Abstract
The activating V617F mutation in Janus kinase 2 (JAK2) has been shown to be the major cause for classic Philadelphia-negative myeloproliferative neoplasms (MPNs). Thus, the development of pharmacologic JAK2 inhibitors is an essential move in combating MPNs. In this study, screening methods examining both ligands and their structures were developed to discover novel JAK2 inhibitors from the ChemDiv database with virtual screening identifying 886 candidate inhibitors. Next, these compounds were further filtered using ADMET, drug likeliness, and PAINS filtering, which reduced the compound number even further. This consolidated list of candidate compounds (n = 49) was then evaluated biologically at molecular level and the highest performing inhibitor with a novel scaffold was selected for further examination. This candidate inhibitor, CD4, was then subjected to molecular dynamics studies, with complex stability, root-mean-square deviation, radius of gyration, binding free energy, and binding properties all examined. The result suggested that CD4 interacts with JAK2 and that the CD4-JAK2 complex is stable. This study was able to identify a candidate inhibitor that warrants further examination and optimization and may potentially serve as a future MPN treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Philadelphia chromosome-negative myeloproliferative neoplasms (MPNs), which include polycythemia vera (PV), essential thrombocytosis (ET), and primary myelofibrosis (PMF), are malignant hematologic diseases characterized by excessive hematopoietic stem cell proliferation [1,2,3]. The clinical features of these MPNs are characterized by excessive proliferation of one or more myeloid cell lineages, including erythroid, granulocytic, and/or megakaryocytic cells. These manifestations are often accompanied by splenomegaly, extramedullary hematopoiesis, arteriovenous thrombosis, hemorrhage, and/or severe physical symptoms such as itching, fatigue, fever, and night sweats. Furthermore, all three diseases may evolve into acute myeloid leukemia (AML) [4]. Currently, clinical treatments comprise both interventional and surgical treatments, such as phlebotomy, radiotherapy, splenectomy, or allogeneic hematopoietic stem cell transplantation [5,6,7]. However, while surgery and radiotherapy have high prognosis risk, they can impose a significant physical burden on patients. As a result, chemical drugs, such as hydroxyurea, interferon α, androgens, alkylating agents, immunomodulators, and anticoagulants, have emerged as a crucial method for treating MPNs [7]. Unfortunately, these drugs can only provide some relief from symptoms, with their effectiveness being limited and temporary. Moreover, many patients develop resistance or intolerance to these drugs, thus limiting their ability to prevent disease progression. Due to these limitations, the use of these drugs in clinical practice is limited, thus leaving patients with an inability to adequately address the clinical requirements associated with MPNs.
In previous studies, MPNs pathogenesis has been associated with JAK2 (Janus kinases 2), MPL (myeloproliferative leukemia virus oncogene), or CALR (calreticulin) gene mutations that subsequently contribute to the overactivation of JAK2 [8, 9]. This overactivation then results in aberrant JAK2/STATs signaling, ultimately resulting in the development of MPNs. The most common MPNs mutation is the V617F mutation that occurs in the JH2 region of JAK2 (JAK2 V617F). This mutation leads to JAK2 hyperactivation and is identified in the blood cells of nearly all PV patients and ∼50% of ET and PMF patients [10,11,12]. Therefore, developing novel JAK2 inhibitors that are effective in treating MPNs has become a research hotspot. Currently, three JAK2 inhibitors (Ruxolitinib, Fedratinib, and Pacritinib) have been approved for the treatment of MPNs [13,14,15]. Furthermore, several additional JAK2 inhibitors have undergone clinical trials and demonstrated promising efficacy and safety profiles. JAK2 inhibitors target for the pathogenic site and treat the disease etiologically, with obvious advantages in improving clinical symptoms, reducing the physical burden on patients, and prolonging survival. However, due to the high degree of JAK2 conservation, clinical JAK2 inhibitors can have off-target effects caused by poor selectivity, which raises potential safety concerns [16]. Furthermore, many patients may develop a tolerance to the JAK2 inhibitors during maintenance therapy, which would result in a reduction in the curative effect. Therefore, it is still necessary to discover new JAK2 inhibitors with a low toxicity and high efficiency to provide alternative options for clinical application.
