Abstract
CRC is the third most common cancer occurring worldwide and the second leading cause of cancer deaths. In the year 2020, 1,931,590 new cases of CRC and 935,173 deaths were reported. The last two decades have witnessed an intensive study of noncoding RNAs and their implications in various pathological conditions including cancer. Noncoding RNAs such as miRNAs, tsRNAs, piRNAs, lncRNAs, pseudogenes, and circRNAs have emerged as promising prognostic and diagnostic biomarkers in preclinical studies of cancer. Some of these noncoding RNAs have also been shown as promising therapeutic targets for cancer treatment. In this review, we have discussed the emerging roles of various types of noncoding RNAs in CRC and their future implications in colorectal cancer management and research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to GLOBOCAN 2020 database, CRC is the third most common cancer (after breast and lung cancer) occurring worldwide and the second (after lung cancer) leading cause of cancer deaths. In men, CRC is the third most commonly diagnosed cancer as well as the cause of cancer-related death, and in women second most commonly diagnosed cancer and the third most common cancer-related death. Males are having a higher incidence and mortality from CRC compared to females and among different continents, Asia is having the highest incidence, 5-year prevalence as well as mortality from CRC. The estimated global burden of CRC by 2030 is projected to increase by 60% that will lead to 2.2 million new cases and 1.1 million annual deaths [1]. Another unexpected finding suggests that CRC is decreasing in the older age group, increasing in the younger age group [2] and it is increasing faster in developing countries compared to developed countries [3]. CRC is having overexpression of the Wnt signaling pathway [4], dominant epithelial-mesenchymal transition (EMT) program [5], epigenetic dysregulations [6], and any drug inhibiting these pathways [7, 8] have the potential to inhibit colon carcinogenesis. Hence, finding some novel prognostic and diagnostic markers and appropriate drug targets is very urgent and needs of the time. RNA plays a very important role in the expression of genes and genetic regulation in the form of protein-coding or noncoding RNAs. Some of the important noncoding RNAs dysregulated in cancer are long noncoding RNAs (lncRNAs), 7SK small nuclear RNAs (snRNA), and microRNAs. Changes in the expression of non-coding RNAs, processing factors, and their mutations have implications in carcinogenesis. N6-methyladenosine modification in RNA has been shown to play an important role in cancer growth and development [9].
Different classes of non-coding RNAs and their roles in cancer
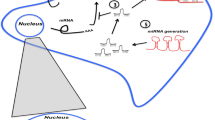
There are multiple types of RNAs in a cell but in recent times non-coding RNAs (ncRNAs) which constitute 90% of RNAs and do not translate into proteins have been shown to play a very important role in physiology, development, and pathogenesis [10]. Non-coding RNAs have been classified into different classes (shown in Fig. 1) based on their size and RNAs involved in cancer pathology are miRNAs, tsRNAs, piRNAs, and lncRNAs. Among various types of non-coding RNAs, miRNAs are the most extensively studied and multiple clinical trials are undergoing which involve some aspects of miRNA biology [11]. miRNA-based drugs that can increase or decrease the target microRNAs (miRNAs) are being extensively investigated in different types of cancers. ncRNAs present in blood can be used for the screening of different diseases including cancer and recently RNAi drug Onpattro™ has been approved and Spinraza®, an RNA targeting oligonucleotide drug has been successfully used in the clinic which hints toward promising therapeutic uses of these classes of molecules in the coming future. Some of the important classes of non-coding RNAs and their functions are summarized in Table 1 and an image of all the representative classes of noncoding RNAs dysregulated in colon cancer is shown in Fig. 1.
Representative classes of non-coding RNAs that are known to be dysregulated in colorectal cancer. Noncoding RNAs shown in green are upregulated, those shown in red are downregulated in CRC. CRC Colorectal cancer, circRNAs Circular RNAs, miRNAs MicroRNAs, lncRNAs Long noncoding RNAs, EV RNAs Extracellular vesicular RNAs, piRNAs PIWI-interacting RNAs, tsRNAs tRNA-derived small RNAs
MicroRNAs
MicroRNAs (miRNAs) are 22 nucleotides in length and regulate mRNAs by binding to their 3′ end with the complementary sequences. miRNA coding genes are transcribed by RNA pol II and they undergo an evolutionarily conserved processing pathway to form the mature and functional miRNAs. Pri-miRNA which has a hairpin structure is recognized by Drosha, a Class 2 ribonuclease III enzyme and DGCR8 (DiGeorge syndrome critical region 8) and cleaved into 60 nucleotides (pre-miRNA). Pre-miRNA thus formed is exported from the nucleus to the cytoplasm by the Ran-GTP complex and Exportin 5 and the Pre-miRNA in the cytoplasm is acted upon by Dicer (an endoribonuclease or helicase with RNase motif) to form a duplex miRNA that has 5′ phosphate and 3′ overhangs of 2 nucleotides. Guide RNA (one of the strands of miRNA duplex) binds with Argonaute protein and forms an RNA-induced silencing complex (RISC), which also contains mature 22 nucleotide miRNA. Mature miRNA binds to 3′ UTR of mRNA and induces its degradation or suppresses its translation [12]. miRNAs are the most intensively studied non-coding RNAs compared to tsRNAs and piRNAs and the miRNAs can act as a tumor suppressor as well as oncogenes (oncomiRs). Each miRNA can regulate multiple mRNAs and each mRNA can be regulated by multiple miRNAs and miRNAs can regulate the expression of thousands of coding and noncoding genes including oncogenes such as RAS, MYC, and EGFR, and tumor suppressors such as TP53, PTEN, and BRCA1. Oncogenic miRNAs can promote cell proliferation and induce tumor formation and it might inhibit the tumor suppressors or it might remove the genetic break on oncogene expression. miR-155 promotes abnormal proliferation of the B cells [21], miR-21 expression is very high in B-cell lymphoma and lung cancer [22], and miR-10b expression is very high in glioblastomas [23]. These miRNAs show oncogene addiction as the survival of the tumor is dependent on the continued expression of these miRNAs. Let-7 miRNA suppresses the RAS oncogenic pathway because there are multiple complementary let-7a binding sites on 3′ UTRs of KRAS, NRAS, and HRAS mRNAs [24]. miR-15a and miR-16-1 are tumor suppressor miRNAs but when it is mutated or deleted it might aggravate chronic lymphocytic leukemia (CLL) [25]. miR-34a regulates several oncogenes and it is a downstream target of p53 [26] and the role of miR-29 can be context-dependent as it prevents indolent B-CLL but it is elevated in acute myeloid leukemia and aggressive form of B-CLL [27]. Truncation mutation of XPO5 (exportin 5) in microsatellite unstable colon tumors can result in miRNA precursor aggregation in the nucleus and reduced miRNA mediated target inhibition. miR-21 and miR-92a are increased in the serum of patients with CRC and advanced adenomas compared to the healthy controls [28]. A higher expression of miR-92a is correlated with the poor survival of CRC patients. Kanaan et al. have suggested a panel of eight miRNAs (miR-652, miR-17, miR-331, miR-195, miR-142-3p, miR-532, miR-15b, and miR-532-3p) to increase the specificity of CRC detection. Another set of three miRNAs (miR-15b, miR-139-3p, and miR-431) were able to distinguish stage IV CRC from the controls [29]. miR-135b was found to have higher expression in CRC and adenomas compared to adjacent normal colon tissues [30]. miR-106a along with fecal blood tests can increase the detection sensitivity of CRCs [31]. Ahmed et al. have shown by global microarray expression of miRNA in the stool that 12 miRNAs (miR-7, miR-17, miR-20a, miR-21, miR-92a, miR-96, miR-106a, miR-134, miR-183, miR-196a, miR-199a-3p, and miR-214) have increased expression in CRC patients compared to the normal controls [32]. Nine miRNAs (including miR-31-3p and miR-31-5p) were found to be differentially expressed when treated with anti-EFGR cetuximab in patients with metastatic CRC [33]. Simmer et al. had shown a better survival of those metastatic CRC patients with lower expression of miR-143 compared to those having higher expression when treated with capecitabine[34]. Perez-Carbonell et al. have shown miR-320e as an indication of poor survival in stage III CRC patients when treated with 5 fluorouracil [35]. miR-220c is higher in the liver metastases of CRCs compared to primary cancers [36] and miRNAs can be further explored to predict the EMT and metastasis in CRCs. miR-224 expression is increased with the cancer burden and the status of the microsatellite stability in CRC [37] and it targets SMAD4 and facilitate metastasis in CRC[38]. miR-214, miR-182, miR-124, miR-30b, and miR-155 have also been reported as potential prognostic markers for CRC patients and these markers need to be further validated by the clinical trials [31].
