Abstract
Visceral Leishmaniasis (VL), the second neglected tropical disease caused by various Leishmania species, presents a significant public health challenge due to limited treatment options and the absence of vaccines. The agent responsible for visceral leishmaniasis, also referred to as “black fever” in India, is Leishmania donovani. This study focuses on L. donovani Minichromosome maintenance 10 (LdMcm10), a crucial protein in the DNA replication machinery, as a potential therapeutic target in Leishmania therapy using in silico and in vitro approaches. We employed bioinformatics tools, molecular docking, and molecular dynamics simulations to predict potential inhibitors against the target protein. The research revealed that the target protein lacks homologues in the host, emphasizing its potential as a drug target. Ligands from the DrugBank database were screened against LdMcm10 using PyRx software. The top three compounds, namely suramin, vapreotide, and pasireotide, exhibiting the best docking scores, underwent further investigation through molecular dynamic simulation and in vitro analysis. The observed structural dynamics suggested that LdMcm10-ligand complexes maintained consistent binding throughout the 300 ns simulation period, with minimal variations in their backbone. These findings suggest that these three compounds hold promise as potential lead compounds for developing new drugs against leishmaniasis. In vitro experiments also demonstrated a dose-dependent reduction in L. donovani viability for suramin, vapreotide, and pasireotide, with computed IC50 values providing quantitative metrics of their anti-leishmanial efficacy. The research offers a comprehensive understanding of LdMcm10 as a drug target and provides a foundation for further investigations and clinical exploration, ultimately advancing drug discovery strategies for leishmaniasis treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Visceral Leishmaniasis (VL), also referred to as Kala-azar, is a neglected tropical disease affecting approximately 500,000 individuals worldwide, with over 90% of cases concentrated in socioeconomically challenged regions, including Brazil, Ethiopia, India, Somalia, and Sudan [1]. The causative agent, Leishmania donovani, transmitted by phlebotomine sand flies, necessitates prompt clinical interventions to avert near-certain fatality [2]. Despite a recent decline in VL incidence in India and Bangladesh attributed to the widespread use of insecticide-treated nets, the epidemiological status in other regions, notably Brazil, remains concerning [3]. In Brazil, urbanization has facilitated the spread of VL, intensifying the public health threat. The emergence of Leishmanial strains resistant to conventional antimonial drugs, the primary frontline defence against VL, as well as their substitutes like Amphotericin B and Miltefosine, further complicates the situation [4, 5]. Current therapeutic options for VL exhibit notable limitations, encompassing severe side effects, restricted applicability to hospital settings, and financial constraints [4]. Addressing these challenges, targeting specific proteins within the L. donovani parasite has gained prominence in drug development. Numerous studies have elucidated that essential proteins are pivotal for the parasite’s survival, replication, and evasion of host immune responses [6, 7]. The rationale for focussing on such proteins lies in the design of drugs that selectively disrupt these essential processes, thereby impeding the parasite’s life cycle. The systematic identification and validation of these proteins as drug targets underscore their significance in advancing therapeutic interventions against VL. Mcm10, a multifunctional protein intricately involved in eukaryotic DNA replication [8], has been recognized as a critical DNA replication protein and a candidate for drug targeting in cancer [9].
Mcm10 consistently engages with DNA, regardless of topological or sequence considerations [10, 11]. Its binding affinities for both single-stranded (ss) and double-stranded (ds) DNA are comparable, with the N-terminal domain (NTD) facilitating protein oligomerization despite modest DNA binding capability [12]. The Mcm10 internal domain (Mcm10-ID) exhibits pronounced sequence homology across diverse species [10]. Crystallographic investigations reveal a unique arrangement of the OB-fold and ZnF domains in xMcm10-ID, presenting a configuration distinct from other DNA binding proteins [10]. This domain tandem forms a continuous DNA binding surface in Mcm10, encompassing specific residues identified through nuclear magnetic resonance chemical shift perturbation, involving a patch of basic lysines and aromatic amino acids that make direct contact with DNA [11]. Despite the commonality of ZnF domains in various proteins, the likelihood of small molecules binding to them and yielding selective drugs is limited. However, it is noteworthy that the C-terminal domain (CTD) ZnF significantly augments the DNA binding affinity of full-length Mcm10 [13].
Consequently, emphasizing the Mcm10 internal domain as the prime target emerges as a judicious strategy for developing specific inhibitors. As there is currently no available vaccine for leishmaniasis, and existing research on the Leishmania parasite lacks insights into potent antigens or immunogens, our focus centres on the internal domain of L. donovani Mcm10 (LdMcm10) as a distinct therapeutic target. Inhibitors targeting LdMcm10 hold promise as robust anti-leishmanial medications for clinical use, addressing leishmaniasis and alleviating associated symptoms. The potential of Mcm10 to sustain DNA replication is critical for all eukaryotes’ survival and existence. The viability of Mcm10 as a possible drug target is based on the fact that Mcm10 is involved in DNA replication regulation, repair mechanisms, and cell proliferation in eukaryotes [11]. As a result, in silico screening was performed against LdMcm10 using drug-like compounds from the DrugBank database.
Using computational techniques in computer-assisted drug design has become a practical approach for exploring new lead compounds [14]. Over the past two decades, academics have created and utilized various computational techniques to enhance efficiency and reduce expenses. Therefore, we have employed virtual screening (VS) and computer simulations to identify lead compounds against LdMcm10 in this study.
Methods
Target selection, molecular modelling, and model validation
Before performing molecular docking, the structure of the target protein was constructed. The sequence of the target protein LdMcm10 was obtained from the KEGG database (https://www.genome.jp/entry)(LDBPK_262410). The received sequence was subsequently cross-verified using UniProt with the accession number A0A3S7X073. To determine the degree of similarity with human homologs, a similarity search was conducted against the human genome using the protein Blast programme available at https://blast.ncbi.nlm.nih.gov/Blast.cgi, and the internal domain of the selected protein was used as the target sequence for homology modelling. Due to the absence of template structures with an identity percentage below 28% for homology modelling of the LdMcm10 internal domain (LdMcm10 ID) sequence, alternative methods such as ab initio and threading were explored to construct a three-dimensional model. RosetaFold, available at http://robetta.bakerlab.org [15], was employed as an online platform offering various computational tools for predicting and analysing protein structures.
Additionally, predictions for the 3D structure of the LdMcm10 ID protein were generated using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [16], ModWeb (https://modbase.compbio.ucsf) [17], and AlphaFold (https://alphafold.ebi.ac.uk/) [18] web servers. Evaluation of the protein’s integrity is crucial to the protein structure prediction procedure. Using the PROCHECK server [19], the accuracy and precision of the model evaluations were determined by Ramachandran plot analysis, which comprises the number of residues in the most favoured, additionally allowed, generously allowed, and disallowed regions and the ERRAT score. The model exhibiting the most favourable PROCHECK score underwent energy minimization for enhanced structural stability, achieved through the YASARA Energy Minimization server [20]. Subsequent validation of the refined model was conducted using PROCHECK [19] to assess the authenticity and structural quality of the LdMcm10 rigorously.
Ligand library preparation
The Drug Bank database (https://www.drugbank.ca/) offered access to a molecular library of nearly 1000 approved molecules in this study. Renowned for reliability in bioinformatics and cheminformatics, Drug Bank is a valuable resource for virtual screening, providing detailed information on drugs, including specific data on targets such as sequence, structure, and pathways. Canonical smiles from Drug Bank (https://www.drugbank.ca/) were utilized for molecular docking, and subsequent conversion to PDB files was achieved using CORINA 3D software (https://www.mn-am.com/online_demos/corina_demo).
