Korban SS, Chen X, Gu M (2004) Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.) Plant Sci 166:357–364
Hao D, Cheng Y, Chen G, Lu H, Shi M, Zhang Z, Huang X, Mao Y, Xue L (2015a) Identification of significant single nucleotide polymorphisms for resistance to maize rough dwarf disease in elite maize (Zea mays L.) inbred lines. Euphytica 203:109–120
Hao D, Zhang Z, Cheng Y, Chen G, Lu H, Mao Y, Shi M, Huang X, Zhou G, Xue L (2015b) Identification of genetic differentiation between waxy and common maize by SNP genotyping. PLoS One 10:e0142585
Hao X, Li X, Yang X, Li J (2014) Transferring a major QTL for oil content using marker-assisted backcrossing into an elite hybrid to increase the oil content in maize. Mol Breeding 34:739–748
Hsu YC, Tseng MC, Wu YP, Lin MY, Wei FJ, Hwu KK, Hsing YI, Lin YR (2014) Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol Breeding 34:655–673
Ketthaisong D, Suriharn B, Tangwongchai R, Lertrat K (2013) Changes in physicochemical properties of waxy corn starches at different stages of harvesting. Carbohydr Polym 98:241–248
Kharabian-Masouleh A, Waters DLE, Reinke RF, Ward R, Henry RJ (2012) SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Sci Rep 2:2016–2016
Lei M, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 121:269–283
Li S, Wang J, Zhang L (2015) Inclusive composite interval mapping of QTL by environment interactions in biparental populations. PLoS One 10:e0132414
Li Z, Yu S, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CHM, Liu GF, Wang GC, Shashidhar HE (2003) QTL × environment interactions in rice. I. Heading date and plant height. Theor Appl Genet 108:141–153
Liang Z, Zhang KP, Liu B, Tian JC (2009) Detection of quantitative trait loci for paste viscosity characteristics based on the doubled haploid progeny from a cross between two Chinese wheat varieties. Can J Plant Sci 89:837–844
Liu C, Hua J, Liu C, Zhang D, Hao Z, Yong H, Xie C, Li M, Zhang S, Weng J, Li X (2016) Fine mapping of a quantitative trait locus conferring resistance to maize rough dwarf disease. Theor Appl Genet. doi:10.1007/s00122-016-2770-7
Liu H, Ramsden L, Corke H (1997) Physical properties and enzymatic digestibility of acetylated ae, wx, and normal maize starch. Carbohydr Polym 34:283–289
Lu D, Lu W (2012) Effects of protein removal on the physicochemical properties of waxy maize flours. Starch-Starke 64:874–881
Lu D, Sun X, Yan F, Wang X, Xu R, Lu W (2013) Effects of high temperature during grain filling under control conditions on the physicochemical properties of waxy maize flour. Carbohydr Polym 98:302–310
Perera C, Lu Z, Sell J, Jane J (2001) Comparison of physicochemical properties and structures of sugary-2 cornstarch with normal and waxy cultivars. Cereal Chem 78:249–256
Raihan MS, Liu J, Huang J, Guo H, Pan Q, Yan J (2016) Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 × SK maize population. Theor Appl Genet 129:1465–1477
Shang L, Wang Y, Wang X, Liu F, Abdugheni A, Cai S, Li Y, Ma L, Wang K, Hua J (2016) Genetic analysis and QTL detection on fiber traits using two recombinant inbred lines and their backcross populations in upland cotton. G3: Genes | Genomes | Genetics 6:2717–2724
Song W, Wang B, Hauck AL, Dong X, Li J, Lai J (2016) Genetic dissection of maize seedling root system architecture traits using an ultra-high density bin-map and a recombinant inbred line population. J Integr Plant Biol 58:266–279
Thanyasiriwat T, Sraphet S, Whankaew S, Boonseng O, Bao J, Lightfoot DA, Tangphatsornruang S, Triwitayakorn K (2014) Quantitative trait loci and candidate genes associated with starch pasting viscosity characteristics in cassava (Manihot esculenta Crantz). Plant Biol 16:197–207
Van Ooijen J (2006) JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations, vol 33. Kyazma BV, Wageningen, p 10.1371
Wang J, Yang J, Mcneil D, Zhou M (2010) Mapping of quantitative trait loci controlling barley flour pasting properties. Genetica 138:1191–1200
Wang LQ, Liu WJ, Xu Y, He YQ, Luo LJ, Xing YZ, Xu CG, Zhang Q (2007) Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor Appl Genet 115:463–476