tRNA fragments
tRNA fragments (tRFs) are small ncRNAs that are derived from transfer RNAs (tRNAs) and are also known as tsRNAs (tRNA-derived small RNAs), and tiRNAs (tRNA-derived stress-induced RNAs). The biogenesis and detailed molecular mechanisms of tsRNAs are still not very clear and a universal nomenclature system is required for the uniformity in the naming of tsRNAs. They are transcribed as pre-tRNAs by the enzyme RNA pol III and are processed to produce the mature tRNAs depending on the cleavage site in pre-tRNA or mature tRNA. Ribonucleases such as Angiogenin, Dicer, and RNase Z are involved in the generation of tsRNAs but the actual mechanism is not known [39]. tsRNAs show some similarities in function with the miRNAs such as involvement of Argonaute proteins, suppression of target mRNAs, binding to 3′ UTR, and having oncogenic as well as tumor suppressor roles. tsRNAs are upregulated in liver cancer and their targeting by Locked nucleic acid (LNA) oligonucleotides leads to the death of liver tumor cells [40]. In a comprehensive analysis by small RNA sequencing in CRC patients, 55 differentially expressed mRNAs were potential targets of differentially expressed tRFs and miRNAs [41]. tRF/miR-1280, a 17 bp fragments of tRNALeu have been shown to regulate Notch signaling in colorectal stem cells and suppress CRC growth and metastasis and the ectopic expression of tRF/miR-1280 decreases cell proliferation and colony formation and its expression is poor in CRC [42]. tiRNAs derived by cleavage of angiogenin are highly expressed in CRC tissues and metastatic cells and help in angiogenin-mediated metastasis and a higher level of 5′-tiRNA derived from tRNA-Val was also observed in CRC and it was correlated with cancer metastasis [43]. 5′tiRNA-His-GTG, upregulated in CRC plays an oncogenic role and targets LATS2 and its suppression induces apoptosis [44].
PIWI-interacting RNAs
PIWI-interacting RNAs (piRNAs) are non-coding RNAs that are approximately 21–35 nucleotides long, associated with the PIWI subfamily of Argonaute proteins, and also involved in transposon silencing during germline development. piRNAs are processed by RNA Pol II in mammals, piRNA precursors are species-specific, sequences are not conserved and 20,000 piRNAs are estimated to be present in the human genome. piRNAs silence transposons by inducing PIWI protein association with transposon promoter or by guiding PIWI complex to the transposon mRNA. piRNA expression is mostly restricted to the gonadal cells but recent findings also suggest a lower level of extragonadal expression in somatic tissues [45]. Dysregulations of piRNAs in cancer are emerging as biomarkers but their role in somatic tissues and cancers are still under intensive investigation [46].
Overexpression of piR-1245 was reported in CRC correlating with the advanced stage and metastasis with shorter overall survival and it acts as an oncogenic noncoding RNA and targets multiple tumor suppressor genes such as ATF3, SESN2, FAS, BTG1, NFKBIA, UPP1, DUSP1, MDX1, and TP53INP1 [47]. piRNA-54265 binds to PIWIL2 protein and other partners to form a complex which induces the STAT3 pathway and activates cell proliferation, chemoresistance, and metastasis in CRC and it is overexpressed in CRC compared to the normal tissues and associated with poor survival of the patients [48]. piRNAs, piR-020619 and piR-020450, are consistently elevated in the CRC patients compared to normal controls but not in lung, breast, and gastric cancers and these can detect small-sized and early-stage CRC and hence these two piRNAs can be used for cancer-specific early detection of CRC [49]. Circulating piR-5937 and piR-28876 has very high sensitivity and specificity to detect the CRC [50] and systematic screening of all known piRNAs revealed piR-015551 to be possibly generated from LNC00964-3 and its involvement in CRC development [51]. Small RNA sequencing revealed that 33 piRNAs are upregulated and 2 piRNAs are downregulated in CRC tissues and piR‑19521, piR‑17724, and piR‑18849 were the top highly expressed piRNAs in the CRC patients. Overexpression of piR‑19521 and piR‑18849 were the indicator of poor differentiation of cells and overexpression of piR‑18849 correlates with metastasis in the lymph nodes. piR‑18849 and piR‑19521 can be utilized as a prognostic biomarker in patients with CRC [52] and piR-54265 level in the serum were shown to be overexpressed only in the patients with CRC compared to the controls and other cancer types hence serum piR-54265 can be a valuable biomarker for the screening of the potential CRC patients as well as early detection and surveillance [53]. Five differentially expressed serum piRNAs (piR-001311, piR-004153, piR-017723, piR-017724, and piR-020365) were suggested to increase the chances of CRC detection and can be used for the prognosis [54]. piR-24000 was found to be overexpressed in the CRC and its expression was correlated with aggressiveness of the disease such as poor differentiation, distant metastasis, and a higher stage [55]. piR-823 is upregulated in CRC and its inhibition leads to inhibition of cell proliferation, cell cycle arrest, induced apoptosis and it was shown to increase the transcription of HSF1 and hence, piR-823 promotes tumor growth and can act as a potential therapeutic target in CRC [56].