Molecular docking
Computational drug design relies heavily on molecular docking, which guarantees that the ligand molecule will fit into the binding pocket of the chosen protein in the correct configuration. Docking investigations in the present study used the PyRx virtual screening programme [21]. PyRx uses the docking applications autodock four and autodock vina. A grid box was built with the following dimensions in X, Y, and Z (120.6361, 115.6029, 116.640) and (26.0286, 44.5394, 43.1279). Almost every residue in the active site is contained inside the grid box. We chose to investigate compounds with the weakest binding energy. Hydrogen bond interactions were studied with LigPlot [22] and visualized in PyMol [23] for docked protein–ligand structures.
Molecular dynamic simulation
Molecular dynamics simulations were executed to prognosticate the ligand binding dynamics with the target protein within a physiological milieu. The docked complex involving the protein target and ligand molecules underwent a 300 ns simulation utilizing Desmond 2020.1 from Schrodinger, LLC [24]. The primary objectives encompassed delineating dynamic behaviour, exploring intermolecular interactions, and assessing the complex’s stability [25]. Employing an orthorhombic simulation box, the complex underwent solvation using the system builder platform with the simple point-charge (SPC) explicit water model. The solvated complex system, established with a salt concentration of 0.15 M, integrated an appropriate number of Na + /Cl − counter ions, maintaining a constant temperature of 300 K and 1 atm pressure throughout the simulation period. The receptor-ligand complex system adopted the OPLS-2005 force field, and a comprehensive solvent model featuring SPC water molecules was implemented in the system [26, 27]. Trajectories were sampled at 300 ps intervals for analysis, with the stability of the protein–ligand complex gauged through metrics such as Root Mean Square Deviation (RMSD), radius of gyration (Rg), root mean square fluctuation (RMSF), and the temporal evolution of hydrogen bonds (H-bonds).
MM-GBSA calculation
We utilized the binding free energy as a strict metric to characterize the binding affinity inside the protein–ligand complex objectively. We examined the binding free energies for LdMcm10 ID and its ligands using the molecular mechanics-generalized born surface area (MM-GBSA) method [26, 27]. The complex calculations were made more accessible by the mmgbsa.py Python script included in the prime module. We calculated the binding free energy (ΔGbind) using the VSGB solvent model, the OPLS 2005 force field, and sophisticated rotamer search methods [28]. To estimate the binding free energy controlling the interaction between ligands and the LdMcm10 ID receptor within the system, use the formula
Essential dynamics (ED) analysis
To elucidate the global motions occurring during the 300-ns simulation of ligand complexes with LdMcm10 ID, we employed principal component analysis (PCA) [29, 30]. Initiated by constructing a covariance matrix, the subsequent PCA computation allowed for extracting fundamental conformational dynamics. The exploration of protein–ligand complex conformational dynamics involved computing the movements of trajectories focussing on ten alternate conformational modes of the major component, with particular attention to comparing the highest mode (PC 2). Throughout the 300-ns molecular dynamics simulation for the protein–ligand complex, we constructed a dynamic cross-correlation matrix (DCCM) encompassing all carbon atoms. This DCCM facilitated an in-depth investigation of domain correlations [31].
In vitro cell-cytotoxicity assay
The cytotoxicity assay utilized the MTT assay [32, 33] against L. donovani (Ld1S) promastigote. In this study, promastigotes were cultivated in Ti75 tissue culture flasks (Thermo Fisher Scientific) and maintained at 25 °C within an incubator. Subsequently, the cells were placed in separate wells of a 96-well microtiter plate (Thermo Fisher Scientific) containing 100 μl of M199 medium. The final cell density in each well was 2 × 106 cells/well. The test compounds, along with amphotericin B utilized as the positive control, were dissolved in a 0.1% dimethyl sulfoxide (DMSO) solution, and the final concentration was made up to (1–20) µM/ml. The cells were treated with Amphotericin B as positive control after an overnight incubation period. In contrast, untreated cells were utilized as the negative control, and cells treated with different concentrations of Suramin, Pasireotide, and Vapreotide were employed as the test sample and kept for overnight incubation at 25 °C. All experiments were conducted in triplicate.
After the incubation period, the cells in the microtiter plate were centrifuged using a Sigma 3–30 K centrifuge at a speed of 4500 revolutions per minute for 45 min. The culture media were then removed, resulting in the formation of a pellet consisting of the cells. Following the process of centrifugation, a volume of 20 μl of MTT reagent with a concentration of 5 mg/ml was introduced into each well. Subsequently, the plate was incubated in the dark for 4 h at 25 °C. Subsequently, the MTT was removed by washing, and the formazan crystals were dissolved in 100 μl of dimethyl sulfoxide (DMSO) per well. The measurement of MTT reduction was conducted by recording the absorbance at a wavelength of 570 nm using a microtiter plate reader. All the reagents were purchased from Thermo Fisher Scientific -IN.
Results
Target selection and homology modelling
The LdMcm10 protein’s amino acid sequence was taken from the KEGG genome database. Protein BLAST confirmed that LdMcm10 does not have any empirically established structures. The protein blast analysis against human Mcm10 yielded insignificant sequence similarity with the target protein. Additionally, superimposing the protein structure onto the human Mcm10 structure revealed a substantial Root Mean Square Deviation (RMSD) of 40.754 Å (Supplementary Fig. 8).
The LdMcm10 protein shares only 28% of the sequence similarities with the internal domain of Xenopus laevis Mcm10 (PDB 3EBE_A), suggesting the need to apply ab initio and threading approaches for model building. Without an appropriate homology modelling template, various web servers were utilized to generate the best 3D model for LdMcm10 ID (Table 1). The model obtained from the Robetta web server was chosen for subsequent investigation, considering its favourable Z score and Ramachandran plot analysis (Supplementary Fig. 1A and B).
Energy minimization was performed using the YASARA Energy minimization server to yield a highly stable protein structure, which was subsequently validated with PROCHECK. Additional ProSa and ERRAT were used to analyse the LdMcm10 ID protein’s integrity and structural quality (Supplementary Fig. 2 A–C).
Prediction of binding site residues
Previous research suggests that multiple sequence alignment (MSA) analysis can accurately identify binding locations [34]. The MSA was utilized to search for conserved LdMcm10 ID amino acid residues, which influence how a protein interacts [35] (Supplementary Fig. 3 B).
Further binding site residues were verified using the https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi server. The amino acid residues from 339–381 were identical to the zinc finger domain of yeast Mcm10 proteins and DnaG-type primases (Supplementary Fig. 3 C). This identified region is deemed a potential binding site for ligands and aligns concordantly with the conserved amino acid residues delineated in the MSA. According to previous studies on X. laevis, the internal domain of the Mcm10 protein offers an entirely novel ssDNA binding platform [36]. However, an earlier study in budding yeast Mcm10 shows that mutation in conserved residues hampers the DNA binding activity of Mcm10 and affects the DNA replication process [37].
In addition, the CASTp web server was used to predict binding sites and select the most likely binding site. CASTp identified 75 possible binding sites; however, binding pockets 2, 3,31 and 6 were selected for our work (Supplementary Table 1). In contrast, the study did not consider those pockets because ligands cannot fit in pockets with limited surface area and volumes [38]. Most of the conserved amino acid residues identified by MSA were inside the anticipated protein model’s selected binding area (supplementary Fig. 3A).
