Abstract
Circular RNAs (circRNAs) are a class of long, non-coding RNAs molecules that shape a covalently closed continuous loop which have no 5′–3′ polarity and contain no polyA tail. CircRNAs also possess relatively jarless framework and are highly tissue-specific expressed in the eukaryotic transcriptome. Emerging evidences have discovered that thousands of endogenous circRNAs are present in mammalian cells and they mediate gene expression at the transcriptional or post-transcriptional level by binding to microRNAs or other molecules and then inhibit their function. Similarly, increasing evidence indicates that circRNAs may play a role in the development of several types of diseases, including atherosclerotic vascular disease risk, neurological disorders, prion diseases, osteoarthritis and diabetes. Furthermore, circRNAs exhibit aberrant expression in multiform types of cancer, including colorectal cancer, hepatocellular carcinoma and pancreatic ductal adenocarcinoma. And based on the function of circRNAs in cancer, we believe that circRNAs may serve as diagnostic or tumor promising biomarkers. Moreover, it will provide a new therapeutic target for the treatment of cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Earlier, circRNAs were found and considered to have no biological function [1]. However, with the development of RNA deep sequencing technology and bioinformatics analyses, it has been reported that parts of circRNAs are endogenous, abundant, conserved and jarless in mammalian cells and own physiological functions [2,3,4,5]. Circular RNAs (circRNAs), a novel type of RNA, are a peculiar group of long, non-coding endogenous RNAs molecules, consisting of at least a few hundred nucleotides [5, 6] that regulate gene expression at the transcriptional or post-transcriptional level by binding to microRNAs (miRNAs) or other molecules [7,8,9]. CircRNAs shape a covalently closed continuous loop which own relatively jarless framework in the eukaryotic transcriptome [6, 10]. Different from linear RNAs, circRNAs are relatively stable and are not easily degraded by endonuclease [6, 11,12,13]. In addition, circRNAs are produced mainly by exons and introns sequences, but the reverse complementary sequences or RNA-binding proteins (RBPs) are essential for circRNA formation [14]. Moreover, two independent groups discovered that circRNAs could sponge miRNAs, which is a new mechanism that circRNAs have the ability to bind to miRNAs and consequently regulate miRNA function [5, 15]. MiRNAs are a class of small, highly conserved, and non-coding RNAs that regulate gene expression at posttranscriptional level by binding to the 3′ untranslated region (UTR) of target mRNAs [16]. Growing evidence has confirmed that miRNAs play a role in multiform biological processes, which have a significant correlation with cancers, consisting of tumor proliferation, apoptosis, differentiation, invasion, and metastasis, as well as tumorigenesis [16,17,18]. Furthermore, increasing evidence also indicates that circRNAs may play a role in the development of some diseases, including atherosclerotic vascular disease risk, neurological disorders, prion diseases, osteoarthritis, diabetes, Parkinson’s disease, Alzheimer’s disease, multiple sclerosis, and schizophrenia [9, 14, 19,20,21,22]. However, recent studies have discovered that circRNAs may be drawn into multiform types of cancer, including colorectal cancer (CRC), hepatocellular carcinoma (HCC), breast cancer and pancreatic ductal adenocarcinoma (PDAC) [6, 9, 23,24,25,26,27]. With the exception of the peculiar expression in tissues, circRNAs are also found in saliva [25] and exosomes [28], and it is believed that circRNAs are also regarded as a biological governor. Altogether, circRNAs have the great potential to regulate the biological function and act as significant biomarkers to predict disease progression and prognosis, especially in tumor. Therefore, in this review, we briefly discuss the latent function of circRNA and its relationship with tumor so that we can provide a better therapeutic target for the treatment of cancer.