Long ncRNAs (lncRNAs)
Long ncRNAs (lncRNAs) were initially identified as factors involved in X chromosome inactivation and embryonic development but later they were shown to be involved in various diseases including cancer. Long ncRNAs are 200 base pairs, transcribed by the RNA Pol II enzyme and the exons of lncRNAs are spliced into mature transcripts that contain 5′ caps and 3′ poly(A) tails. Long ncRNAs are evolutionarily not conserved, have a lower number of exons, level of expression is low and they can influence gene expression at epigenetic, transcriptional, and post-transcriptional levels. Long ncRNAs can bind to chromatin-modifying complexes and guide them to the promoter to regulate transcription and by binding to transcription factors they can affect the downstream processes. Long ncRNAs can bind to RNA binding proteins (RBPs) and regulate mRNA processing and stability and can also directly bind to the mRNA and DNA and regulate their functions [57]. Long ncRNAs have a wide range of regulatory mechanisms and downstream effects and they can act as a tumor suppressor, oncogene, or might have context-dependent roles [58]. Some of the important emerging oncogenic long ncRNAs are THOR, ARLNC1, DSCAM-AS1, lncARSR, CamK-A, EPIC1 [11], and the overexpression of HOTAIR is associated with poor outcomes in breast cancer [59] and SAMMSON is associated with melanoma [60]. REG1CP is upregulated in colon cancer and it promotes the growth of the colon cancer xenograft by associating FANCJ to the promoter of REG3A leading to enhanced cell proliferation [61]. MEG3 is a well-known long ncRNA acting as a tumor suppressor and it downregulates MDM2 and upregulates p53 proteins and regulates the TGF beta pathway [62, 63]. GAS5 is downregulated in various cancers more importantly in breast cancer and glioblastoma [64, 65], and the antisense transcript of SATB2-AS1 inside the nucleus of CRC cells acts as a tumor suppressor [66]. CCAT1 (colon cancer-associated transcript 1) generates multiple lncRNA transcripts and affects MYC expression in CRC and a long transcript of CCAT1, CCAT1-L is expressed in CRC and interacts with CTCF leading to chromosome looping [67]. CCAT1-5L interacts with HNRNPK and non-coding RNAs generated from the MYC promoter and PVT1 enhancer leading to increased MYC transcription by chromatin looping [68]. SATB2 represses Snail and slows down the cell proliferation, invasion, and migration of CRC cells and long ncRNA NKILA has been shown to act as an oncogene as well as a tumor suppressor in a context-dependent manner. lncRNA GAS5 interacts with YAP and inhibits CRC progression and its expression is negatively correlated with the expression of YAP and YTHDF3 in CRC patients [69]. lnc273-31 and lnc273-34 were shown to maintain CSC stemness and its depletion leads to suppression of migration, invasion, and stem cell self-renewal and chemoresistance [70]. LINRIS is upregulated in CRC and correlates with poor survival and the LINRIS-IGF2BP2-MYC axis induces the CRC progression and promotes aerobic glycolysis in CRC [71]. lncRNA RP11 is highly expressed in CRC and it can induce the CRC progression by upregulation of Zeb 1 [72] and SNHG1 is upregulated in CRC and it is associated with poor survival [73]. SNHG1 reduces the level of miR-154-5p leading to an increase in Cyclin D2 expression [74] and FEZF1-AS1 is overexpressed in CRC and correlates with poor survival of the patients. FEZF1-AS1 was shown to increase cell proliferation and metastasis and increase the stability of PKM2 proteins [75] and lncRNA NEAT1 is highly expressed in CRC and regulates proliferation, migration, and invasion. NEAT1 activates Wnt/β-catenin signaling through DDX5 protein and promotes CRC progression and metastasis [76]. SNHG5 is overexpressed in CRC and promotes CRC cell survival and the overexpression of SNHG5 diminishes the oxaliplatin induced apoptosis [77]. CRNDE, H19, UCA1, and HOTAIR are involved in the induction of resistance against oxaliplatin or irinotecan treatment in CRC [78]. LncRNA MALAT1 can facilitate CRC cell proliferation, invasion, and migration by the downregulation of miR-145 and upregulation of SOX9 [79] and overexpression of SNHG11 has been shown to promote cell proliferation and metastasis in CRC by regulating the Hippo pathway [80]. CASC9 is overexpressed in CRC and correlates with poor survival and it shows oncogenic activity by upregulation of TGFβ2 and TERT [81]. The whole-transcriptome analysis resulted in the finding of 27 upregulated and 22 downregulated lncRNAs in CRC [82] and lncRNA SNHG14 was shown to facilitate CRC metastasis by targeting EZH2-regulated EPHA7 [83]. lncCMPK2 expression was found to be highly expressed in CRC and it positively relates with clinical stage and lymphatic metastasis and its overexpression leads to cell proliferation and activates the FUBP3-c-Myc axis [84]. lncCCAT1-L plays a very important role in MYC transcriptional regulation as it is located 515 kb upstream of MYC within a strong super-enhancer and promotes long-range looping of chromatin. It has also been shown to interact with CTCF to modulate the conformation of the loop [67]. SP1 induced ZFAS1 upregulates VEGFA and contributes to CRC progression [85] and SNHG6 interacts with miR-26a, miR-26b, and miR-214 and controls EZH2 to promote CRC [86]. lncRNA UICLM is upregulated in CRC and induces liver metastasis [87] and lncRNA SLCO4A1-AS1 is overexpressed in CRC and correlates with poor prognosis and metastasis. SLCO4A1-AS1 enhances the stability of β-catenin leading to activation of the Wnt/β-catenin signaling pathway [88] and the oncogenic SNHG6 activates the TGF-β/Smad signaling pathway by targeting UPF1 and induces the EMT pathway by regulating ZEB1 in CRC [89]. lnc OCC-1 suppresses CRC tumor by binding and destabilizing HuR protein [90]. LINC02418 is highly expressed in CRC, and LINC02418-miR-1273 g-3p-MELK axis plays an important role in the progression of CRC [91]. lncH19 mediates 5Fu resistance in CRC through induction of SIRT1 mediated autophagy and hence it can act as a prediction marker for the response to the 5 Fu treatment in CRC [92]. lncCCAT1 promotes CRC by regulating the miR-181a-5p expression and it is positively related to cancer stages [93]. Overexpression of SNHG7 induces the tumor progression and liver metastasis of SW480 cells and it regulates GALNT1 by sponging the miR-216b and hence it plays an oncogenic role in tumor progression [94]. Upregulation of lncRP11-468E2.5 in CRC inhibits JAK/STAT signaling, suppresses cell proliferation, and promotes apoptosis [95].
Pseudogenes
Pseudogenes are a special category of lncRNA transcripts that are inactivated and cannot produce functional proteins otherwise very similar to the coding genes. They act as a decoy and divert the essential components of the molecular processes. Pseudogenes are now emerging as an important component in cancer initiation and progression and it requires an in-depth study and analysis to elucidate their involvement in disease development. PTENP1 is a pseudogene that is very similar to PTEN and acts as a sponge that diverts the miRNAs targeting the PTEN mRNAs and hence PTENP1 is considered as a tumor suppressor pseudogene [96]. BRAFP1 pseudogene is involved in the development of aggressive B-cell lymphoma in mice model [97] and VEGFR1 pseudogene, FLT1P1, modulates VEGFR1 protein expression in CRC cells [98]. The pseudogene DUXAP8 is having increased expression in CRC and it induces cancer development by negatively regulating E-cadherin by interacting with EZH2 and H3K27me3 [99].