The drug development process is risky and costly, with an average of 10 to 15 years required to develop a new drug at a cost of more than $2 billion [17]. At present, computer-aided methods are widely used in drug design and development to reduce cost, shorten the development time, and reduce the occurrence of drug toxicity and side effects [18,19,20]. In 2017, Hinton et al. proposed a new deep learning algorithm called capsule neural network (CapsNet) that aimed to solve the problem of information loss between multiple neural network layers [21]. Additionally, CapsNet was shown to exhibit excellent performance for small sample sizes and was able to accurately identify tiny differences in compounds based on previous research [22, 23]. Herein, to identify novel JAK2 inhibitors against MPNs that possess a low toxicity and high efficiency, a multitask CapsNet model was employed. This enabled virtual ligand-based screening to predict JAK2 inhibitor compounds based on the ChemDiv compound libraries that contain about 1.68 million compounds. Subsequently, structure-based molecular docking was applied to further explore associations between the candidate inhibitors and JAK2. After analyzing the results of two rounds of virtual screening, less than 50 compounds were selected and purchased. The purchased compounds were then subjected to an in vitro bioactivity evaluation. First, the screened compounds were verified at the protein level using an ADP-Glo™ kinase assay. Next, the anti-JAK2 compound with the highest antiproliferative effect was selected and evaluated in JAK2 V617F mutant human erythroleukemia (HEL) cells. Finally, the interaction mechanisms between JAK2 and the candidate compound were explored using a molecular dynamics simulation, with the observed interactions compared to the clinically used JAK2 inhibitor Ruxolitinib. Ultimately, this screening approach (Fig. 1) successfully identified CD4 as a promising JAK2 inhibitor with a binding activity that future studies could focus on optimizing to generate a potentially new clinical MPN treatment.
Materials and methods
Virtual screening based on multitask CapsNet
To ensure an accurate depiction of the bioactivity for each compound, four JAKs (JAK1, JAK2, JAK3, and TYK2) were annotated based on experimental IC50 values from an open-access database for drug discovery (https://www.ebi.ac.uk/chembl/). Duplicated compounds and compounds without IC50 values were removed with Pipeline Pilot, and the compounds indicated “Not Determined” or “Not Active” were deleted by manual inspection. The selected compounds were annotated as JAK inhibitors if their IC50 values was < 10 μM and JAK non-inhibitors if their IC50 values was > 10 μM. The final dataset for the JAKs consisted of 14,249 molecules, of which 11,879 molecules were classified as inhibitors and 2370 molecules were classified as non-inhibitors. Molecular fingerprints, as well as descriptors, were used simultaneously as characterization of compounds descriptors for model input. To characterize the structural features of the chemicals accurately, a total of five molecular fingerprints were generated using the chemical development kit (CDK), molecular access system (MACCS), extended connectivity fingerprint 4 and 6 (ECFP4 and ECFP6) and PubChem, and calculated using the PaDEL-Descriptor toolkit (v 2.21) [24]. In addition, a set of molecular descriptors was calculated by DS 3.1, namely 13 molecular descriptors (13MD), to represent the physicochemical features of chemicals. The detailed list of these properties is given in Table S1.
In this study, a multitask CapsNet framework, which had been developed by our team in 2021 [25], was used to build a ligand-based virtual screening model for the discovery of novel JAK2 inhibitors. The algorithms and training procedures are summarized in Table S2. In the output file, JAK family activity was indicated as follows: “1” = JAK1, “2” = JAK2, “3” = JAK3, and “4” = TYK2 activity, with a result of “0” indicating that the predicted compound does not have JAK family activity. Only compounds with an output result of “2” were retained for further examination.
Molecular docking
To improve active compound identification, DS 3.1 was used to further screen the compounds that were identified above. The crystal structure of JAK2 was downloaded from the PDB database (https://www.rcsb.org/; PDB ID: 6VGL) [26], along with the small molecule ligand Ruxolitinib. Ruxolitinib was the first JAK inhibitor to be marketed, and it is also the most used inhibitor in the treatment of MPNs [27]. This drug acts by competitively inhibiting the ATP-binding catalytic site within the kinase domain of JAK2; thus, it was used as a positive control. Prior to performing docking analysis, the protein crystal structures were pre-processed to remove water molecules, small molecule ligands, and polypeptide chains that do not participate in docking. Next, hydrogenates were added and incomplete amino acid residues were corrected. Then, the coordinates for the Ruxolitinib binding site were set as the docking site, with a site readjustment radius of 11 Å to ensure the entire ligand binding site was covered. Finally, biomolecular simulations were carried out using the CHARMM force field [28]. Ligand preparation modules were used to optimize small molecule energetics and generate reasonable conformations in conjunction with CDOCK. The obtained docking results were filtered and only the top 10% of candidate compounds were retained for further examination.
In silico ADMET, PAINS, and drug-likeness assessments for JAK2 inhibitors
For the compounds of interest, various predictive ADMET (absorption, distribution, metabolism, excretion, and toxicity) parameters were assessed by using ADMETlab 2.0 (https://admetmesh.scbdd.com/) [29]. This online platform provides accurate a total of 23 ADME endpoints and 27 toxicity endpoints, such as Caco-2 permeability, human intestinal absorption, plasma protein binding, blood–brain barrier, CYP450 1A2 inhibitor, CYP450 1A2 substrate, clearance, hERG blockers, and human hepatotoxicity.