Structure-based virtual screening
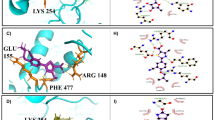
We screened a 1000-compound library against LdMcm10 and identified three lead compounds with favourable docking scores. Vapreotide (DB04894) exhibited a substantial docking score of − 11.3 kcal/mol, characterized by 19 hydrophobic interactions with amino acids. Additionally, it established seven hydrogen bonds with Gln 426, Ser 349, His 320, Ala 342, and Ser 401 within binding pockets 2, 3, and 6. Suramin (DB04786) exhibited a robust interaction profile, with a docking score of − 10.8 kcal/mol featuring twelve hydrophobic interactions with amino acid residues in binding pockets 2, 3, and 6. It formed eight hydrogen bonds with Ser 401, Arg 313, Ser 316, Ala 315, Thr 346, Ala 341, Gln 368, and Asn 363. Meanwhile, Pasireotide (DB06663) showed a docking score of -9.8 kcal/mol for the target protein. This compound engaged in fifteen hydrophobic interactions within pockets 2, 3, and 6, along with six hydrogen bonds involving Ser 403, Asn 362, Ser 349, Tyr 352, and Ala 315. A comprehensive depiction of all hydrogen bonds and hydrophobic interactions is delineated in Table 2. All three lead molecules interact with amino acid residues in the protein’s internal domain (LdMcm10), essential for ss DNA binding [37]. Hence, these specific ligand molecules and the corresponding protein–ligand complexes were selected for molecular dynamics (MD) simulation and subsequent in vitro studies.
Mcm10, a protein involved in DNA replication, serves as a scaffold protein and coordinator for different proteins in the replication process. Unlike an enzyme with a specific substrate, Mcm10 interacts with other proteins and complexes essential for DNA replication, enabling the replication process to proceed smoothly [11]. Along with other replisomes, Mcm10 was found to interact with DNA polymerase α, PCNA, and single-stranded DNA [11]. Therefore, we also docked DNA polymerase α, PCNA, and single-stranded DNA as positive control against Mcm10. Docking interaction and analysis with the help of HADDOCK 2.4 server exhibits Z score of − 2, 0, & 2 for DNA polymerase α, PCNA, and single-stranded DNA, respectively. Further 2D and 3D interaction analysis revealed DNA polymerase α, PCNA, and single-stranded DNA formed H-bonds with residues Arg 357, Arg 382, Lys 379, Ser 383, Glu 390, Glu 356, Tyr 352, Thr 386 (Supplementary Fig. 7 A–F) suggesting that these are potential amino acid residues involved in interaction with other replisome proteins. Thus, selected drugs might also utilize these amino acid residues to hinder protein’s function.
The generation of three-dimensional (3D) and two-dimensional (2D) protein–ligand interaction graphs was accomplished using PyMol and LigPlot software. These graphical representations visually depict hydrogen bond interactions between amino acid residues within the active site and the chosen ligand molecules, as illustrated in Figs. 1 and 2.
LigPlot 2D visualizations show the interactions between active site residues and the top ligands Vapreotide (A), Suramin (B), and Pasireotide (C). Hydrogen bonds are shown as green dashed lines with distances (Å), whereas hydrophobic interactions are represented by red arcs. Carbon (black), nitrogen (blue), and oxygen (red) are the three atomic hues
Structural dynamics
A molecular docking study reveals that suramin, pasireotide, and vapreotide had the strongest binding affinity with the target protein LdMcm10 ID. However, the chemical interactions between proteins and ligands are crucial to the structural stability of a protein–ligand complex. However, it also depends on the simulated environment [39, 40]. Recent investigations indicate that drugs with high binding affinity and docking scores may have escaped the binding pocket [41, 42]. Thus, it is crucial to study the conformational changes. In order to understand the role of ligands in protein dynamics, MD simulation for 300 ns was run for protein–ligand docked complexes along with the apo structure of LdMcm10 ID to have a detailed understanding of the conformational changes in the protein.
Root mean square fluctuation (RMSD) analysis
RMSD quantifies the protein–ligand complex’s average atom departure from the original frame after a defined time. The calculation assessed complex stability over time for each simulation trajectory frame. Figure 3A represents the RMSD development of the protein and protein–ligand complexes. When compared to the structure of the protein in its apo state, the flexibility of the protein is decreased when it is bound to a ligand. In LdMcm10ID-vapreotide, an initial rise in RMSD was seen up to 50 ns. However, after that, there was a decline with an average RMSD of 7.02 Å. The results remained steady throughout the simulation, suggesting that the ligand stayed connected to the protein’s binding pocket [43, 44]. From 50 ns onwards, the average RMSD for the LdMcm10ID-suramin complex was 5.7 Å, with only a modest drift of 0.2 Å to 0.5 Å.
The 300 ns molecular dynamic simulation results of top three protein–ligand complexes (LdMCM10-Vapreotide shown in yellow, LdMCM10-Suramin shown in green, LdMCM10-Pasireotide shown in blue, and LdMCM10 shown in red). A RMSD value of carbon alpha atoms of the complex. B RMSF of carbon alpha of complex structure. C Rg of backbone atoms. D SASA of the ligands. E Total number of H-bonds between ligands and Protein. F The binding free energy of LdMCM10 inhibitors has been assessed through the MM-GBSA approach
On the other hand, it was discovered that LdMcm10ID-pasireotide fluctuated with an average value of 6.8 Å from 150 ns to the end. In contrast, many drifts were seen during the first initial phase of the simulation. These conformational flips are caused mainly by the ligand’s torsion angle [44], which suggests that the observed change may be attributable to a conformational shift in the ligand’s rotatable bonds. The drug binding provided stability to the LdMcm10 protein.
Root mean square fluctuation (RMSF) analysis
In order to observe the local fluctuation in protein structure [45], the RMSF of each amino acid residue of LdMcm10 ID that interacted with the ligands were calculated for 300 ns (Fig. 3B). The figure indicates that conformation was maintained throughout the simulation. In LdMcm10ID-vapreotide, the average RMSF value was below 2.10 Å. However, some amino acid residues, such as Ser 319 (0.882 Å), Ala 335 (0.834 Å), Arg 337 (0.888 Å), Gln 338 (0.982 Å), Gln 339 (0.986 Å), Val 341 (0.89 Å), Leu 342 (0.758 Å), and Tyr 348 (0.861 Å), had lower RMSF values, indicating tight ligand binding within the binding pocket (Fig. 3) [46]. LdMcm10ID-Suramin had an average RMSF of 2.77 Å, whereas LdMcm10ID- pasireotide had 3.10 Å. The study on RMSF demonstrates that the stability of molecular connections between ligands and LdMcm10 ID in complexes exhibits little fluctuations. However, stable secondary structures like alpha-helices and beta-sheets stay the same throughout the simulation. The primary cause for the fluctuations is that the protein’s N– and C– ends have loops and low-stable beta-sheets [47].
Secondary structure analysis
The simulation predicted the protein’s secondary structure by plotting alpha-helices and beta-strands against the residual index. LdMcm10ID-Suramin has 3.87% alpha-helices and 31.6% beta-sheets, compared to 5.95% and 26.37% in LdMcm10ID-pasireotide and 0.85% and 33.73% in LdMcm10ID-vapreotide (Supplementary Fig. 4). The alpha–beta helices-sheet ratio impacts protein RMSD. Protein residues in rigid structures have lower RMSD than coils and loops [48, 49].
Radius of Gyration analysis
To assess LdMcm10ID-ligand-bound complex structural compactness and stability, the radius of gyration (Rg) is studied [50]. Figure 3C displays the Rg of all protein–ligand complexes and apoprotein. The Rg figure for LdMcm10ID-vapreotide indicates that the structure stabilized around 16.24 Å throughout the simulation, with modest drifts of roughly 2 Å in the first 50 ns, indicating changes in structure. However, the structure remained stable after 50 ns, suggesting the structural compactness of these complexes throughout the simulation. The Rg figure for LdMcm10ID-pasireotide shows a little trajectory increase in the first half of the simulation, but this drift reduces, and the structure stabilizes. After 150 ns, the simulation stays constant with an average value of 16.94 Å for 300 ns. Compared to LdMcm10ID-vapreotide and LdMcm10ID-pasireotide, LdMcm10ID-Suramin exhibits moderate perturbation with an average value of 17.27 Å, indicating less stable structural compactness and integrity.