Circular RNAs
Circular RNAs (circRNAs) are the class of noncoding RNAs that are single-stranded and covalently closed structure that is formed by back splicing but the detailed mechanisms involved in their generation are not known. Important features of circRNAs include longer flanking introns, a higher level of intronic repetitive and reverse complement elements, a covalently closed continuous loop without 5′-3′ polarity, and a polyA tail. They act as sponges to divert miRNAs, stabilize the miRNA binding molecules, and act as a scaffold to bind to various regulatory proteins [100]. CircACC1 overexpressed in CRC modulates fatty acid β-oxidation and glycolysis and the upregulated Circ-Erbin induces metastasis, proliferation, and angiogenesis [101]. The upregulation of circRNA_0001178 and circRNA_0000826 in CRC was identified as a potential indicator of liver metastasis [102] and circITGA7 was shown to inhibit CRC growth and metastasis through the Ras pathway and upregulation of ITGA7 [103]. ciRS-7 is upregulated in CRC and it correlates with poor survival of the patients [104]. CircCCDC66 is overexpressed in polyps and colon cancer and it is correlated with poor survival and controls cell proliferation, cell dissemination, and stemness character [105]. GLIS2 was shown to enhance CRC cell motility and activate the NF-κB pathway by sponging miR-671 [106]. CircHIPK3 has been shown to sponge miR7 and activate CRC growth and metastasis [107]. The decreased plasma levels of circ-CCDC66, circ-ABCC1, and circ-STIL in CRC patients [108], and the up-regulated serum level of exosomal circ-PNN (hsa_circ_0101802) in CRC can be used as diagnostic markers in the pathogenesis of CRC [102]. Circ-SMAD7 is having lower expression in CRC and it suppresses the process of EMT [109] while circVAPA is overexpressed in CRC and plays an oncogenic role by sponging miR-101 [110]. Exosomal circPACRGL derived from CRC cells has been shown to play an oncogenic role and it induces proliferation and metastasis [111] and the CircRNA circDENND4C has been shown to induce cell proliferation, glycolysis, and migration in CRC cells by sponging miR-760 and regulating GLUT1 [112]. Circ-0004277 promotes cell proliferation of CRC cells by sponging off miR-512-5p and upregulation of PTMA expression [113] and hsa_circ_0000523 regulates cell proliferation, and apoptosis by sponging of miR-31, and regulation of Wnt/β-catenin signaling pathway [114]. Circular RNA hsa_circ_0008285 inhibits PI3K/AKT pathway through miR-382-5p/PTEN axis and acts as a tumor suppressor [115] and CircPIP5K1A is overexpressed in CRC and regulates cell proliferation and dissemination through circPIP5K1A-miR-1273a axis [116]. CircPTK2 plays an important role in CRC growth and metastasis [117] and circCAMSAP1/miR-328-5p/E2F1 axis is involved in CRC carcinogenesis [118]. CircMAT2B is upregulated in CRC and promotes cell proliferation by the regulation of miR-610/E2F1 axis [119] and circ-NSD2 plays an oncogenic role and is involved in cancer metastasis through circ-NSD2/miR-199b-5p/DDR1/JAG1 axis [120]. hsa_circRNA_102958 plays an oncogenic role in CRC through miR-585/CDC25B axis and promotes tumorigenesis of CRC [121] while circ_0002138 is downregulated in CRC and inhibits cell proliferation in vitro [122]. Circular RNA circ_0007142 modulates CDC25A expression through miR-122-5p and facilitates CRC progression [123] and circular RNA, NOX4 promotes CRC development through miR‑485‑5p/CKS1B axis [124]. hsa_circ_0053277 upregulates MMP14 through miR-2467-3p and promotes CRC development [113]. circ-FARSA is upregulated in CRC and its suppression leads to inhibition of cell proliferation and dissemination and it acts as a sponge for miR-330-5p and regulates the expression of LIM and LASP1 proteins [125]. Circ-001971/miR-29c-3p axis regulates CRC cell dissemination, and angiogenesis [126], and circCTNNA1 is shown to sponge miR-149-5p and regulate the expression of FOXM1 leading to CRC progression [127]. Circular RNA hsa_circ_0007142 is upregulated in CRC and promotes tumorigenesis by targeting miR-103a-2-5p [123] and circular RNA CDR1‑AS has been shown to increase the expression of PD‑L1 in CRC cells [128]. The ratio of circRNAs and linear RNA is lower in CRC tissues as suggested by RT-qPCR and the data set analysis studies suggest that there are 39 circRNAs that have significant abnormal expression, 11 circRNAs are overexpressed and 28 are downregulated in cancers which include CRC [129]. circRNAs are also present in exosomes of CRC cell lines and the number of circRNAs is higher in the exosomes compared to the cells. Some of the important circRNAs having abnormal expressions in exosomes are circRTN4, circARHGAP5, circFAT1, circHIPK3, and circMAN1A2. Hence the circRNAs have promising potential to be used as biomarkers in the future [130].
Extracellular vesicular miRNAs as CRC biomarkers
National Cancer Institute defines biomarker as “A biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease”. Biomarkers are used to monitor the risk assessment, evaluation of the response to the drugs, screening of the disease, prognosis determination, evaluation of disease progression, and differential diagnosis. Since the biomarkers play a very important role in all the stages of the disease, they must undergo a rigorous evaluation before their application in the clinic. The screening methods most commonly used for CRC diagnosis are colonoscopy, fecal occult blood testing, and fecal immunochemical testing [131] which are limited in sensitivity in early stages of carcinogenesis and hence the development of more sensitive, and less invasive methods for the detection of early stages of CRC might be very helpful for the disease management. Liquid biopsy can be a promising screening and diagnostic method for the detection of CRC in the early stages and the extracellular vesicular (EV) miRNAs that are specific to the tumors can be a good candidate for the detection of early stages of cancer compared to the total circulating miRNAs. EVs can be isolated based on the expression of the surface markers which might also increase their specificity as cancer biomarkers. The EVs that have CD147 expression on their membrane are increased in the blood circulation of CRC patients compared to the normal controls and EpCAM and A33 expressions on the EVs are also increased in CRC patients. Lower levels of miR-4772 are associated with CRC recurrence, miR-27a and miR-130a are associated with poor outcomes in terms of 5-year survival, and miR30 in EVs is associated with the metastatic progression of the CRC tumors [132]. Some of the important EV-miRNAs associated with CRC are summarized in Table 2.
Noncoding RNAs other than miRNAs have also been reported in the EVs of CRC patients such as long non-coding RNA CRNDE and CCAT1 are upregulated in CRC patients [139] compared to the normal mucosa. circRNA (circ_001988) is dysregulated in CRC tissues compared to the normal mucosa and in the different stages of CRC. SNORA42 is highly expressed in CRC tumors and is associated with poor survival and piR-019825 in the EVs along with other miRNAs can differentiate the early stages of CRC from healthy individuals. Sometimes, a single non-coding RNA cannot be a useful diagnostic or prognostic marker yet combining several miRNAs or EV-miRNAs as a panel of biomarkers provides a better result.