To reduce potential false positives and ensure the successful identification of a candidate drug, pan assay interference compounds (PAINS) were filtered out using RDKit (https://www.rdkit.org) [30, 31]. To further ensure the quality candidate compounds, Lipinski’s rule-of-five was applied to eliminate molecules with potentially poor permeation or poor oral bioavailable using SwissADME (http://www.swissadme.ch) [32]. Additionally, to further optimize the list of potential drug candidates, a QED (Quantitative Estimate of Drug-likeness) analysis was performed using RDkit, with scores ranging from 0 (all properties unfavorable) to 1 (all properties favorable) [33].
Biological validation
Preparation of compounds
All purchased candidate compounds, as well as the positive control Ruxolitinib (TargetMol, Shanghai, China), were prepared by adding an amount of DMSO based on their respective relative molecular masses, and then allowing the compounds to fully dissolve to form a 10 mM masterbatch to be set aside.
ADP-Glo™ kinase assay
To verify the JAK2 inhibitory effect of the identified candidate compounds, an ADP-Glo™ kinase assay (Promega, Madison, WI, USA) was employed according to the manufacture’s protocols. Their inhibiting capacities were determined based on their IC50 values at the concentrations of 50 μM, 25 μM, 12.5 μM, 6.25 μM, and 3.125 μM. Briefly, 1 μL of the candidate compounds and the positive control Ruxolitinib (TargetMol, Shanghai, China) were transferred to a 384-well plate, ensuring that the final concentration of the compounds in the kinase reaction system was 200 μM. Then, 10 µM JAK2 (Sigma-Aldrich, Darmstadt, Germany) was prepared with 1 × Kinase buffer (40 mM Tris–HCl, pH 7.5; 20 mM MgCl2; and 0.1 mg/mL bovine serum albumin), and 1 μL of JAK2 was added to a 384-well plate and incubated for 15 min at room temperature. Next, 2 μL of a mixture containing ATP and kinase substrate was added to initiate the kinase reaction and incubated for 2 h at room temperature. 4 μL of ADP-Glo reagent was added and allowed to incubate for 1.5 h. Following incubation, 8 µL of kinase detection reagent was added and incubated at room temperature for 1 h to promote ADP to ATP conversion and generate luminescence that was then quantified on a microplate reader to establish kinase activity. Negative control wells contained all experimental components, but no compound was added, and the blank control wells contained neither inhibitor compound nor JAK2.
The inhibiting capacity of compounds obtained from the above screening would be verified in depth by ADP-Glo™ kinase Assay, and their IC50 values are calculated. The concentration gradient of the compound in the system is set as follows: 50 μM, 25 μM, 12.5 μM, 6.25 μM, and 3.125 μM. Then, ADP-Glo™ is applied to evaluate the inhibition of JAK2 kinase activity by the hit. The levels of JAK2 kinase inhibition were then determined based on the luminescence (L) as follows: inhibition rate (%) = (Lcontrol − Lsample)/(Lcontrol − Lblank) × 100%.
CCK-8 assay cell proliferation assay
To evaluate the antiproliferative effect of the candidate compounds, a CCK-8 assay was employed to evaluate the effect of the inhibitor in human erythroleukemia (HEL) cells carrying the JAK2 V617F mutation (Pricella Life Technologies Co., Ltd,). Cells were cultured in RPMI-1640 complete medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin mixture (P/S) in a 37 °C incubator with 5% CO2. Then, 100 μL of HEL cellular suspension was inoculated into a 96-well plate and samples were divided into experimental (HEL + CD4), control (only HEL), and blank (only media) groups. Next, 10 μL of CCK-8 solution was added to each well, with absorbance values (OD450) determined at 24 h and 48 h using a microplate reader. The cellular proliferation inhibition rate for each group was calculated according to the formula: cellular proliferation inhibition rate (%) = (ODcontrol − ODexperiment)/(ODcontrol − ODblank) × 100%, with all experiments performed in triplicate.
Molecular dynamics simulation and analysis
To further investigate the interactions between the candidate compounds and JAK2, molecular dynamics simulations using the JAK2 crystal 6VGL structure were conducted using Gromacs2018 [34]. Prior to performing the molecular dynamics simulation, JAK2 protein and small molecule structures were hydrogenated using MOE software (https://www.chemcomp.com), and the OPLS force field was applied to the JAK2 protein crystal structure and small molecules [35]. At the start of the simulation, TIP3P water was added to the system using the TIP3P model to create a water box with a size of 13 × 13 × 13 nm3, and the edges of the water box were at least 1.2 nm from the protein [36]; followed by the addition of Na+ or Cl− to ensure the system is electrically neutral. Next, the simulation process was divided into three steps. First, 50,000 steps of energy optimization were performed on the complex system using the steepest descent method. Next, the system was equilibrated under NVT ensemble for 1000 ps at a constant temperature of 300 K. Finally, the MD simulations were carried out in the NPT ensemble for 100 ns at a constant pressure of 1 bar and a constant temperature of 300 K. The simulation was repeated three times for all systems.