The biogenesis and function of circRNAs
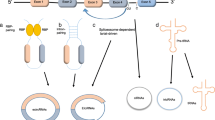
Latest studies have shown that the biogenesis of circRNAs occurs by backsplicing unlike the canonical linear RNAs splicing [29], and circRNAs are considered as co-transcriptional products. Recent studies have shown that the circRNAs are ubiquitous in eukaryotic cells and mostly transcribed from protein-coding genes by RNA polymerase II [30, 31]. Precursor messenger RNAs (pre-mRNAs) contain exons and introns, and they are first synthesized in the nucleus [32], and then pre-mRNAs transfer to the cytoplasm that they are cut into different complete separate introns or exons. However, circRNAs can be engendered mostly by a realignment of exons due to the diversity of splicing, which is called “head-to-tail” or backsplicing [2, 5, 7]. The backsplicing process comprises RNA circularization that is facilitated via covalent attachment between a downstream splice donor site (5′ splice site) and an upstream acceptor splice site (3′ splice site). In addition, researchers find that the RNA-binding protein Muscleblind can bind to circMbl flanking introns to stimulate circRNA biogenesis [31, 33]. Furthermore, a recent study also shows that the major circRNA biogenesis is mediated by epithelial mesenchymal transition (EMT) [34]. The EMT could lose tumor cells epithelial features transiently, including the loss of apico-basal polarity, disassembly of tight junctions, and then obtain mesenchymal traits, therefore, circRNA may be correlated with cell invasion and metastasis [35]. Besides, it is also found that circRNAs can act as miRNA and latent ceRNAs (competitive endogenous RNAs) molecules [5, 7, 36], the ceRNAs contain miRNA response elements, such as mRNAs, pseudogenes and long non-coding RNAs (lncRNAs), and they also compete with miRNA binding sites. Furthermore, Circular RNAs (circRNAs) consist of intronic circRNA and exonic circRNA; intronic circRNA has the capability to affect RNA-mediated inheritance and epigenetics in the cytoplasm; exonic circRNAs could interact with miRNA sponges in the cytoplasm and mediate miRNAs function [34], such as, circRNAs ciRS-7/CDR1as (for circular RNA sponge for miR-7 or CDR1 antisense) and Sry may serve as miRNAs sponges [37]. The cerebellar degeneration related protein 1 (CDR1) gene could translate a natural circular antisense transcript, which is called antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as) [38]. CDR1as expression is high in the brain and contains at least 60 binding sites for miR-7 [7, 39]. Furthermore, it is also shown that CDR1as may be connected with Parkinson’s disease, Alzheimer’s disease (AD) and brain development [21, 39, 40]. The sex-determining region Y (Sry) gene could mediate the mammalian sex determination and contains 16 conserved binding sites for miR-138 [7, 34, 41]. Most importantly, miR-7 and Sry have been confirmed playing a role in the occurrence and progression of cancer; consequently, it implies that circRNAs may mediate physiological and pathological processes by binding to miRNAs (Fig. 1).
The biogenesis and function of circRNAs. a The circRNAs are widespread existence in eukaryotic cells and mostly transcribed from protein-coding genes by RNA polymerase II. The backsplicing process comprises RNA circularization promoted via covalent joining between a downstream splice donor site (5′ splice site) and an upstream acceptor splice site (3′ splice site). Moreover, circRNAs could function as miRNA sponges in the cytoplasm, such as, CDR1as contains over 60 binding sites for miR-7, or ciR-Sry contains 16 conserved binding sites for miR-138. b Taking hsa_circ_0045324 and hsa_circ_0069626 qPCR primers for example to detect their back-splice junction
Circular RNA in alimentary canal tumor
Esophageal cancer is the eighth most common cancer and the sixth leading cause of mortality in the word [42,43,44], and one of the subtypes is esophageal squamous cell carcinoma (ESCC). Based on the latest study, it is found that circ_0067934 expression is significantly higher in ESCC than normal tissues. The circRNA hsa_circ_0067934 is located at chromosomal region 3q26.2 that consists of two exons joining through back-splicing [45]. Significantly, it is also found that circ_0067934 expression is concerned with ESCC differentiation, T stage, and TNM stage, and the lower differentiated tumors, the higher expression levels of circ_0067934. However, the circ_0067934 expression levels are not concerned with tumor location or size and lymph node metastasis [45]. Furthermore, circ_0067934 could boost ESCC cell proliferation and migration in vitro and si-circ_0067934 could arrest cell cycle in G2 phase. In addition, another research shows that hsa-circ_002059 expression is significantly lower in gastric cancer than normal tissues [6]. Furthermore, circ_002059 expression levels in the plasma of postoperative gastric cancer patients are significantly higher than those of preoperative gastric cancer patients. More importantly, it is further found that circ_002059 expression levels are significantly concerned with gastric cancer patients’ distant metastasis, TNM stage, gender and age [6, 46]. Colorectal cancer (CRC) is the third most common cancer in the world and its prognosis rests with tumor stage. By using RT-qPCR, it is found that the ratios of circRNA/linear RNA are lower in colorectal cancer tissues. Furthermore, through data set analysis, it is found that there are 39 circRNAs significant, abnormal expressions, and eleven circRNAs of them are overexpressions and twenty-eight of them are downexpressions in cancer [23]. In addition, several studies also show that circRNAs could be detected in the exosomes of colon cancer cell lines [25, 28]; surprisingly, circRNAs tend to be more enriched in exosomes than cells. Subsequently, it is revealed that circFAT1, circHIPK3, circARHGAP5, circRTN4, and circMAN1A2 are proved to be existent in exosomes and abnormal expression [47]. Therefore, it is suggested that circRNAs may have the potential ability to participate in oncogenesis (see Fig. 2).