In CRC, NEAT_v2 non-coding RNA, SEPT9 methylated DNA, and SDC2 methylated DNA are the very potent nucleic acid biomarkers, interleukin 8 is the most promising cytokine marker, and CA11-19 glycoprotein and DC-SIGN/DC-SIGNR are the most promising protein biomarkers. By combining these emerging markers with the fecal immunochemical test, the sensitivity can be highly enhanced to detect CRC. SIGN/DC-SIGNR protein, methylated SDC2, and methylated SEPT9 had better specificity and sensitivity compared to CEA or CA 19–9 but these markers need to be studied in a larger population to strongly establish and implement them in the clinics [140]. Bioinformatic analysis of The Cancer Genome Atlas (TCGA) has predicted a set of five miRNAs, hsa-miR-5091, hsa-miR-10b-3p, hsa-miR-9-5p, hsa-miR-187-3p, and hsa-miR-32-5p as a prognostic biomarker which was associated with the survival of the patients [141]. In a study consisting of six patients with and without recurrent CRC, miRNAs from the exosomes of the serum were extracted and miRNA microarray was performed to be found that exosomal miR-17-92a was correlated with the recurrence of CRC. Exosomal miR-19a was significantly increased in the serum of the CRC patients [142] and MicroRNA-92a was predicted to be a novel and potential diagnostic biomarker in CRC [143].
Therapeutic targeting of ncRNAs
miRNAs can act as a promising therapeutic target for cancer treatment by manipulating the tumor suppressor miRNAs that regulate the cell signaling networks [144]. Tumor suppressor miRNAs (mimics) have been designed to manage the developing tumors. The transfection of let-7 mimic in CRC cells has shown an increase in apoptosis and a decrease in the proliferation of cancer cells [145]. Transfection of miR-34a into HCT 116 and RKO cells leads to inhibition of cell proliferation and induction of senescence in the cells. [146]. miRNA replacement therapy by the efficacious delivery of PEI/miR-145 and PEI/miR-33a complexes in colon carcinoma model of mice has proved it to be a robust and safe preclinical strategy and it holds a lot of potential in humans as the systemic and local introduction of miRNA has resulted in the delivery of the intact miRNAs to the tumors [147].
Conclusions
Non-coding RNAs are emerging as important prognostic and diagnostic tools for the management of CRC. miRNAs and piRNAs are the most well-explored noncoding RNAs in cancer compared to other non-coding RNAs, and the biology of tsRNAs and pseudogenes is yet to be fully understood. A better understanding of the biological mechanism of synthesis and interactions of these noncoding RNAs will open new windows for their better utilization in cancer therapy and personalized medicines. Because of sample-to-sample variations, differences in the efficiency of different protocols for miRNA isolation, and the inherent instability and small content of noncoding RNAs, utilizing a panel of noncoding RNAs or noncoding RNAs combined with traditional methods of screening and detection might give better efficiency and efficacy in the management of CRC.
Data availability
Not applicable.
Code availability
Not applicable.
References
Arnold M, Sierra MS, Laversanne M et al (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut 66:683–691. https://doi.org/10.1136/gutjnl-2015-310912
Dozois EJ, Boardman LA, Suwanthanma W et al (2008) Young-onset colorectal cancer in patients with no known genetic predisposition: can we increase early recognition and improve outcome? Medicine (Baltimore) 87:259–263. https://doi.org/10.1097/MD.0b013e3181881354
Arnold M, Soerjomataram I, Ferlay J, Forman D (2015) Global incidence of oesophageal cancer by histological subtype in 2012. Gut 64:381–387. https://doi.org/10.1136/gutjnl-2014-308124
Schatoff EM, Leach BI, Dow LE (2017) Wnt signaling and colorectal cancer. Curr Colorectal Cancer Rep 13:101–110. https://doi.org/10.1007/s11888-017-0354-9
Loboda A, Nebozhyn MV, Watters JW et al (2011) EMT is the dominant program in human colon cancer. BMC Med Genomics 4:9. https://doi.org/10.1186/1755-8794-4-9
Kishore C (2021) Epigenetic regulation and promising therapies in colorectal cancer. Curr Mol Pharmacol 14:5. https://doi.org/10.2174/1874467214666210126105345
Kishore C, Sundaram S, Karunagaran D (2019) Vitamin K3 (menadione) suppresses epithelial-mesenchymal-transition and Wnt signaling pathway in human colorectal cancer cells. Chem Biol Interact 309:108725. https://doi.org/10.1016/j.cbi.2019.108725
Kishore C, Bhadra P (2021) Current advancements and future perspectives of immunotherapy in colorectal cancer research. Eur J Pharmacol 893:173819. https://doi.org/10.1016/j.ejphar.2020.173819
Deng X, Su R, Weng H et al (2018) RNA N 6 -methyladenosine modification in cancers: current status and perspectives. Cell Res 28:507–517. https://doi.org/10.1038/s41422-018-0034-6
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12:861–874. https://doi.org/10.1038/nrg3074
Slack FJ, Chinnaiyan AM (2019) The role of non-coding RNAs in oncology. Cell 179:1033–1055. https://doi.org/10.1016/j.cell.2019.10.017
MacFarlane L-A, Murphy PR (2010) MicroRNA: biogenesis, function and role in cancer. Curr Genomics 11:537–561. https://doi.org/10.2174/138920210793175895
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. https://doi.org/10.1038/nature02871
Yu X, Xie Y, Zhang S et al (2021) tRNA-derived fragments: mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 11:461–469. https://doi.org/10.7150/thno.51963
Yu M, Lu B, Zhang J et al (2020) tRNA-derived RNA fragments in cancer: current status and future perspectives. J Hematol Oncol 13:121. https://doi.org/10.1186/s13045-020-00955-6
Huang Y, Bai JY, Ren HT (2014) PiRNAs biogenesis and its functions. Bioorg Khim 40:320–326
Fang Y, Fullwood MJ (2016) Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 14:42–54. https://doi.org/10.1016/j.gpb.2015.09.006
Tutar Y (2012) Pseudogenes. Comp Funct Genomics 2012:e424526. https://doi.org/10.1155/2012/424526
Xiao-Jie L, Ai-Mei G, Li-Juan J, Jiang X (2015) Pseudogene in cancer: real functions and promising signature. J Med Genet 52:17–24. https://doi.org/10.1136/jmedgenet-2014-102785
Meng S, Zhou H, Feng Z et al (2017) CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 16:94. https://doi.org/10.1186/s12943-017-0663-2
Cui B, Chen L, Zhang S et al (2014) MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 124:546–554. https://doi.org/10.1182/blood-2014-03-559690
Liu K, Du J, Ruan L (2017) MicroRNA-21 regulates the viability and apoptosis of diffuse large B-cell lymphoma cells by upregulating B cell lymphoma-2. Exp Ther Med 14:4489–4496. https://doi.org/10.3892/etm.2017.5021
Gabriely G, Yi M, Narayan RS et al (2011) Human glioma growth is controlled by MicroRNA-10b. Cancer Res 71:3563–3572. https://doi.org/10.1158/0008-5472.CAN-10-3568
Roncarati R, Lupini L, Shankaraiah RC, Negrini M (2019) The importance of microRNAs in RAS oncogenic activation in human cancer. Front Oncol. https://doi.org/10.3389/fonc.2019.00988