Binding free energy calculation
To evaluate the inhibitor/JAK2 binding strengths of the candidate inhibitor, the g_mmpbsa program was used to calculate the binding free energy of the complex [37]. For analysis, the trajectory was evaluated for the last 10 ns, with nodes created at every 100 ps to generate a total of 100 snapshots. Finally, the binding free energy (ΔGbind) was calculated using the molecular mechanics/Poisson-Boltzmann and surface area solvation (MM/PBSA) method [38, 39] as follows:
where ΔGcomplex, ΔGreceptor, and ΔGlignad represent the free energy of the complex, protein receptor, and ligand, respectively. The free energy of each component is composed of a total meteorological energy (EMM), solvation free energy (Gsol), and entropy contribution (TS). Additionally, EMM is composed of intramolecular energy (Eint), electrostatic energy (Eele), and van der Waals force (Evdw), and Gsol includes non-polar solvation energy (Gnonpol, sol) and polar solvation energy (Gpol, sol).
Statistical analysis
All biological experiments were repeated at least three times. When comparing two groups for statistical significance, a one-way ANOVA was performed with p < 0.05 deemed significant. All graphs were generated using GraphPad Prism 9 software.
Results and discussion
Virtual screening based on multitask CapsNet and molecular docking
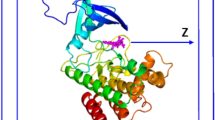
Following multitask CapsNet analysis, a total of 886 compounds were predicted to modulate JAK2 (6VGL) bioactivity. To establish that the applied molecular docking settings were reasonable, the JAK2 protein was redocked with Ruxolitinib, with a root-mean-square deviation (RMSD) value < 2 Å required when comparing the original and docked conformations. Following re-docking with Ruxolitinib, a RMSD value of 0.3091 Å was noted, thus indicating that the docking parameters are accurate and reliable (Fig. 2). To further evaluate the bioactivity of the selected compounds, CDOCKER was employed and only the top 10% of ranked compounds (n = 89) were selected for further analysis.
Enrichment of the candidate compounds via ADMET, PAINS, and QED assessments
The remaining compounds (n = 89) were evaluated in ADMET, with a total of 23 ADME endpoints and 27 toxicity endpoints examined. For the endpoints, the prediction probability values are represented by six symbols: 0–0.1(− − −), 0.1–0.3(− −), 0.3–0.5( −), 0.5–0.7( +), 0.7–0.9(+ +), and 0.9–1.0(+ + +). Usually, the symbol ‘ + + + ’ or ‘ + + ’ indicates the molecule is more likely to be toxic or defective, while ‘ − − − ’ or ‘ − − ’ represents nontoxic or appropriate. In addition, the predictions of each property are visualized as different colored dots; green, yellow, and red indicate that the ADME/T property of the compound is excellent, medium, and poor, respectively. Following analysis, a total of 67 compounds with probable values in the range of 0–0.5 and with green dots remained for further examination.
PAINS screening was done to filter out false positive compounds having PAINS patterns. A KNIME workflow was utilized to implement PAINS filtering in RDKit to remove ligands with matching patterns, thereby reducing the number of candidate compounds to 64.
Herein, to be considered a drug-like molecule, each candidate compound had to meet the conditions of Lipinski’s rule-of-five and obtain a QED value > 0.115. These requirements reduced the list of interest to 59 compounds. The chemical diversity of the space was analyzed using MACCS-based Tanimoto similarity to compare each compound with its nearest neighbor. The analysis of the scaffolds for 59 compounds yielded an average Tanimoto similarity of 0.26, indicating a significant level of structural diversity. Further, ligand efficiency (LE) was employed as a guiding principle for lead discovery, with the criterion that the initial hit should have a LE score > 0.3, indicating the presence of potent drug-like compounds [40, 41]. After that, the number of compounds determined to have an acceptable modulation of JAK2 inhibitive activity was reduced to 49 (Table S3). Thus, these inhibitor compounds were purchased for biological validation.
Biological validation
Kinase activity assays
To validate the bioactivity of the candidate compounds (n = 49), an ADP-Glo™ kinase assay was employed, with luminescence quantified to determine JAK2 kinase activity. Of the examined compounds, CD4, CD10, CD31, CD48, and CD49 showed the highest levels of JAK2 kinase inhibition (Fig. S1). Since CD4 was associated with the lowest level of JAK2 kinase activity, thus suggesting a strong inhibitory affect, this compound was selected for further examination. To determine the optimal concentration of CD4, the ADP-Glo™ kinase assay was repeated with CD4 concentrations of 50 μM, 25 μM, 12.5 μM, 6.25 μM, and 3.125 μM. A concentration curve was then constructed and the IC50 value was determined to be 15.22 μM (Fig. 3). These results of virtual screening can be influenced by several important factors, including the protein structure chosen for screening, the size and diversity of the compound library, the docking strategy employed, and the manual selection of candidate compounds. Thus, the hits obtained by virtual screening are generally at the micromolar level. The aim of virtual screening is to obtain novel hits rapidly and low-costly. To improve the activity and ADME/T properties of hits, further structural optimization following this molecular design is an essential work.