Solvent-accessible surface (SASA) analysis
The solvent-accessible surface area (SASA) is a metric quantifying the conformational changes induced by the ligand throughout the simulation. SASA data are crucial in discerning whether the ligand remains within the binding pocket or is expelled from the binding cavity [51]. SASA was calculated for both LdMcm10 ID (apo state) and LdMcm10ID-ligand complexes to analyse the behavioural changes. The average SASA values for LdMcm10 ID (apo state) and its complexes with suramin, pasireotide, and vapreotide were determined as 9296.05 Å2, 8959.2 Å2, 9265.98 Å2, and 9241.76 Å2, respectively. Suramin and vapreotide complexes with LdMcm10ID exhibited slightly higher SASA values than the LdMcm10 ID (apo form) (Fig. 3D). Conversely, the pasireotide complexes displayed a lower SASA pattern than the apo form. Elevated and relatively stable SASA values in a protein–ligand complex indicate that the ligand can interact with solvents without causing significant structural alterations to the protein [52].
H-bond analysis
The overall stability of the protein–ligand complex is intricately influenced by numerous hydrogen bond (H-bond) interactions occurring within the active site of the protein. In their respective bound states, pasireotide, suramin, and vapreotide exhibit multiple H-bond acceptors and donor sites that directly interact with the predicted binding sites of LdMcm10ID. The temporal evolution of H-bond interactions in these three complexes over the 300 ns simulation is illustrated in Fig. 3E. Figure 3E depicts an average of five, four, and three H-bond contacts in the LdMcm10ID-pasireotide, LdMcm10ID-Suramin, and LdMcm10ID-vapreotide complexes throughout the simulation period. Notably, the number of H-bond interactions exhibits fluctuations. The presence of robust H-bonds between the ligands and LdMcm10 ID implies the formation of enduring protein–ligand complexes, underscoring their structural stability.
This study evaluated the hydrogen bond interactions between Mcm10 and DNA polymerase α, PCNA, and single-stranded DNA (ssDNA) as a positive control. Through a meticulous 50 ns simulation, we observed a substantial formation of hydrogen bonds within the Mcm10-PCNA, Mcm10-DNA polymerase α, and Mcm10-ssDNA complexes. The Mcm10-ssDNA complex displayed approximately 8–10 hydrogen bonds, whereas interactions between Mcm10 and DNA polymerase α and Mcm10 and PCNA exhibited a range of 4–6 and 2–4 hydrogen bonds, respectively. (Fig. 4D). These findings elucidate the intricate and dynamic nature of protein–protein and protein-DNA interactions, affirming their significance throughout the simulation.
50 ns molecular dynamic simulation findings for the three protein–substrate complexes (LdMCM10 with three substrates i.e. ssDNA, PCNA, and DNA pol α). A RMSD value of carbon alpha atoms of the complex. B RMSF of carbon alpha of complex structure. C Rg of backbone atoms. D Total number of H-bonds between ligands and Protein. E SASA of the ligands
Additionally, we analyse the structural stability and compactness of the protein–protein and protein–DNA complexes through rigorous analysis of RMSD, RMSF, the radius of gyration, and solvent-accessible surface area (SASA). These metrics serve as indicators of the stability and dynamic behaviour of the complexes over time, as illustrated in (Fig. 4 A, B, C, E).
The RMSD values for Mcm10 remained consistently stable throughout the simulations. Notably, when Mcm10 formed complexes with other replisome proteins and DNA, its backbone exhibited heightened stability, as indicated by minimal RMSD values. Moreover, the binding of PCNA and ssDNA to Mcm10 did not induce substantial alterations in its backbone RMSD. This finding suggests that Mcm10 maintains a stable backbone conformation when interacting with these proteins and DNA entities, which is crucial for its functional role within the replisome machinery. Similarly, a notable decrease in Root Mean Square Fluctuation (RMSF) values was observed upon the binding of Mcm10 with PCNA, DNA polymerase α, and ssDNA, compared to its unbound form (Apo-Mcm10). This reduction signifies increased stability of the protein–protein and protein–DNA complexes. Furthermore, analysis of the Radius of Gyration (Rg) and Solvent Accessible Surface Area (SASA) values for these complexes revealed consistent and persistent interactions between the protein and DNA components throughout the simulation period. These findings highlight the robustness and compact nature of the formed complexes, indicating their functional significance in replication machinery.
Ligand dynamics
To understand the role of ligands on protein binding affinities throughout simulations. Total seven properties of ligand, including (1) RMSD properties: Root mean square deviation of a ligand from the reference conformation (usually the first frame at time t = 0); (2) Radius of gyration (rGyr): Measures ligand “extendedness” and is comparable to its primary moment of inertia; (3) intramolecular hydrogen bond (intraHB): Number of ligand molecule internal HB. Calculated molecular surface area (MolSA) using 1.4 A probe radius; 5. Solvent-accessible surface area (SASA): Water-accessible molecular surface area; (6) Polar surface area (PSA): A molecule’s solvent-accessible surface area is solely oxygen and nitrogen [53] was assessed to analyse ligand stability about the protein and binding pocket. Supplementary Fig. 5 A, B, and C displays the RMSD of suramin, pasireotide, and vapreotide values. Suramin maintained an average RMSD value of ~ 3.8 Å throughout the simulation, followed by vapreotide and pasireotide, with 5.09 Å and 5.8 Å. Suramin and vapreotide stay constant after 100 ns of simulation, unlike pasireotide. Although there is a minor change between 50 and 90 ns, suramin remains stable and compact with an average rGyr value of 10.90 Å throughout the simulation. Despite slight deviations, vapreotide maintains compactness throughout the simulation, with an average rGyr value of 7.88 Å. Pasireotide had more rGyr variation than the other two compounds across the process. Finally, rGyr values show the “extendedness” or compactness of ligands in the binding pocket during simulation. Pasireotide transitioned from compact to stable extended conformation, suramin showed terminal stability followed by early oscillations, while vapreotide remained compact during 300 ns of simulation. The intra HB, MolSA, SASA, and PSA graphs showed the ligand’s character during the simulation. Suramin’s MolSA, SASA, and PSA graphs showed consistency throughout the simulation procedure, with just a few tiny drifts between 200 and 250 ns. MolSA, SASA, and PSA plots for pasireotide and vapreotide revealed inconsistency during the first simulation stage. Overall, there is a tendency for various properties to fluctuate at first, then become more consistent and steadier as the simulation progresses. These observations analyse the ligand’s behaviour and interaction pattern with protein (Supplementary Fig. 5 A, B, and C).
Binding free energy analysis
Protein–ligand complex stability is indicated by binding free energy [54]. The binding free energy for 300 ns of simulated trajectories was computed to measure stability. Electrostatic, covalent, hydrogen bonding, hydrophobic, Van der Waals, and polar solvation energies contributed to the binding free energy. MM-GBSA determined the total binding free energy of suramin, pasireotide, and vapreotide against the LdMcm10ID binding pocket. The binding free energy for LdMcm10ID-suramin, LdMcm10ID-pasireotide, and LdMcm10ID-vapreotide complexes was − 101.67 ± 4.12, − 93.72 ± 1.01, and − 96.39 ± 1.32, respectively (Supplementary Table 2). LdMcm10-suramin has the lowest binding free energy of the three complexes. Electrostatic binding and van der Waals energy contributed most to the interactions. H-bond interaction for LdMcm10-suramin, pasireotide, and vapreotide was − 5.99 ± 0.91, − 11.15 ± 2.11, and − 5.80 ± 1.87 kcal/mol (Supplementary Table 2). Figure 3F compares the energies at which the inhibitors suramin, pasireotide, and vapreotide bind to the binding pocket in the LdMcm10ID.