Calin GA, Cimmino A, Fabbri M et al (2008) MiR-15a and miR-16-1 cluster functions in human leukemia. PNAS 105:5166–5171. https://doi.org/10.1073/pnas.0800121105
Okada N, Lin C-P, Ribeiro MC et al (2014) A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 28:438–450. https://doi.org/10.1101/gad.233585.113
Pekarsky Y, Croce CM (2010) Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget 1:224–227
Liu G-H, Zhou Z-G, Chen R et al (2013) Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol 34:2175–2181. https://doi.org/10.1007/s13277-013-0753-8
Kanaan Z, Roberts H, Eichenberger MR et al (2013) A plasma MicroRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 258:400–408. https://doi.org/10.1097/SLA.0b013e3182a15bcc
Valeri N, Braconi C, Gasparini P et al (2014) MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 25:469–483. https://doi.org/10.1016/j.ccr.2014.03.006
Hollis M, Nair K, Vyas A et al (2015) MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World J Gastroenterol 21:8284–8292. https://doi.org/10.3748/wjg.v21.i27.8284
Ahmed FE, Ahmed NC, Vos PW et al (2013) Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I proof of principle. Cancer Genomics Proteomics 10:93–113
Mlcochova J, Faltejskova-Vychytilova P, Ferracin M et al (2015) MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget 6:38695–38704
Hiyoshi Y, Akiyoshi T, Inoue R et al (2017) Serum miR-143 levels predict the pathological response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer. Oncotarget 8:79201–79211. https://doi.org/10.18632/oncotarget.16760
Perez-Carbonell L, Sinicrope FA, Alberts SR et al (2015) MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer 113:83–90. https://doi.org/10.1038/bjc.2015.168
Nassar FJ, Msheik ZS, Itani MM et al (2021) Circulating miRNA as biomarkers for colorectal cancer diagnosis and liver metastasis. Diagnostics (Basel). https://doi.org/10.3390/diagnostics11020341
Fassan M, Cui R, Gasparini P et al (2019) miR-224 is significantly upregulated and targets caspase-3 and caspase-7 during colorectal carcinogenesis. Trans Oncol 12:282–291. https://doi.org/10.1016/j.tranon.2018.10.013
Zhou J, Hu M, Wang F et al (2017) miR-224 controls human colorectal cancer cell line HCT116 proliferation by targeting Smad4. Int J Med Sci 14:937–942. https://doi.org/10.7150/ijms.19565
Kumar P, Kuscu C, Dutta A (2016) Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci 41:679–689. https://doi.org/10.1016/j.tibs.2016.05.004
Zuo Y, Chen S, Yan L et al (2021) Development of a tRNA-derived small RNA diagnostic and prognostic signature in liver cancer. Genes & Diseases. https://doi.org/10.1016/j.gendis.2021.01.006
Xiong W, Wang X, Cai X et al (2019) Identification of tRNA-derived fragments in colon cancer by comprehensive small RNA sequencing. Oncol Rep 42:735–744. https://doi.org/10.3892/or.2019.7178
Huang B, Yang H, Cheng X et al (2017) tRF/miR-1280 suppresses stem cell–like cells and metastasis in colorectal cancer. Cancer Res 77:3194–3206. https://doi.org/10.1158/0008-5472.CAN-16-3146
Li S, Shi X, Chen M et al (2019) Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer 145:1395–1407. https://doi.org/10.1002/ijc.32245
Tao E-W, Wang H-L, Cheng WY et al (2021) A specific tRNA half, 5’tiRNA-His-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. J Exp Clin Cancer Res 40:67. https://doi.org/10.1186/s13046-021-01836-7
Iwasaki YW, Siomi MC, Siomi H (2015) PIWI-Interacting RNA: its biogenesis and functions. Annu Rev Biochem 84:405–433. https://doi.org/10.1146/annurev-biochem-060614-034258
Weng W, Li H, Goel A (2019) Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochimica et Biophysica Acta (BBA) 1871:160–169. https://doi.org/10.1016/j.bbcan.2018.12.005
Weng W, Liu N, Toiyama Y et al (2018) Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. https://doi.org/10.1186/s12943-018-0767-3
Mai D, Ding P, Tan L et al (2018) PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 8:5213–5230. https://doi.org/10.7150/thno.28001
Wang Z, Yang H, Ma D et al (2020) Serum PIWI-interacting RNAs piR-020619 and piR-020450 are promising novel biomarkers for early detection of colorectal cancer. Cancer Epidemiol Biomarkers Prev 29:990–998. https://doi.org/10.1158/1055-9965.EPI-19-1148
Vychytilova-Faltejskova P, Stitkovcova K, Radova L et al (2018) Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev 27:1019–1028. https://doi.org/10.1158/1055-9965.EPI-18-0318
Chu H, Xia L, Qiu X et al (2015) Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer 121:2044–2052. https://doi.org/10.1002/cncr.29314
Yin J, Qi W, Ji C-G et al (2019) Small RNA sequencing revealed aberrant piRNA expression profiles in colorectal cancer. Oncol Rep 42:263–272. https://doi.org/10.3892/or.2019.7158
Mai D, Zheng Y, Guo H et al (2020) Serum piRNA-54265 is a new biomarker for early detection and clinical surveillance of human colorectal cancer. Theranostics 10:8468–8478. https://doi.org/10.7150/thno.46241
Qu A, Wang W, Yang Y et al (2019) <p>A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer</p>. CMAR 11:3703–3720. https://doi.org/10.2147/CMAR.S193266
Iyer DN, Wan TM-H, Man JH-W et al (2020) Small RNA profiling of piRNAs in colorectal cancer identifies consistent overexpression of pir-24000 that correlates clinically with an aggressive disease phenotype. Cancers 12:188. https://doi.org/10.3390/cancers12010188
Yin J, Jiang X-Y, Qi W et al (2017) piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci 108:1746–1756. https://doi.org/10.1111/cas.13300
Rinn JL, Chang HY (2020) Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem 89:283–308. https://doi.org/10.1146/annurev-biochem-062917-012708
Jiang M-C, Ni J-J, Cui W-Y et al (2019) Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res 9:1354–1366
Xue X, Yang YA, Zhang A et al (2016) LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 35:2746–2755. https://doi.org/10.1038/onc.2015.340
Leucci E, Vendramin R, Spinazzi M et al (2016) Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531:518–522. https://doi.org/10.1038/nature17161
Yari H, Jin L, Teng L et al (2019) LncRNA REG1CP promotes tumorigenesis through an enhancer complex to recruit FANCJ helicase for REG3A transcription. Nat Commun 10:5334. https://doi.org/10.1038/s41467-019-13313-z
Zhou Y, Zhong Y, Wang Y et al (2007) Activation of p53 by MEG3 Non-coding RNA*. J Biol Chem 282:24731–24742. https://doi.org/10.1074/jbc.M702029200
Mondal T, Subhash S, Vaid R et al (2015) MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat Commun 6:7743. https://doi.org/10.1038/ncomms8743
Zhao X, Wang P, Liu J et al (2015) Gas5 Exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol Ther 23:1899–1911. https://doi.org/10.1038/mt.2015.170
Pickard MR, Williams GT (2014) Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat 145:359–370. https://doi.org/10.1007/s10549-014-2974-y
Xu M, Xu X, Pan B et al (2019) LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer 18:135. https://doi.org/10.1186/s12943-019-1063-6