Effect of CD4 on HEL/HEK-293 T cells
Herein, DMSO was diluted to the same concentration as the drug and added to HEL cells and assayed using the CCK-8 assay, Fig. S2 shows that low concentrations of DMSO do not have an effect on subsequent compound activity assays. The inhibitory effect of CD4 on proliferation was assessed by using a CCK-8 analysis. HEL cells were cultured with CD4 at concentrations of 0.5 μM, 1 μM, 5 μM 10 μM, 15 μM, 20 μM, 40 μM, 80 μM, and 100 μM. The results showed that the anti-proliferation rate in the CD4 treated HEL cells was significantly higher than that of the control (Fig. 4a, p < 0.05), with the inhibition rate (%) significantly increased in a dose-dependent manner. Furthermore, at 24 h, CD4 had an IC50 = 37.24 μM, and at 48 h, the IC50 = 46.73 μM, thus suggesting a relatively consistent inhibition ability.
To evaluate CD4 toxicity, human embryonic kidney 293 T cells (HEK-293 T) were treated with CD4 at concentrations of 6.25 μM, 12.5 μM, 25 μM, 50 μM, and 100 μM. After 48 h, HEK-293 T cellular proliferation was detected using a CCK-8 assay. The results showed that CD4 had no inhibitory effect on the growth of the HEK-293 T cells, thus indicating that CD4 may have low toxicity (Fig. 4b).
Novel scaffold of CD4
To further characterize CD4 and its properties, its scaffold structure was compared to that of the three clinically used JAK2 inhibitors, Ruxolitinib, Fedratinib, and Pacritinib. As shown in Fig. 5, it is clear that CD4 consists of the following two main substructures: benzothiazole and methoxybenzamide. And the chemical structures of Ruxolitinib, Fedratinib, and Pacritinib mainly include pyrimidine, pyrazole, pyrrole, and benzenesulfonamide. Moreover, we calculated the Tanimoto coefficient (Tc) between the four compounds through Morgan fingerprints to evaluate the similarity between CD4 and the other three JAK2 inhibitors. The Tanimoto coefficient measures the similarity between two compounds. A higher coefficient value, closer to 1, indicates a higher similarity between the compounds [42]. The results showed that CD4 does possess a novel scaffold when compared to Ruxolitinib (Tc = 0.074), Fedratinib (Tc = 0.151), and Pacritinib (Tc = 0.119).
Molecular dynamics simulation analysis
To examine the compactness of three systems, the radius of gyration (Rg) value was calculated during the molecular dynamics simulation. A smaller Rg value indicates better compactness and a more stable structure. As shown in Fig. 6, at the beginning of the simulation, the Rg values of the three systems slightly decrease and then gradually stabilize. At equilibrium, the Rg values of all three systems fluctuated around 2.15 nm. Furthermore, to investigate the overall stability of the CD4-JAK2 and Ruxolitinib-JAK2, the root-mean-square deviation (RMSD) of the three systems relative to the initial conformation during the simulation were calculated (Fig. 7). The RMSD of the three systems showed an increasing trend in the early stage of the simulation and reached about 20 ns; each system basically reached stability, and the RMSD values of the three systems stabilized at about 0.5 nm. The RMSD and Rg values suggest that the conformation of stabilizes as the system gradually equilibrates over the 100 ns simulation. Both complex systems closely parallel the trends observed for the JAK2 protein in the absence of small molecules, with the differences being minimal. Therefore, the JAK2 protein–ligand complexes reached a converged state in the 100 ns simulation. Additionally, CD4 interacts with JAK2 protein as effectively as Ruxolitinib, and the binding of either small molecule does not significantly alter the structural integrity of JAK2. The system’s consistency confirms its stability.
In addition, root-mean-square function (RMSF) values of JAK2 amino acid residues were calculated, which can reflect the volatility and flexibility of amino acid sites. As depicted in Fig. 8, the RMSF values for the majority of amino acid residues in the JAK2 protein were found to be less than 0.5 nm, suggesting that these residues exhibit relative stability throughout the protein. With the exception of the head and tail residues, which typically exhibit normal large fluctuations in RMSF values, only a narrow range of residues from 1070 to 1080 displayed slight fluctuations. The relatively small RMSF values observed for the CD4-JAK2 and Ruxolitinib-JAK2 complexes in the P-loop, hinge, and DFG motif regions, indicating that both CD4 and Ruxolitinib are capable of forming stable interactions with the respective binding pockets of JAK2.