Essential dynamics and principal component analysis
Principal Component Analysis (PCA) is a robust method for characterizing protein dynamics within molecular simulations [55]. In this study, we applied PCA to scrutinize the conformational dynamics of LdMcm10 ID and its protein–ligand complexes. PCA scatter plots were constructed by projecting simulated trajectories onto the two-dimensional subspace defined by the first three eigenvectors (PC1, PC2, and PC3), with the colour spectrum representing the temporal evolution from blue (initiation) to white (intermediate) to red (termination). The outcomes of the PCA analysis for both the apoprotein and the protein–ligand complexes are shown in Supplementary Fig. 6A–D.
Comparative scrutiny of PCA plots for LdMcm10ID-pasireotide (Supplementary Fig. 6B), LdMcm10ID-suramin (Supplementary Fig. 6C), and LdMcm10ID-vapreotide (Supplementary Fig. 6D) underscores the pronounced variability within the PC1 cluster, contributing 35.08%, 55.88%, and 35.64% of the respective variances. The PC2 cluster manifests variabilities of 21.23%, 17.33%, and 13.58%, while the PC3 cluster demonstrates minimal variability, accounting for merely 5.42% of the variance in the LdMcm10ID-vapreotide complex (Supplementary Fig. 6D). The restricted variability in PC3 for LdMcm10ID-vapreotide, relative to PC1 and PC2 (Supplementary Fig. 6D), indicates a notably stable binding of vapreotide to the protein, resulting in a compact structural conformation. Examination of clustered regions within the PC subspace unveils distinctive conformational variations across all groups, where the blue region signifies pronounced movements, the white region denotes intermediate motions, and the red region indicates the least flexible movements [56].
Dynamic cross-correlation matrix (DCCM) analysis
Dynamic Cross-Correlation Matrix (DCCM) analysis was conducted on the trajectories of Cα backbone atom positions in both apo and ligand-bound independent simulations to discern the presence of correlated motions. Figure 5A–D shows positive correlations in the upper left triangle, while negative correlations are illustrated in the lower right triangle. The intensity of colour reflects the degree of correlation or anticorrelation, with deeper colours indicating a more robust correlation or anticorrelation. The correlation coefficient, ranging from − 1 to 1, signifies that a positive correlation implies movement in the same direction, while a negative correlation indicates movement in opposite directions. Examination of DCCM diagrams across all systems revealed notable correlations, with selected ligand molecules displaying high pairwise cross-correlation coefficient values on the cross-correlation map. The dark blue colour, indicating correlated residues (~ 1.0), suggests many pairwise correlated residues between the target protein and the chosen ligands [57]. Notably, DCCM analysis of Ligand-stand indicates a marked reduction in correlated motions upon ligand binding, underscoring the impact of the ligand on the protein’s dynamic behaviour.
Effect of lead molecules on the cell viability of L. donovani promastigotes
The effect of selected compounds on L. donovani Ld1S promastigotes was examined using an MTT assay to determine their biological implications. The results of the MTT test are reported as a percentage of cell viability, which was determined by applying the following equation while performing the calculation [33].
For this purpose, promastigote cells are incubated with different concentrations of selected compounds (1-30uM/mL). The study found that increasing drug concentration decreased L. donovani percentage viability, suggesting a dose-dependent lethal effect of selected drug molecules (Fig. 6). In order to verify the deterrent effect of suramin, vapreotide, and pasireotide, a negative control experiment was carried out in which the cells were not subjected to any treatment with the selected compounds. With a sample size of three, the proportion of viable cells in this particular instance was 99.10 ± 2.2%, showing that the cell death in the prior instance was solely attributed to the lethal action of the drugs. Further, we also calculated the IC50 of the selected compounds. Suramin is a primary agent in treating African trypanosomiasis (commonly referred to as African sleeping sickness) caused by Trypanosoma brucei gambiense or Trypanosoma brucei rhodesiense [58]. Suramin possesses an IC50 value of – 11.05 ± 0.29 µM (R2 = 0.88). It is worthwhile to discuss that the IC50 value obtained in our case (LDBPK1S) for suramin is moderately less in comparison with the IC50 value of Suramin against L. donovani (AG83) [59]. Vapreotide is a somatostatin analogue used as an antiparasitic agent against cryptosporidium-associated diarrhoea in HIV-infected patients [60]. At the same time, pasireotide is used to treat Cushing’s disease [61]. In our study, both the compound shows potent anti-leishmanial activity with IC50values of − 16.71 ± 0.15 µM (R2 = 0.89) and 12.59 ± 0.39 µM (R2 = 0.86), respectively. To our knowledge, this is the first report of these two compounds as anti-leishmanial agents.
A dose-response inhibition plot was constructed against Leishmania donovani promastigotes using Amphotericin B (the positive control), pasireotide, suramin, and vapreotide. The half maximal inhibitory doses (IC50) for Amphotericin B, pasireotide, suramin, and vapreotide were 8.73±0.89 μM, 12.59±0.39 μM, 11.05±0.29 μM, and 16.71±0.15 μM, respectively
Discussion
Leishmaniasis is a group of parasitic diseases caused by at least 20 species of the protozoan parasite Leishmania. It includes illnesses ranging from modest cutaneous blemishes to catastrophic visceral forms [7]. Because there are no effective leishmaniasis vaccinations, chemotherapy is used to manage the illness. Initial therapy with pentavalent antimonial is limited by resistance. Alternative chemotherapies, including amphotericin B, miltefosine, and paromomycin, are hazardous or expensive. Leishmaniasis chemotherapy faces medication shortages, resistance, toxicity, and expense. To address these problems and enhance leishmaniasis treatment, new, effective medications and pharmacological targets are needed [62]. Finding targets in a biological system is one of the most essential parts of drug research discovery. A pathogen target should be either absent or considerably distinct from the host homolog to be used as a therapeutic target. Secondly, the target selected should be necessary for the pathogen’s survival.
Due to its pivotal role in cellular proliferation, the DNA replication machinery is a promising therapeutic target for diverse medical conditions, including infections, autoimmune diseases, and cancer [63]. Mcm10, a versatile protein, plays a crucial role in eukaryotes’ initiation and elongation phases of DNA replication. Its involvement in licencing origins, interaction with the MCM2-7 complex, coordination with DNA polymerase α-primase, single-stranded DNA binding, and regulation of replication fork progression underscores its significance in ensuring accurate DNA replication in eukaryotic cells [11]. Therefore, this study employs a combination of in silico and in vitro approaches to explore novel therapeutic strategies targeting the Mcm10 protein in L. donovani.
New drug selection comprises numerous crucial steps to identify compounds with fewer side effects and interactions with other drugs. Bioinformatics and in silico drug design significantly affect cutting-edge drug discovery and design. These methods are used to look at target structures for binding active sites, guess what medicinal chemicals will work, and study the 3D structures of proteins [30, 31]. Predicting a protein’s 3D structure is a fundamental step that bridges the gap between sequence information and functional insights. It is pivotal in advancing our understanding of cellular processes, facilitating drug discovery, and guiding experimental studies to modify or inhibit protein function [33].
This study’s BLAST search results revealed the absence of host homologues for the target protein. The target sequence exhibited less than 28% sequence identity with available templates. This unique combination of factors underscores the target’s significance as a potential drug target. It emphasizes the need for alternative computational approaches in structure prediction due to its limited sequence similarity with existing templates. It signifies the necessity of using a threading-based method to predict its 3D structure accurately [64]. In this research, various bioinformatics tools were systematically applied to predict the three-dimensional structure of the internal domain of LdMcm10. Notably, the model generated by RosetaFold demonstrated the highest Z score (Table 1). This compelling outcome led to the selection of the RosetaFold-generated model as the optimal candidate for subsequent in-depth analyses.
A notable drawback in computational protein structure prediction is the divergence of predicted models from experimentally derived native structures, limiting their application in biochemical assignment and drug design, which demand high structural precision. This disparity hinders the effective utilization of computational methods in scenarios where accuracy is paramount [65]. Therefore, refining protein structures has become essential for enhancing knowledge-based predictions and advancing therapeutic investigations [66]. Energy was minimized via the YASSARA web server to improve the predicted model’s quality. In our study, the Z score of the selected model rose from − 5.80 to − 5.89 after refining, indicating improvement in the model’s quality (Supplementary Fig. 2B).