Wang T, Wang M, Hu S, Xiao Y, Hao T, Pan Q, Xue J, Yan J, Li J, Yang X (2015) Genetic basis of maize kernel starch content revealed by high-density single nucleotide polymorphism markers in a recombinant inbred line population. BMC Plant Biol 15:288. doi:10.1186/s12870-015-0675-2
Wilson LM, Whitt SR, Ibáñez AM, Rocheford TR, Goodman MM, Buckler ES (2004) Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16:2719–2733
Xie X, Xue L, Shi M, Cai Z, Huang X, Chen G, Lu J, Yuan C, Tang M (1997) Breeding of a waxy maize hybrid of Suyunuo 1 with excellent characteristics of good quality, high yield and multiple resistance. Maize Sci 5:11–15
Xie XY, Xue L, Hu-Hua LU, Chen GQ, Xie LM, Dong-Mei LU (2009) Study on heterotic groups and patterns of high amylopectin maize. Maize Sci 17:35–38
Xu F, Bao J, He Q, Park Y-J (2016) Genome-wide association study of eating and cooking qualities in different subpopulations of rice (Oryza sativa L.) BMC Genomics 17:663. doi:10.1186/s12864-016-3000-z
Xu F, Sun C, Huang Y, Chen Y, Tong C, Bao J (2015) QTL mapping for rice grain quality: a strategy to detect more QTLs within sub-populations. Mol Breeding 35:105. doi:10.1007/s11032-015-0296-3
Xu F, Zhang G, Tong C, Sun X, Corke H, Sun M, Bao J (2013) Association mapping of starch physicochemical properties with starch biosynthesizing genes in waxy rice (Oryza sativa L.) J Agr Food Chem 61:10110–10117
Xu Y, Crouch J (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48:391–407
Yacouba NT, Bo Y, Gao GJ, Zhang QL, Xu CG, Xiao JH, He YQ (2013) QTL analysis of eating quality and cooking process of rice using a new RIL population derived from a cross between Minghui 63 and Khao Dawk Mali105. Aust J Crop Sci 7:2036–2047
Yan B, Yacouba NT, Chen J, Wang Y, Gao G, Zhang Q, Liu X, He Y (2014) Analysis of minor quantitative trait loci for eating and cooking quality traits in rice using a recombinant inbred line population derived from two indica cultivars with similar amylose content. Mol Breeding 34:2151–2163
Yan CJ, Tian ZX, Fang YW, Yang YC, Li J, Zeng SY, Gu SL, Xu CW, Tang SZ, Gu MH (2011) Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.) Theor Appl Genet 122:63–76
Yang Z, Zhang E, Jiang Y, Xu S, Pan L, Chen Q, Xu C (2014) Sequence polymorphisms in Zmisa2 gene are significantly associated with starch pasting and gelatinization properties in maize (Zea mays L.) Mol Breeding 34:1833–1842
Zhang CQ, Bing HU, Zhu KZ, Zhang H, Leng YL, Tang SZ, Ming-Hong GU, Liu QQ (2013) QTL mapping for rice RVA properties using high-throughput re-sequenced chromosome segment substitution lines. Rice Sci 20:407–414
Zhang D, Liu LJ, Xue L, Li J, Xu C (2010) Association analysis of starch pasting characteristic traits with SSR markers in waxy corn in Jiangsu province. J Yangzhou Univ 31:36–42 (in chinese)
Zhang H, Miao H, Li C, Wei L, Duan Y, Ma Q, Kong J, Xu F, Chang S (2016) Ultra-dense SNP genetic map construction and identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci Rep 6:31556
Zhao L, Zhang K, Liu B, Tian J (2009) Detection of quantitative trait loci for paste viscosity characteristics based on the doubled haploid progeny from a cross between two Chinese wheat varieties. Can J Plant Sci 89:837–844
Zhou Z, Zhang C, Zhou Y, Hao Z, Wang Z, Zeng X, Di H, Li M, Zhang D, Yong H (2016) Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines. BMC Genomics 17:178. doi:10.1186/s12864-016-2555-z
Acknowledgements
This work was funded by the Natural Science Foundation of Jiangsu Province, China (BK20141241), the National Natural Science Foundation of China (91535103), the Six Major Talent Project of Jiangsu Province, China (2014-NY-032, 2016-NY-143), the Scientific and Technological Project of Nantong City, China (MS32015003), and the Agricultural San-xin Project of Jiangsu Province, China [SXGC(2014)088, SXGC(2015)105].
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hao, D., Xue, L., Yuan, J. et al. Genetic dissection of starch paste viscosity characteristics in waxy maize revealed by high-density SNPs in a recombinant inbred line population. Mol Breeding 37, 50 (2017). https://doi.org/10.1007/s11032-017-0652-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0652-6