Binding free energy analysis
To further analyze the interactions between the inhibitor compounds and JAK2, the last 10 ns of the molecular dynamics was investigated. A total of 100 snapshots were taken at 100 ps and the binding free energy was calculated using the MM/PBSA method, with a lower energy value indicating a greater stability. The binding free energy is a measure of the stability of the complex, with lower energy values indicating greater stability. Table 1 lists the binding free energies of CD4-JAK2 and Ruxolitinib-JAK2 complexes, respectively. As can be seen from the Table 1, the binding free energies of CD4-JAK2 and Ruxolitinib-JAK2 were − 38.34 ± 4.39 kcal/mol and − 36.21 ± 3.49 kcal/mol, respectively. The lower binding free energies of the complexes of CD4 with JAK2 suggest that the affinity of CD4 with JAK2 may be stronger. Moreover, of the other three energies that were examined, the van der Waals force was significantly greater for both of the complexes. These findings suggest that binding is primarily driven by the van der Waals force and shape complementarity.
Complex interactions analysis
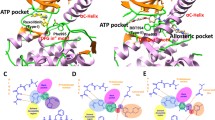
Non-covalent bond interactions, such as hydrogen bonds, hydrophobic interactions, and salt bridges, play a crucial role in stabilizing the spatial conformation of a protein. These interactions are essential for the biological functions of proteins and for the interactions between proteins and ligands. Thus, the mode of interaction between JAK2 and two inhibitors (CD4 and Ruxolitinib) was analyzed using PLIP [43]. These structures revealed that both CD4 and Ruxolitinib can interact with amino acid residues in the ATP pocket of JAK2, thus confirming that JAK2 active-site binding for both inhibitors. Furthermore, the structures also show that hydrophobic interactions formed between the inhibitors and JAK2 are also crucial for ligand–receptor binding. In CD4, the benzothiazole structure extends deeply into the ATP pocket and forms robust hydrophobic interactions with the Leu855 residue in the P-loop region and the main chain Leu983 residue. The core structures of Ruxolitinib, pyrrolopyrimidine and a five-membered carbon ring, form hydrophobic interactions with residues Val863, Ala880, and Leu983.
Additionally, the ligand binding stability is also reliant on hydrogen bond formation. In CD4, the main hydrogen bond is formed between the benzamide nitrogen atom Asp994 residue in the DFG motif. The DFG motif plays a crucial role in regulating kinase activity, and it has been previously shown that interaction with the DFG motif results in more potent inhibition of kinase activity [44]. Therefore, it is possible that CD4 inhibits kinase activity by interacting with the DFG motif. In the CD4 structure, the oxygen atom in the benzyloxy group forms a hydrogen bond with Lys882, creating a ‘water bridge’. This interaction strengthens the binding between the ligand and JAK2. In Ruxolitinib, three hydrogen bonds are formed between Ruxolitinib and specific regions of JAK2, namely the P-loop region Leu855, the hinge region Leu932, and the main chain Glu930. These hydrogen bonds serve to anchor Ruxolitinib in the ATP pocket, with each bond connecting to a different part of the pyrrolopyrimidine portion of the compound. Moreover, the hydrogen bond formation with the JAK2 hinge region Leu932 residue is a prevalent binding characteristic that is observed in most JAK2 ATP competitive inhibitors [45]. When comparing the two complex binding models, the CD4-JAK2 complex has one less hydrophobic interaction and one less hydrogen bond when compared to the Ruxolitinib–JAK2 complex. Furthermore, the interacting residues between CD4 and JAK2 are all different, with CD4 failing to form a hydrogen bond with the important hinge residue Leu932. In future, priority should be given to optimizing the structure of CD4 to enhance the inhibitory function of CD4 on the basis of this study, so as to strengthen the interaction between CD4 and the key residues in the JAK2 active site (Fig. 9).
Conclusions
Using various computer-aided drug design platforms, especially multitask CapsNet and molecular docking, we identified 49 candidate inhibitor compounds from a pool of 1.68 million small molecule compounds within the ChemDiv database. Following in vitro analysis of the 49 compounds, CD4 exhibited the most potent inhibitory effect on JAK2, as determined via an ADP-Glo™ kinase assay. Moreover, in HEL cells carrying the JAK2 V617F mutation, the effect of various CD4 concentrations on cell proliferation was determined using a CCK-8 assay and showed a successful inhibitory affect at an IC50 value of 32.57 μM. To examine potential similarities between CD4 and three marketed JAK2 inhibitors, Tanimoto coefficients were determined and showed that the structure of CD4 is dissimilar from the others and it possessed a unique framework. Moreover, CD4 was found to have a high JAK2 inhibitor activity and a low toxicity. Finally, a molecular dynamics simulation showed that the CD4-JAK2 complex is stable but does not have as low of a binding free energy as Ruxolitinib. Overall, these findings provide a reliable theoretical basis for the subsequent optimization of the chemical structure of CD4 as a means to improve its inhibitory activity and increase its potential as MPNs therapeutic agent.