Additionally, we utilized multiple sequence alignment (MSA) to forecast conserved amino acid residues (Supplementary Fig. 3B). Prior studies indicate the significant contribution of MSA in accurately analysing binding site prediction [34]. In this context, the internal domain of the protein was identified as the binding site (Supplementary Fig. 3A).
Moreover, we employed molecular docking to predict and identify the optimal conformation of small-molecule ligands within the target binding site. This technique assesses scoring functions that estimate the energy of anticipated ligand-receptor complexes. The scoring function calculates the total energy of the ligand–protein interaction combined with the ligand’s energy; a lower energy value signifies a more effective protein–ligand binding and a more stable structure [67]. Virtual screening, a swift and cost-effective in silico approach, involves the evaluation of vast compound libraries for their potential to bind specific regions on target molecules like proteins [68]. In our current study, three compounds have emerged as promising therapeutic candidates against L. donovani, displaying the highest affinity for the active site of LdMcm10 and possessing the lowest binding energy, as delineated in Table 2. Serine 349 is identified as a common interacting residue for both vapreotide and pasireotide, whereas Serine 401 is shared between suramin and vapreotide. It is noteworthy that these interactions are, to the best of our knowledge, reported here for the first time in the context of L. donovani Mcm10. These compounds establish hydrogen bonds with critical amino acids pivotal for interactions and binding with various replisome proteins and single-stranded DNA, thus facilitating seamless DNA replication processes, as evidenced in supplementary Fig. 7A–F.
Molecular dynamics simulations were subsequently employed to assess these protein–ligand complexes’ stability and conformational dynamics. The findings of our study align with previous research, emphasizing the importance of understanding the structural dynamics and stability of protein–ligand complexes for effective drug discovery. Similar studies, such as those conducted by Chandra et al. [69], have underscored the significance of solvent-accessible surface area (SASA) analysis in determining ligand behaviour within the binding pocket. Our observations of ligand-induced changes in SASA complement their insights, suggesting a consistent approach to studying ligand–protein interactions (Fig. 3D). Additionally, the assessment of Root Mean Square Deviation (RMSD) in our study aligns with the work of Pieroni et al. 2023 [70], who emphasized the importance of evaluating ligand–receptor complex stability over time. The declining RMSD values in our simulations (Fig. 3A) indicate a stable protein–ligand interaction, supporting the notion that lower energy values correspond to more effective binding, as suggested by Pieroni et al. 2023 [70]. The Root Mean Square Deviation (RMSD) analysis indicated that the ligands remained consistently bound to the protein, demonstrating stability over the simulation period.
Root Mean Square Fluctuation (RMSF) analysis highlighted specific amino acid residues with reduced fluctuations, suggesting rigid ligand binding within the protein’s binding pocket (Fig. 3B). An observable shift in the Root Mean Square Fluctuation (RMSF) values is noted within the LdMcm10ID-Pasireotide complex, specifically across residues spanning from 300 to 400 and 400 to 500, displaying a range of variation from 4 to 8 Å. This discrepancy is presumed to be attributable to conformational alterations occurring within the rotatable bonds of the ligand. This inference is substantiated by visualising these bonds in the two-dimensional representation of the protein–ligand interactions (Fig. 2C). These fluctuations result from perturbations in the torsion angles of the ligand, indicative of dynamic conformational changes within the complex [71]. Secondary structure analysis illustrated the persistence of stable secondary conformations throughout the simulation (Supplementary Fig. 4).
Furthermore, our study’s emphasis on Hydrogen bond (H-bond) analysis is consistent with research by Liao et al. [52], which highlighted the critical role of H-bond interactions in stabilizing protein–ligand complexes. Our complexes’ average number of H-bond contacts resonates with their findings, further reinforcing that durable complexes rely on H-bond solid interactions (Fig. 3E). In contrast, our observations regarding the Radius of Gyration (Rg) align with earlier studies [50, 62], emphasizing Rg analysis for assessing structural compactness and stability. The varying Rg values for different complexes in our study correlate with their findings, emphasizing the importance of this metric in gauging structural integrity (Fig. 3C). Our study corroborates and extends previous research findings, providing a comprehensive understanding of the structural dynamics and stability of LdMcm10ID-ligand complexes. These consistent observations across various studies strengthen the validity and reliability of our findings, offering valuable insights for developing effective therapeutics against L. donovani.
While our study provides a detailed examination of ligand behaviour during simulations, it aligns with previous research in certain aspects. The utilization of RMSD analysis, as discussed in our work, is consistent with earlier findings [53], emphasizing its importance in evaluating ligand stability. As noted in our study, the observation of consistent RMSD values for suramin resonates with findings in literature where ligands exhibit stable conformations throughout simulations. Our work goes into more depth about how ligands behave during simulations, but in some ways, it is similar to other studies that have been done. We used RMSD analysis in our work, similar to earlier works [53], which shows how important it is for checking how stable ligands are. The fact that the RMSD values for suramin stayed the same throughout our work is similar to what other research has found: that ligands keep their stable shapes during models. The use of rGyr values to measure ligand “extendedness” or “compactness” makes it easy to make comparisons with other studies [53]. Our study’s different rGyr trends for suramin, pasireotide, and vapreotide align with other studies that stress how important this parameter is for understanding how ligand shape changes during simulations.
Additionally, the analysis of intra HB, MolSA, SASA, and PSA, as conducted in our study, shares similarities with previous works [53], where these properties indicate ligand stability and interaction patterns. The trend of initial fluctuations followed by stabilization, as observed in our research, is consistent with observations in the literature, indicating a typical pattern in ligand behaviour during simulation progress. Moreover, the binding free energy determined through MM-GBSA calculation indicated suramin, pasireotide, and vapreotide efficiency against LdMcm10 (Supplementary Table 2).
While each study contributes unique insights, the alignment of our findings with established methodologies and patterns in ligand behaviour, as demonstrated in previous research, reinforces the robustness and reliability of our approach. These comparisons strengthen the validity of our observations and provide a foundation for future studies to build upon, fostering a cumulative understanding of ligand–protein interactions in computational simulations.
Our study’s observations on the dose-dependent reduction in L. donovani viability, as influenced by suramin, vapreotide, and pasireotide, align with established antiparasitic research in the field. The inclusion of negative controls adheres to rigorous experimental standards, ensuring that any observed cellular responses are attributable exclusively to the administered compounds. The computed IC50 values for each compound provide quantitative metrics of their anti-leishmanial efficacy. The dose-response study through MTT assay evaluated Suramin with an IC50 value of 11.05 ± 0.29 µM against L. donovani promastigotes (LDBPK1S) which was comparatively less in comparison with the IC50 value of Suramin against L. donovani (AG83) [59]. Also, the study first reports the evaluation of vapreotide and pasireotide as anti-leishmanial as both the compounds showed potent anti-leishmanial activity with IC50values of 16.71 ± 0.15 µM and 12.59 ± 0.39 µM, respectively, against LDBPK1S (Fig. 6). Our study confirms Suramin’s known effectiveness against African trypanosomiasis [58], and the IC50 values obtained suggest a consistent yet strain-dependent efficacy against L. donovani. This aligns with existing literature on suramin’s broad-spectrum antiparasitic activity.
The novel contribution of our study lies in elucidating the anti-leishmanial activity of vapreotide and pasireotide, both somatostatin analogues. While somatostatin analogues have been explored for parasitic infections [60], their specific efficacy against L. donovani is a noteworthy finding. Our research aligns with established antiparasitic principles, reinforcing dose-dependent responses and emphasizing the therapeutic potential of suramin, vapreotide, and pasireotide. The unique contribution stems from confirming their efficacy against L. donovani strains, warranting further comparative investigations and clinical exploration.