Xiang J-F, Yin Q-F, Chen T et al (2014) Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 24:513–531. https://doi.org/10.1038/cr.2014.35
Cai Z, Cao C, Ji L et al (2020) RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature 582:432–437. https://doi.org/10.1038/s41586-020-2249-1
Ni W, Yao S, Zhou Y et al (2019) Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer 18:143. https://doi.org/10.1186/s12943-019-1079-y
Zhao Y, Li Y, Sheng J et al (2019) P53–R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J Exp Clin Cancer Res 38:379. https://doi.org/10.1186/s13046-019-1375-9
Wang Y, Lu J-H, Wu Q-N et al (2019) LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer 18:174. https://doi.org/10.1186/s12943-019-1105-0
Wu Y, Yang X, Chen Z et al (2019) m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer 18:87. https://doi.org/10.1186/s12943-019-1014-2
Avazpour N, Hajjari M, Kazemi Nezhad SR, Tahmasebi Birgani M (2020) SNHG1 long noncoding RNA is potentially up-regulated in colorectal adenocarcinoma. Asian Pac J Cancer Prev 21:897–901. https://doi.org/10.31557/APJCP.2020.21.4.897
Xu M, Chen X, Lin K et al (2018) The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer 17:141. https://doi.org/10.1186/s12943-018-0894-x
Bian Z, Zhang J, Li M et al (2018) LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res 24:4808–4819. https://doi.org/10.1158/1078-0432.CCR-17-2967
Zhang M, Weng W, Zhang Q et al (2018) The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol 11:113. https://doi.org/10.1186/s13045-018-0656-7
Damas ND, Marcatti M, Côme C et al (2016) SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun 7:13875. https://doi.org/10.1038/ncomms13875
Sun F, Liang W, Qian J (2019) The identification of CRNDE, H19, UCA1 and HOTAIR as the key lncRNAs involved in oxaliplatin or irinotecan resistance in the chemotherapy of colorectal cancer based on integrative bioinformatics analysis. Mol Med Rep 20:3583–3596. https://doi.org/10.3892/mmr.2019.10588
Xu Y, Zhang X, Hu X et al (2018) The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med 24:52. https://doi.org/10.1186/s10020-018-0050-5
Xu W, Zhou G, Wang H et al (2020) Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer 146:2901–2912. https://doi.org/10.1002/ijc.32747
Luo K, Geng J, Zhang Q et al (2019) LncRNA CASC9 interacts with CPSF3 to regulate TGF-β signaling in colorectal cancer. J Exp Clin Cancer Res 38:249. https://doi.org/10.1186/s13046-019-1263-3
Yamada A, Yu P, Lin W et al (2018) A RNA-sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci Rep 8:575. https://doi.org/10.1038/s41598-017-18407-6
Di W, Weinan X, Xin L et al (2019) Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis 10:514. https://doi.org/10.1038/s41419-019-1707-x
Gao Q, Zhou R, Meng Y et al (2020) Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3-c-Myc axis. Oncogene 39:3926–3938. https://doi.org/10.1038/s41388-020-1266-8
Chen X, Zeng K, Xu M et al (2018) SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis 9:982. https://doi.org/10.1038/s41419-018-0962-6
Xu M, Chen X, Lin K et al (2019) lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol 12:3. https://doi.org/10.1186/s13045-018-0690-5
Chen D-L, Lu Y-X, Zhang J-X et al (2017) Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics 7:4836–4849. https://doi.org/10.7150/thno.20942
Yu J, Han Z, Sun Z et al (2018) LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J Exp Clin Cancer Res 37:222. https://doi.org/10.1186/s13046-018-0896-y
Wang X, Lai Q, He J et al (2019) LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci 16:51–59. https://doi.org/10.7150/ijms.27359
Lan Y, Xiao X, He Z et al (2018) Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res 46:5809–5821. https://doi.org/10.1093/nar/gky214
Zhao Y, Du T, Du L et al (2019) Long noncoding RNA LINC02418 regulates MELK expression by acting as a ceRNA and may serve as a diagnostic marker for colorectal cancer. Cell Death Dis 10:568. https://doi.org/10.1038/s41419-019-1804-x
Wang M, Han D, Yuan Z et al (2018) Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis 9:1149. https://doi.org/10.1038/s41419-018-1187-4
Shang A, Wang W, Gu C et al (2020) Long non-coding RNA CCAT1 promotes colorectal cancer progression by regulating miR-181a-5p expression. Aging (Albany NY) 12:8301–8320. https://doi.org/10.18632/aging.103139
Shan Y, Ma J, Pan Y et al (2018) LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis 9:722. https://doi.org/10.1038/s41419-018-0759-7
Jiang L, Zhao X-H, Mao Y-L et al (2019) Long non-coding RNA RP11–468E2.5 curtails colorectal cancer cell proliferation and stimulates apoptosis via the JAK/STAT signaling pathway by targeting STAT5 and STAT6. J Exp Clin Cancer Res 38:465. https://doi.org/10.1186/s13046-019-1428-0
Haddadi N, Lin Y, Travis G et al (2018) PTEN/PTENP1: ‘regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol Cancer 17:37. https://doi.org/10.1186/s12943-018-0803-3
Karreth FA, Reschke M, Ruocco A et al (2015) The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161:319–332. https://doi.org/10.1016/j.cell.2015.02.043
Ye X, Fan F, Bhattacharya R et al (2015) VEGFR-1 pseudogene expression and regulatory function in human colorectal cancer cells. Mol Cancer Res 13:1274–1282. https://doi.org/10.1158/1541-7786.MCR-15-0061
He W, Yu Y, Huang W et al (2020) The pseudogene DUXAP8 promotes colorectal cancer cell proliferation, invasion, and migration by inducing epithelial-mesenchymal transition through interacting with EZH2 and H3K27me3. Onco Targets Ther 13:11059–11070. https://doi.org/10.2147/OTT.S235643
Kristensen LS, Andersen MS, Stagsted LVW et al (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691. https://doi.org/10.1038/s41576-019-0158-7
Chen L-Y, Wang L, Ren Y-X et al (2020) The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer 19:164. https://doi.org/10.1186/s12943-020-01272-9
Xu H, Wang C, Song H et al (2019) RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer 18:8. https://doi.org/10.1186/s12943-018-0932-8
Li X, Wang J, Zhang C et al (2018) Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol 246:166–179. https://doi.org/10.1002/path.5125
Weng W, Wei Q, Toden S et al (2017) Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 23:3918–3928. https://doi.org/10.1158/1078-0432.CCR-16-2541
Hsiao K-Y, Lin Y-C, Gupta SK et al (2017) Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77:2339–2350. https://doi.org/10.1158/0008-5472.CAN-16-1883
Chen J, Yang X, Liu R et al (2020) Circular RNA GLIS2 promotes colorectal cancer cell motility via activation of the NF-κB pathway. Cell Death Dis 11:788. https://doi.org/10.1038/s41419-020-02989-7
Zeng K, Chen X, Xu M et al (2018) CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 9:417. https://doi.org/10.1038/s41419-018-0454-8
Lin J, Cai D, Li W et al (2019) Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin Biochem 74:60–68. https://doi.org/10.1016/j.clinbiochem.2019.10.012
Wang D-K, Chong R-F, Song B-L et al (2020) Circular RNA circ-SMAD7 is downregulated in colorectal cancer and suppresses tumor metastasis by regulating epithelial mesenchymal transition. Eur Rev Med Pharmacol Sci 24:9241. https://doi.org/10.26355/eurrev_202009_23002