References
Grinfeld J, Nangalia J, Green AR (2017) Molecular determinants of pathogenesis and clinical phenotype in myeloproliferative neoplasms. Haematologica 102:7–17. https://doi.org/10.3324/haematol.2014.113845
Grinfeld J, Nangalia J, Baxter EJ et al (2018) Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med 379:1416–1430. https://doi.org/10.1056/NEJMoa1716614
Silvennoinen O, Hubbard SR (2015) Molecular insights into regulation of JAK2 in myeloproliferative neoplasms. Blood 125:3388–3392. https://doi.org/10.1182/blood-2015-01-621110
Bose P, Masarova L, Verstovsek S (2020) Novel concepts of treatment for patients with myelofibrosis and related neoplasms. Cancers. https://doi.org/10.3390/cancers12102891
Samuelson Bannow BT, Salit RB, Storer BE et al (2018) Hematopoietic cell transplantation for myelofibrosis: the dynamic international prognostic scoring system plus risk predicts post-transplant outcomes. Biol Blood Marrow Transplant 24:386–392. https://doi.org/10.1016/j.bbmt.2017.09.016
Guglielmelli P, Vannucchi AM (2020) Current management strategies for polycythemia vera and essential thrombocythemia. Blood Rev 42:100714. https://doi.org/10.1016/j.blre.2020.100714
Tefferi A (2018) Primary myelofibrosis: 2019 update on diagnosis, risk-stratification and management. Am J Hematol 93:1551–1560. https://doi.org/10.1002/ajh.25230
Greenfield G, McMullin MF, Mills K (2021) Molecular pathogenesis of the myeloproliferative neoplasms. J Hematol Oncol 14:103. https://doi.org/10.1186/s13045-021-01116-z
How J, Hobbs GS, Mullally A (2019) Mutant calreticulin in myeloproliferative neoplasms. Blood 134:2242–2248. https://doi.org/10.1182/blood.2019000622
Li B, Rampal RK, Xiao Z (2019) Targeted therapies for myeloproliferative neoplasms. Biomarker Res. https://doi.org/10.1186/s40364-019-0166-y
Helbig G (2018) Classical philadelphia-negative myeloproliferative neoplasms: focus on mutations and JAK2 inhibitors. Med Oncol 35:119. https://doi.org/10.1007/s12032-018-1187-3
Torres DG, Paes J, da Costa AG et al (2022) JAK2 variant signaling: genetic, hematologic and immune implication in chronic myeloproliferative neoplasms. Biomolecules. https://doi.org/10.3390/biom12020291
Lph Y, Gm K (2012) Ruxolitinib in the treatment of myelofibrosis. Drugs 72:2117–2127
Talpaz M, Kiladjian JJ (2021) Fedratinib, a newly approved treatment for patients with myeloproliferative neoplasm-associated myelofibrosis. Leukemia 35:1–17. https://doi.org/10.1038/s41375-020-0954-2
Lamb YN (2022) Pacritinib: first approval. Drugs 82:831–838. https://doi.org/10.1007/s40265-022-01718-y
McLornan DP, Pope JE, Gotlib J et al (2021) Current and future status of JAK inhibitors. Lancet 398:803–816. https://doi.org/10.1016/S0140-6736(21)00438-4
Lavecchia A (2019) Deep learning in drug discovery: opportunities, challenges and future prospects. Drug Discov Today 24:2017–2032. https://doi.org/10.1016/j.drudis.2019.07.006
Vamathevan J, Clark D, Czodrowski P et al (2019) Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 18:463–477. https://doi.org/10.1038/s41573-019-0024-5
Schneider P, Walters WP, Plowright AT et al (2019) Rethinking drug design in the artificial intelligence era. Nat Rev Drug Discov 19:353–364. https://doi.org/10.1038/s41573-019-0050-3
Moingeon P, Kuenemann M, Guedj M (2022) Artificial intelligence-enhanced drug design and development: toward a computational precision medicine. Drug Discov Today 27:215–222. https://doi.org/10.1016/j.drudis.2021.09.006
Sabour S, Frosst N, Hinton G (2017) Dynamic routing between capsules. Comput Vision Pattern Recognit. https://doi.org/10.48550/arXiv.1710.09829
Wang Y, Huang L, Jiang S et al (2019) Capsule networks showed excellent performance in the classification of hERG blockers/nonblockers. Front Pharmacol 10:1631. https://doi.org/10.3389/fphar.2019.01631
Wang YW, Huang L, Jiang SW et al (2020) CapsCarcino: a novel sparse data deep learning tool for predicting carcinogens. Food Chem Toxicol 135:110921. https://doi.org/10.1016/j.fct.2019.110921
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32:1466–1474. https://doi.org/10.1002/jcc.21707
Wang Y, Wang B, Jiang J et al (2021) Multitask CapsNet: an imbalanced data deep learning method for predicting toxicants. ACS Omega 6:26545–26555. https://doi.org/10.1021/acsomega.1c03842