Conclusion
Overall, our research aimed to assess the possibility that the Mcm10 protein found in L. donovani may serve as a treatment target for leishmaniasis. We could predict the unique three-dimensional structure of LdMcm10 by using the in silico method. Suramin, vapreotide, and pasireotide were discovered as viable candidates through molecular docking, and further simulations revealed that these compounds formed stable complexes. It was shown that the compounds exhibited dose-dependent decreases in the viability of L. donovani. The effectiveness of suramin was consistent with the research that had been done previously, but vapreotide and pasireotide demonstrated unique anti-leishmanial activity, which broadened the range of possible treatments. In conclusion, the findings of our study give insights into the structural dynamics of the internal domain of LdMcm10 and identify prospective therapeutic options against L. donovani. These findings also have significant clinical implications for the treatment of leishmaniasis.
Data availability
All data generated or analysed during this study are included in this article [and its supplementary information files].
References
Karunaweera ND, Ferreira MU (2018) Leishmaniasis: current challenges and prospects for elimination with special focus on the South Asian region. Parasitology 145:425–429
Sinha M, Jagadeesan R, Kumar N et al (2022) In-silico studies on Myo inositol-1-phosphate synthase of Leishmania donovani in search of anti-leishmaniasis. J Biomol Struct Dyn 40:3371–3384. https://doi.org/10.1080/07391102.2020.1847194
Ghorbani M, Farhoudi R (2018) Leishmaniasis in humans: drug or vaccine therapy? Drug Des Devel Ther 12:25–40
Singh OP, Singh B, Chakravarty J, Sundar S (2016) Current challenges in treatment options for visceral leishmaniasis in India: a public health perspective. Infect Dis Poverty 5:1–5
Sindermann H, Engel KR, Fischer C, Bommer W (2004) Oral miltefosine for leishmaniasis in immunocompromised patients: compassionate use in 39 Patients with HIV infection. Clin Infect Dis 39(10):1520–1523
Brindha J, Balamurali MM, Chanda K (2021) An overview on the therapeutics of neglected infectious diseases—leishmaniasis and chagas diseases. Front Chem 9:622286
Chawla B, Madhubala R (2010) Drug targets in Leishmania. J Parasit Dis 34:1–13
Baxley RM, Bielinsky AK (2017) Mcm10: a dynamic scaffold at eukaryotic replication forks. Genes (Basel) 8:73
Chen J, Wu S, Wang J et al (2023) MCM10: An effective treatment target and a prognostic biomarker in patients with uterine corpus endometrial carcinoma. J Cell Mol Med 27:1708–1724. https://doi.org/10.1111/jcmm.17772
Warren EM, Vaithiyalingam S, Haworth J et al (2008) Structural basis for DNA Binding by replication initiator Mcm10. Structure 16:1892–1901. https://doi.org/10.1016/j.str.2008.10.005
Thu YM, Bielinsky AK (2013) Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci 38:184–194
Robertson PD, Chagot B, Chazin WJ, Eichman BF (2010) Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif. J Biol Chem 285:22942–22949. https://doi.org/10.1074/jbc.M110.131276
Cassandri M, Smirnov A, Novelli F et al (2017) Zinc-finger proteins in health and disease. Cell Death Discov. https://doi.org/10.1038/cddiscovery.2017.71
Yu W, Mackerell AD (2017) Computer-aided drug design methods. In: Sass P (ed) Methods in molecular biology. Humana Press Inc., Totowa, pp 85–106
Baek M, DiMaio F, Anishchenko I et al (2021) Accurate prediction of protein structures and interactions using a three-track neural network 1979. Science 373:871–876. https://doi.org/10.1126/science.abj8754
Zhou X, Zheng W, Li Y et al (2022) I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nat Protoc 17:2326–2353. https://doi.org/10.1038/s41596-022-00728-0
Pieper U, Webb BM, Barkan DT et al (2011) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. https://doi.org/10.1093/nar/gkq1091
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/s0021889892009944
Krieger E, Joo K, Lee J et al (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins: Struct Funct Bioinform 77:114–122
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8(2):127–134. https://doi.org/10.1093/protein/8.2.127
Lill MA, Danielson ML (2011) Computer-aided drug design platform using PyMOL. J Comput Aided Mol Des 25:13–19. https://doi.org/10.1007/s10822-010-9395-8
Bowers KJ, Chow E, Xu H, et al (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC’06
Hildebrand PW, Rose AS, Tiemann JKS (2019) Bringing molecular dynamics simulation data into view. Trends Biochem Sci 44:902–913
Rasheed MA, Iqbal MN, Saddick S et al (2021) Identification of lead compounds against scm (Fms10) in enterococcus faecium using computer aided drug designing. Life 11:1–15. https://doi.org/10.3390/life11020077
Shivakumar D, Williams J, Wu Y et al (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the opls force field. J Chem Theory Comput 6:1509–1519. https://doi.org/10.1021/ct900587b
Li J, Abel R, Zhu K et al (2011) The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins: Struct Funct Bioinform 79:2794–2812. https://doi.org/10.1002/prot.23106
Palma J, Pierdominici-Sottile G (2023) On the uses of PCA to characterise molecular dynamics simulations of biological macromolecules: basics and tips for an effective use. ChemPhysChem. https://doi.org/10.1002/cphc.202200491
Kitao A (2022) Principal component analysis and related methods for investigating the dynamics of biological macromolecules. J (Basel) 5:298–317. https://doi.org/10.3390/j5020021
Avti P, Chauhan A, Shekhar N et al (2022) Computational basis of SARS-CoV 2 main protease inhibition: an insight from molecular dynamics simulation based findings. J Biomol Struct Dyn 40:8894–8904. https://doi.org/10.1080/07391102.2021.1922310
van Meerloo J, Kaspers GJL, Cloos J (2011) Cell sensitivity assays: the MTT assay. In: Cree IA (ed) Cancer cell culture: methods and protocols. Humana Press, Totowa, pp 237–245
Baranwal A, Chiranjivi AK, Kumar A et al (2018) Design of commercially comparable nanotherapeutic agent against human disease-causing parasite Leishmania. Sci Rep. https://doi.org/10.1038/s41598-018-27170-1
Chakrabarti S, Lanczycki CJ (2007) Analysis and prediction of functionally important sites in proteins. Protein Sci 16:4–13. https://doi.org/10.1110/ps.062506407
Rodina A, Godson GN (2006) Role of conserved amino acids in the catalytic activity of Escherichia coli primase. J Bacteriol 188:3614–3621. https://doi.org/10.1128/JB.188.10.3614-3621.2006
Mayle R, Langston L, Molloy KR et al (2019) Mcm10 has potent strand-annealing activity and limits translocase-mediated fork regression. Proc Natl Acad Sci U S A 116:798–803. https://doi.org/10.1073/pnas.1819107116
Perez-Arnaiz P, Kaplan DL (2016) An Mcm10 mutant defective in ssDNA binding shows defects in DNA replication initiation. J Mol Biol 428:4608–4625. https://doi.org/10.1016/j.jmb.2016.10.014
Broni E, Kwofie SK, Asiedu SO et al (2021) A molecular modeling approach to identify potential antileishmanial compounds against the cell division cycle (Cdc)-2-related kinase 12 (crk12) receptor of leishmania donovani. Biomolecules 11:1–32. https://doi.org/10.3390/biom11030458
Pandey P, Prasad K, Prakash A, Kumar V (2020) Insights into the biased activity of dextromethorphan and haloperidol towards SARS-CoV-2 NSP6: in silico binding mechanistic analysis. J Mol Med 98:1659–1673. https://doi.org/10.1007/s00109-020-01980-1