The function of circ-Foxo3. Circ-Foxo3 is highly expressed in non-cancer cells and associated with cell proliferation, apoptosis and cell cycle. Circ-Foxo3 regulates cell cycle by combining with CDK2 and p21, which hinders the combination between CDK2 and cyclin E. Meanwhile, circ-Foxo3 decreases p53 expression and increases Foxo3 expression, which contributes to cell apoptosis and inhibiting proliferation
Circular RNA in hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the third leading cause of mortality in the word, and most HCC patients have a poor prognosis because of its high rate of metastasis and recurrence [42, 48,49,50]. However, circRNAs are connected with HCC according to latest research. It is found that circ_0001649 expression is lower in HCC than the adjacent tissues, and its expression is related to tumor size and tumor embolus. Significantly, the larger the tumor size, the lower the expression of circ_0001649. Meanwhile, the expressions of MMP-9, 10, and 13 are significantly increased when si-circ_0001649 is transfected into HCC cells. Therefore, the implication is that circ_0001649 may be involved in tumor growth and metastasis [26]. Similarly, we also find that circ_0000520, circ_0005075 and circ_0066444 are also significantly and abnormally expressed in HCC tissues through high-throughput circRNA microarray. However, only circ_0005075 expression is upregulated [51]. Furthermore, circ_0005075 expression is also associated with tumor size. Notably, it is found that circHIPK3 is significantly overexpressed in liver cancer than normal tissues [27]. CircHIPK3 is abundant and constant in the cytoplasm, which only contains a large second exon from the HIPK3 gene. Furthermore, circHIPK3 is found to sponge to 9 miRNAs, including miR-124, miR-152, miR-193a, miR-29a, miR-29b, miR-338, miR-379, miR-584 and miR-654. Notably, it is shown that circHIPK3 directly combines with miR-124 and suppresses miR-124 function [27]. MicroRNA-7 (miR-7) is one of the endogenous non-coding RNA molecules, containing 23 nucleotides and is regarded as a crucial tumor suppressor. Furthermore, miR-7 is often down-regulated in various tumors and connected with the tumor cell proliferation, invasion and apoptosis [52,53,54,55,56]. With the progress of exploring circRNAs, it is found that circRNA-ITCH is the sponge for miR-7, miR-17, and miR-214 [20]. CiRS-7 (also termed as Cdr1as) is the sponge for miR-7 in the embryonic zebrafish midbrain and islet cells [5]. Meanwhile, it is also shown that ciRS-7 expression is significantly higher in HCC than normal tissues, and overexpression of ciRS-7 could increase the HCC cells’ proliferation [57]. However, according to another research, there is no significant difference of ciRS-7 expression levels between HCC and normal tissues, which needs to be further gone into [58]. Yet ciRS-7 expression is significantly connected with the serum AFP, and hepatic MVI of HCC patients. Furthermore, ciRS-7 expression with concurrent MVI is negatively related to miR-7 and synergistically related to PIK3CD and p70S6K, which are the targets of miR-7. Therefore, the implication is that ciRS-7 will become a novel biomarker of hepatic MVI [58].
The function of circ-Foxo3
The Foxo3 gene encodes circular Foxo3 (circ-Foxo3) and linear Foxo3 [59]. Previous research has confirmed that Foxo3 acts as a tumor suppressor and correlates with cancer progression [60]. Furthermore, Foxo3 could regulate cell function by mediating Akt, phosphatase and tensin homolog (PTEN) and cyclins and cyclin-dependent kinases (CDKs) [61, 62]. The transcript of the Foxo3 comprises of 7341 nucleotides, but the nucleotides Foxo3 circular RNA only comprises of 1435 nucleotides. However, there are 25 identical binding sites among circ-Foxo3, Foxo3P and Foxo3. It implies that these sequences may play a crucial biological role [63]. Based on further experiments, it is found that the circ-Foxo3 is highly expressed in non-cancer cells and associated with cell proliferation and cell cycle. Ectopic expression of circ-Foxo3 inhibits cell proliferation, promotes cell apoptosis and arrests cell cycle in the G1 phase. Furthermore, we also find that circ-Foxo3 inhibits cell cycle progression by combining with the cell cycle proteins, cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (or p21). Cyclins and cyclin-dependent kinases (CDKs) are two types of cell cycle regulators. CDK2 is an essential regulator in the G1-S phase transition. P21Cip1/Waf1 and p27Kip1 are two very important cyclin-dependent kinase (CDK) inhibitors, which negatively mediate G0/G1 phase progression [64]. Circ-Foxo3 acts on p21 and CDK2 to form circ-Foxo3–p21–CDK2 ternary complex which hinders the combination between CDK2 and cyclin E, which hinders cell transition from G1 into S phase [65]. Meanwhile, it is also found that circ-Foxo3 could induce cell apoptosis. It is well known that p53 acts as an admitted mediator to induce cell cycle arrest and contributes to DNA repair or DNA damage caused apoptosis [66]. Circ-Foxo3 promotes the combination between MDM2 and p53, which contributes to MDM2-induced p53 ubiquitination and degradation, leading to inhibiting the expression of p53. Similarly, circ-Foxo3 decreases MDM2-induced Foxo3 ubiquitination and degradation, which leads to increasing Foxo3 expression that serves as a trigger for apoptosis by up-regulating the expression of Puma gene [67, 68]. To sum up, circ-Foxo3 could inhibit tumor cell proliferation, induce apoptosis and arrest cell cycle in G1 phase.