Davis RR, Li B, Yun SY et al (2021) Structural insights into JAK2 inhibition by Ruxolitinib, Fedratinib, and derivatives thereof. J Med Chem 64:2228–2241. https://doi.org/10.1021/acs.jmedchem.0c01952
Palandri F, Bartoletti D, Iurlo A et al (2022) Peripheral blasts are associated with responses to ruxolitinib and outcomes in patients with chronic-phase myelofibrosis. Cancer 128:2449–2454. https://doi.org/10.1002/cncr.34216
Malolepsza E, Strodel B, Khalili M et al (2010) Symmetrization of the AMBER and CHARMM force fields. J Comput Chem 31:1402–1409. https://doi.org/10.1002/jcc.21425
Xiong G, Wu Z, Yi J et al (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49:W5–W14. https://doi.org/10.1093/nar/gkab255
Baell J, Holloway G (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740. https://doi.org/10.1021/jm901137j
Saubern S, Guha R, Baell JB (2011) KNIME workflow to assess PAINS filters in SMARTS format. Comparison of RDKit and indigo cheminformatics libraries. Mol Inform 30:847–850. https://doi.org/10.1002/minf.201100076
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. https://doi.org/10.1038/srep42717
Bickerton GR, Paolini GV, Besnard J et al (2012) Quantifying the chemical beauty of drugs. Nat Chem 4:90–98. https://doi.org/10.1038/nchem.1243
Van Der Spoel D, Lindahl E, Hess B et al (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Jorgensen WL, Chandrasekhar J, Madura JD et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. https://doi.org/10.1063/1.445869
Kumari R, Kumar R, Open Source Drug Discovery C et al (2014) g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Wang E, Sun H, Wang J et al (2019) End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev 119:9478–9508. https://doi.org/10.1021/acs.chemrev.9b00055
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10:1–13
Hopkins AL, Keserü GM, Leeson PD et al (2014) The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov 13:105–121. https://doi.org/10.1038/nrd4163
Schultes S, de Graaf C, Haaksma EEJ et al (2010) Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discov Today Technol 7:e157–e162. https://doi.org/10.1016/j.ddtec.2010.11.003
Butina D (1999) Unsupervised data base clustering based on daylight’s fingerprint and Tanimoto similarity: a fast and automated way to cluster small and large data sets. J Chem Inf Comput Sci 39:747–750. https://doi.org/10.1021/ci9803381
Adasme MF, Linnemann KL, Bolz SN et al (2021) PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res 49:W530–W534. https://doi.org/10.1093/nar/gkab294
Peng YH, Shiao HY, Tu CH et al (2013) Protein kinase inhibitor design by targeting the Asp-Phe-Gly (DFG) motif: the role of the DFG motif in the design of epidermal growth factor receptor inhibitors. J Med Chem 56:3889–3903. https://doi.org/10.1021/jm400072p
Dymock BW, Yang EG, Chu-Farseeva Y et al (2014) Selective JAK inhibitors. Future Med Chem 6:1439–1471. https://doi.org/10.4155/fmc.14.92
Acknowledgements
Financial support from Sichuan Science and Technology Program of China (2023NSFSC1692 and 2022YFS0614), Luzhou Science and Technology Program of China (2022-SYF-46), and Southwest Medical University Applied Basic Research Program Project (2021ZKMS006).
Funding
Funding was provided by Southwest Medical University (Grant No. 2021ZKMS006), Sichuan Provincial Department of Science and Technology (Grant Nos. 2022YFS0614, 2023NSFSC1692). Luzhou Science and Technology Bureau (Grant No. 2022-SYF-46).
Author information
Authors and Affiliations
Contributions
JW and YW contributed to the study’s conception and design. BW conducted the experiments and wrote the first draft of the manuscript. BC and YJ conducted a portion of the biological validation experiments, while JG and JW were involved in the data analysis. YW provided supervision throughout the process and also revised the manuscript. Additionally, all authors participated in reviewing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, B., Guo, J., Chen, B. et al. Combination of ligand‑based and structure‑based virtual screening for the discovery of novel Janus kinase 2 inhibitors against philadelphia-negative myeloproliferative neoplasms. Mol Divers (2024). https://doi.org/10.1007/s11030-024-10938-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-024-10938-1