Amadei A, Linssen ABM, Berendsen HJC (1993) Essential Dynamics of Proteins. Proteins: Struct Funct Bioinform 17(4):412–425
Macip G, Garcia-Segura P, Mestres-Truyol J et al (2022) Haste makes waste: a critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition. Med Res Rev 42:744–769
Omoboyowa DA, Iqbal MN, Balogun TA et al (2022) Inhibitory potential of phytochemicals from Chromolaena odorata L. against apoptosis signal-regulatory kinase 1: a computational model against colorectal cancer. Computational Toxicology 23:100235. https://doi.org/10.1016/j.comtox.2022.100235
Hollingsworth SA, Dror RO (2018) Molecular dynamics simulation for all. Neuron 99:1129–1143
Hao MH, Haq O, Muegge I (2007) Torsion angle preference and energetics of small-molecule ligands bound to proteins. J Chem Inf Model 47:2242–2252. https://doi.org/10.1021/ci700189s
Mitra S, Dash R (2018) Structural dynamics and quantum mechanical aspects of shikonin derivatives as CREBBP bromodomain inhibitors. J Mol Graph Model 83:42–52. https://doi.org/10.1016/j.jmgm.2018.04.014
De Vita S, Chini MG, Bifulco G, Lauro G (2021) Insights into the ligand binding to bromodomain-containing protein 9 (BRD9): a guide to the selection of potential binders by computational methods. Molecules. https://doi.org/10.3390/molecules26237192
Kuldeep J, R K, Kaur P, et al (2021) Identification of potential anti-leishmanial agents using computational investigation and biological evaluation against trypanothione reductase. J Biomol Struct Dyn 39:960–969. https://doi.org/10.1080/07391102.2020.1721330
Zhang G, Su Z (2012) Inferences from structural comparison: flexibility, secondary structure wobble and sequence alignment optimization. BMC Bioinformatics 13:S12. https://doi.org/10.1186/1471-2105-13-S15-S12
Carugo O, Pongor S (2001) A normalized root-mean-spuare distance for comparing protein three-dimensional structures. Protein Sci 10:1470–1473. https://doi.org/10.1110/ps.690101
Prakash A, Kumar V, Meena NK, Lynn AM (2018) Elucidation of the structural stability and dynamics of heterogeneous intermediate ensembles in unfolding pathway of the N-terminal domain of TDP-43. RSC Adv 8:19835–19845. https://doi.org/10.1039/c8ra03368d
Bagewadi ZK, Yunus Khan TM, Gangadharappa B et al (2023) Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of Micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach. Saudi J Biol Sci. https://doi.org/10.1016/j.sjbs.2023.103753
Liao KH, Chen KB, Lee WY et al (2014) Ligand-based and structure-based investigation for Alzheimer’s disease from traditional Chinese medicine. Evid-Based Complement Altern Med. https://doi.org/10.1155/2014/364819
Raj U, Kumar H, Gupta S, Varadwaj PK (2015) Novel DOT1L receptornatural inhibitors involved in mixed lineage leukemia: a virtual screening, molecular docking and dynamics simulation study. Asian Pac J Cancer Prev 16:3817–3825. https://doi.org/10.7314/APJCP.2015.16.9.3817
Hata H, Tran DP, Sobeh MM et al (2021) Binding free energy of protein/ligand complexes calculated using dissociation Parallel Cascade Selection Molecular Dynamics and Markov state model. Biophys Physicobiol 18:305–316. https://doi.org/10.2142/biophysico.bppb-v18.037
David CC, Jacobs DJ (2014) Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol Biol 1084:193–226. https://doi.org/10.1007/978-1-62703-658-0_11
Ashraf N, Asari A, Yousaf N et al (2022) Combined 3D-QSAR, molecular docking and dynamics simulations studies to model and design TTK inhibitors. Front Chem. https://doi.org/10.3389/fchem.2022.1003816
Yousaf N, Jabeen Y, Imran M et al (2023) Exploiting the co-crystal ligands shape, features and structure-based approaches for identification of SARS-CoV-2 Mpro inhibitors. J Biomol Struct Dyn 41:14325–14338. https://doi.org/10.1080/07391102.2023.2189478
Bouteille B, Buguet A (2012) The detection and treatment of human African trypanosomiasis. Res Rep Trop Med. https://doi.org/10.2147/rrtm.s24751
Khanra S, Juin SK, Jawed JJ et al (2020) In vivo experiments demonstrate the potent antileishmanial efficacy of repurposed suramin in visceral leishmaniasis. PLoS Negl Trop Dis 14:1–20. https://doi.org/10.1371/journal.pntd.0008575
Khan SM, Witola WH (2023) Past, current, and potential treatments for cryptosporidiosis in humans and farm animals: a comprehensive review. Front Cell Infect Microbiol 13:1115522
Simeoli C, Ferrigno R, De Martino MC et al (2020) The treatment with pasireotide in Cushing’s disease: effect of long-term treatment on clinical picture and metabolic profile and management of adverse events in the experience of a single center. J Endocrinol Invest 43:57–73. https://doi.org/10.1007/s40618-019-01077-8
Saha S, Srivastava R, Sarma P et al (2023) Identification of potential inhibitors of Leishmania donovani Sterol 24-C- methyltransferase: in silico and in vitro studies. Mol Simul 49:1311–1323. https://doi.org/10.1080/08927022.2023.2227288
Gardner AF, Kelman Z (2019) Editorial: the DNA replication machinery as therapeutic targets. Front Mol Biosci 6:35
Adinehbeigi K, Razi Jalali MH, Shahriari A, Bahrami S (2017) In vitro antileishmanial activity of fisetin flavonoid via inhibition of glutathione biosynthesis and arginase activity in Leishmania infantum. Pathog Glob Health 111:176–185. https://doi.org/10.1080/20477724.2017.1312777
Bhattacharya D, Cheng J (2013) 3Drefine: consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization. Proteins: Struct. Funct. Bioinform. 81:119–131. https://doi.org/10.1002/prot.24167
Feig M (2017) Computational protein structure refinement: almost there, yet still so far to go. Wiley Interdiscip Rev Comput Mol Sci. https://doi.org/10.1002/wcms.1307
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321. https://doi.org/10.1021/jm051197e
Karthick V, Nagasundaram N, Doss CGP et al (2016) Virtual screening of the inhibitors targeting at the viral protein 40 of Ebola virus. Infect Dis Poverty. https://doi.org/10.1186/s40249-016-0105-1
Chandra A, Chaudhary M, Qamar I et al (2022) In silico identification and validation of natural antiviral compounds as potential inhibitors of SARS-CoV-2 methyltransferase. J Biomol Struct Dyn 40:6534–6544. https://doi.org/10.1080/07391102.2021.1886174
Pieroni M, Madeddu F, Di Martino J et al (2023) MD–ligand–receptor: a high-performance computing tool for characterizing ligand-receptor binding interactions in molecular dynamics trajectories. Int J Mol Sci. https://doi.org/10.3390/ijms241411671
Hao MH, Haq O, Muegge I (2007) Torsion angle preference and energetics of small-molecule ligands bound to proteins. J Chem Inf Model 47:2242–2252
Acknowledgements
Deep Bhowmik acknowledges the financial support of ICMR (Fellowship/95/2022-ECD-II, dated 17/05/2022, respectively). Satabdi Saha recognizes the financial support from the Inspire Fellowship (IF180806).
Author information
Authors and Affiliations
Contributions
" SS, DB, AS and DK carried out the experiment. SS, DB and DK wrote the manuscript. SS, DB, AS and DK contributed to the analysis of the results. DK supervised the project and conceived the original idea."
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saha, S., Sharma, A., Bhowmik, D. et al. Investigation into in silico and in vitro approaches for inhibitors targeting MCM10 in Leishmania donovani: a comprehensive study. Mol Divers (2024). https://doi.org/10.1007/s11030-024-10876-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-024-10876-y