Li X-N, Wang Z-J, Ye C-X et al (2019) Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother 112:108611. https://doi.org/10.1016/j.biopha.2019.108611
Shang A, Gu C, Wang W et al (2020) Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer 19:117. https://doi.org/10.1186/s12943-020-01235-0
Zhang Z-J, Zhang Y-H, Qin X-J et al (2020) Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur Rev Med Pharmacol Sci 24:2387–2400. https://doi.org/10.26355/eurrev_202003_20506
Yang L, Sun H, Liu X et al (2020) Circular RNA hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-512-5p to upregulate the expression of PTMA. J Cell Physiol. https://doi.org/10.1002/jcp.29484
Jin Y, Yu LL, Zhang B et al (2018) Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res 51:e7811. https://doi.org/10.1590/1414-431X20187811
Wang J, Luo J, Liu G, Li X (2020) Circular RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation and migration via the miR-382-5p/PTEN axis. Biochem Biophys Res Commun 527:503–510. https://doi.org/10.1016/j.bbrc.2020.03.165
Zhang Q, Zhang C, Ma J-X et al (2019) Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol 25:5300–5309. https://doi.org/10.3748/wjg.v25.i35.5300
Yang H, Li X, Meng Q et al (2020) CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 19:13. https://doi.org/10.1186/s12943-020-1139-3
Zhou C, Liu H-S, Wang F-W et al (2020) circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol Ther 28:914–928. https://doi.org/10.1016/j.ymthe.2019.12.008
Zhao JP, Chen LL (2020) Circular RNA MAT2B induces colorectal cancer proliferation via sponging miR-610, resulting in an increased E2F1 expression. Cancer Manag Res 12:7107–7116. https://doi.org/10.2147/CMAR.S251180
Chen L-Y, Zhi Z, Wang L et al (2019) NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol 248:103–115. https://doi.org/10.1002/path.5238
Li R, Wu B, Xia J et al (2019) Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res 11:6887–6893. https://doi.org/10.2147/CMAR.S212180
Ruan H, Deng X, Dong L et al (2019) Circular RNA circ_0002138 is down-regulated and suppresses cell proliferation in colorectal cancer. Biomed Pharmacother 111:1022–1028. https://doi.org/10.1016/j.biopha.2018.12.150
Yin W, Xu J, Li C et al (2020) Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR-122-5p. Onco Targets Ther 13:3689–3701. https://doi.org/10.2147/OTT.S238338
Wang X, Tao G, Huang D et al (2020) Circular RNA NOX4 promotes the development of colorectal cancer via the microRNA-485-5p/CKS1B axis. Oncol Rep 44:2009–2020. https://doi.org/10.3892/or.2020.7758
Lu C, Fu L, Qian X et al (2020) Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330-5p/LASP1 axis. Arch Biochem Biophys 689:108434. https://doi.org/10.1016/j.abb.2020.108434
Chen C, Huang Z, Mo X et al (2020) The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res 39:91. https://doi.org/10.1186/s13046-020-01594-y
Chen P, Yao Y, Yang N et al (2020) Circular RNA circCTNNA1 promotes colorectal cancer progression by sponging miR-149-5p and regulating FOXM1 expression. Cell Death Dis 11:557. https://doi.org/10.1038/s41419-020-02757-7
Tanaka E, Miyakawa Y, Kishikawa T et al (2019) Expression of circular RNA CDR1-AS in colon cancer cells increases cell surface PD-L1 protein levels. Oncol Rep 42:1459–1466. https://doi.org/10.3892/or.2019.7244
Geng Y, Jiang J, Wu C (2018) Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. https://doi.org/10.1186/s13045-018-0643-z
Zhang H, Jiang L-H, Sun D-W et al (2018) CircRNA: a novel type of biomarker for cancer. Breast Cancer 25:1–7. https://doi.org/10.1007/s12282-017-0793-9
Strul H, Arber N (2007) Screening techniques for prevention and early detection of colorectal cancer in the average-risk population. Gastrointest Cancer Res 1:98–106
Desmond BJ, Dennett ER, Danielson KM (2019) Circulating extracellular vesicle microRNA as diagnostic biomarkers in early colorectal cancer—a review. Cancers (Basel). https://doi.org/10.3390/cancers12010052
Liu Y, Liu R, Yang F et al (2017) miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer 16:53. https://doi.org/10.1186/s12943-017-0625-8
Zhu M, Huang Z, Zhu D et al (2017) A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 8:17081–17091. https://doi.org/10.18632/oncotarget.15059
Karimi N, Ali Hosseinpour Feizi M, Safaralizadeh R et al (2019) Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J Chin Med Assoc 82:215–220. https://doi.org/10.1097/JCMA.0000000000000031
Peng Z-Y, Gu R-H, Yan B (2018) Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem. https://doi.org/10.1002/jcb.27291
Wang J, Yan F, Zhao Q et al (2017) Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep 7:4150. https://doi.org/10.1038/s41598-017-04386-1
Shirafkan N, Mansoori B, Mohammadi A et al (2018) MicroRNAs as novel biomarkers for colorectal cancer: new outlooks. Biomed Pharmacother 97:1319–1330. https://doi.org/10.1016/j.biopha.2017.11.046
Ding J, Li J, Wang H et al (2017) Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis 8:e2997–e2997. https://doi.org/10.1038/cddis.2017.328
Nikolaou S, Qiu S, Fiorentino F et al (2018) Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol 22:481–498. https://doi.org/10.1007/s10151-018-1820-3
Yang G, Zhang Y, Yang J (2019) A five-microRNA signature as prognostic biomarker in colorectal cancer by bioinformatics analysis. Front Oncol 9:1207. https://doi.org/10.3389/fonc.2019.01207
Matsumura T, Sugimachi K, Iinuma H et al (2015) Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 113:275–281. https://doi.org/10.1038/bjc.2015.201
Yang X, Zeng Z, Hou Y et al (2014) MicroRNA-92a as a potential biomarker in diagnosis of colorectal cancer: a systematic review and meta-analysis. PLoS ONE 9:e88745. https://doi.org/10.1371/journal.pone.0088745
Henry JC, Azevedo-Pouly ACP, Schmittgen TD (2011) MicroRNA replacement therapy for cancer. Pharm Res 28:3030–3042. https://doi.org/10.1007/s11095-011-0548-9
Akao Y, Nakagawa Y, Naoe T (2006) let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 29:903–906. https://doi.org/10.1248/bpb.29.903
Tazawa H, Tsuchiya N, Izumiya M, Nakagama H (2007) Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA 104:15472–15477. https://doi.org/10.1073/pnas.0707351104
Ibrahim AF, Weirauch U, Thomas M et al (2011) MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res 71:5214–5224. https://doi.org/10.1158/0008-5472.CAN-10-4645
Acknowledgements
Not applicable.
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
CK wrote the original draft of the manuscript and conducted the literature research, DK critically revised the final draft and supervised the manuscript write-up.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kishore, C., Karunagaran, D. Non-coding RNAs as emerging regulators and biomarkers in colorectal cancer. Mol Cell Biochem 477, 1817–1828 (2022). https://doi.org/10.1007/s11010-022-04412-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04412-5