Conclusion
In this review, it is learned that circRNAs could be regarded as a novel biomarker in cancer development and progression. Indeed, it has been confirmed that circRNAs are involved in different types of cancer, including esophageal cancer, gastric cancer and hepatocellular carcinoma. Furthermore, circRNAs could regulate the biological function by sponge for miRNA. However, further research will be needed to reveal the biological functions of the majority of circRNAs in terms of both physiological and pathological processes so that it can be applied to clinical use in future.
References
Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–60.
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733.
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777.
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–6.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409.
Nair AA, Niu N, Tang X, et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7(49):80967–79.
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–8.
Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32(7):923–5.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57.
Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63.
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–8.
Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51.
Zhang HD, Jiang LH, Sun DW, Li J, Tang JH. MiR-139-5p: promising biomarker for cancer. Tumour Biol. 2015;36(3):1355–65.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Liu M, Huang F, Zhang D, Ju J, Wu XB, Wang Y, et al. Heterochromatin protein HP1γ promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75(21):4593–604.
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233.
Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–13.
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283.
Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–12.
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–80.
Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–30.
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–9.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4.
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47.
Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer. Oncol Rep. 2015;33(6):2669–74.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Zhou H, Arcila ML, Li Z, Lee EJ, Henzler C, Liu J, et al. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012;40(13):5864–75.
Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159(1):13–4.
Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2015;9:1–8.
Chang CW, Yu JC, Hsieh YH, Yao CC, Chao JI, Chen PM, et al. MicroRNA-30a increases tight junction protein expression to suppress the epithelial-mesenchymal transition and metastasis by targeting Slug in breast cancer. Oncotarget. 2016;7(13):16462–78.
Taulli R, Loretelli C, Pandolfi PP. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol. 2013;20(5):541–3.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61.
Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–22.
Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–65.
Lukiw WJ, Circular RNA. circRNA) in Alzheimer’s disease (AD. Front Genet. 2013;4:307.
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–30.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–52.
Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14(1):225.
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:3.
Zhang M, Du X. Noncoding RNAs in gastric cancer: research progress and prospects. World J Gastroenterol. 2016;22(29):6610–8.
Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982.
Sasaki Y, Yamada T, Tanaka H, Peng YF, Liu WR, Shi GM, et al. Risk of recurrence in a long-term follow-up after surgery in 417 patients with hepatitis B or hepatitis C related hepatocellular carcinoma. Ann Surg. 2006;244(5):771–80.
Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–55.
Llovet JM, Zucman-Rossi J, Pikarsky E, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018.
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore). 2016;95(22):e3811.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, Kolokythas A, et al. microRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432(1):199–205.
Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68(10):3566–72.
Kong D, Piao YS, Yamashita S, Oshima H, Oguma K, Fushida S, et al. Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene. 2012;31(35):3949–60.
Li J, Zheng Y, Sun G, Xiong S. Restoration of miR-7 expression suppresses the growth of Lewis lung cancer cells by modulating epidermal growth factor receptor signaling. Oncol Rep. 2014;32(6):2511–6.
Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347.
Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2016.
Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human fork head genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47(2):187–99.
Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–59.
Cho EC, Kuo ML, Liu X, Yang L, Hsieh YC, Wang J, et al. Tumor suppressor FOXO3 regulates ribonucleotide reductase subunit RRM2B and impacts on survival of cancer patients. Oncotarget. 2014;5(13):4834–44.
Du WW, Yang W, Fang L, Xuan J, Li H, Khorshidi A, et al. miR-17 extends mouse lifespan by inhibiting senescence signaling mediated by MKP7. Cell Death Dis. 2014;5:e1355.
Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919–31.
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014;13:124.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58.
Tokino T, Nakamura Y. The role of p53-target genes in human cancer. Crit Rev Oncol Hematol. 2000;33(1):1–6.
Ekoff M, Kaufmann T, Engstrom M, Motoyama N, Villunger A, Jönsson JI, et al. The BH3-only protein Puma plays an essential role in cytokine deprivation induced apoptosis of mast cells. Blood. 2007;110:3209–17.
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–70.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article does not contain informed consent from all individual participants.
About this article
Cite this article
Zhang, Hd., Jiang, Lh., Sun, Dw. et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer 25, 1–7 (2018). https://doi.org/10.1007/s12282-017-0793-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-017-0793-9