Abstract
The methicillin-resistant Staphylococcus aureus (MRSA) causes serious health problems such as community-acquired infections and nosocomial infections, posing a significant public health threat worldwide. The ability of MRSA to adhere to host tissues and medical implants form a mature biofilm that effectively covers host cells in a polymer-based matrix leads to the deduction in the host-immune defences, antimicrobial therapy and difficulty in disease eradication. The present investigation mainly focusing the isolation of the bacteriocin producing microbes from soil and testify their biofilm inhibition ability against MRSA. Bacteriocin-producing bacteria are identified by utilizing nutritious agar media and the agar well-diffusion bioassay in the presence of MRSA as an indicator organism. The 16S rDNA gene sequencing, molecular weight identification and antibiofilm activity showed the four isolates (PU1, PU2, PU3, and PU4) produce bacteriocin and have a wide-ranging antagonistic activity against MRSA. Among them, PU3 exhibits maximum inhibitory activity against MRSA at 100AU/mL concentration using agar well diffusion method. The biochemical assay and 16S rDNA sequencing confirm the isolate PU3 affirmed as Bacillus cereus with a molecular weight of ~ 3 kDa. In situ zymogram assay showed a zone of inhibition corresponding to the estimated protein band size and Time-kill study also displayed that B. cereus producing metabolites (bacteriocin) comprises a bactericidal effect and 90% reduction in biofilm biomass toward MRSA. This study showed that soil isolated bacteria-based antimicrobial compounds control microbial infections and may instruct future antibacterial research.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biofilm-associated infections are linked to a variety of health problems in both animal and human. The biofilm detachment allows the cells to spread the other body parts by blood and body fluids (David et al. 2013). Most biofilm-forming bacterial strains are highly resistant to antibiotics and host-innate defences (Alejandro et al. 2013). In addition, Klebsiella pneumoniae (Liu et al. 2020), Staphylococcus aureus (Gomes 2019), and Pseudomonas aeuroginosa (Eladawy et al. 2019) are among the most common biofilm-producing bacteria. Gram-positive methicillin-resistant Staphylococcus aureus (MRSA), a causative agent for nosocomial infections anchored within the extracellular hydrated polymeric matrix and promoted increases death rates in patients (Tong et al. 2015). The antibiofilm reasons for antimicrobial substance mostly speak to in two viewpoints, for instance, to repress the improvement of biofilm and dispense with biofilm arrangement and expelling the made biofilm. Fundamentally, the examines reports that antimicrobial substances can suppress the biofilm arrangements since they can successfully execute planktonic microscopic organisms (Pinto et al. 2019; Strempel et al. 2014). Recently, certain bacteriocins consist of antibiofilm capabilities used to deconstruct biofilm development, impede grip, and prevent biofilm formation (Harsh et al. 2018). The newly developed bacteriocin, named Auranofin (AF), is a blended ligand gold compound to treat the antimicrobial and hostile to parasitic action from biofilm formation (She et al. 2019). Bacteriocins are a bacterial source, heat-stable peptides with low sub-atomic weight and sensitivity to proteolytic agents (Karthikeyan and Santhosh 2009; O’sullivan et al. 2003). Bacteriocin ordinarily shows its antimicrobial activity by meddling with the cell divider or film of target living beings, either restraining the cell divider blend or premise pore arrangement, resulting in death (Lopez and Belloso 2008). Bacteriocins are proteins or protein mixes with bactericidal movement coordinated against common species, especially with maker microorganisms. They are heterogeneous and exacerbate that shift in atomic weight, biochemical properties, movement spectra, and activity instrument (Klaenhammer 1988). The activity component of lantibiotic nisin and pediocin-like bacteriocins relies on the cell film protein, layer potential, pH, lipid arrangement, and explicit amino acids and essential spaces (Montvillle and Chen 1998). The bactericidal activities of bacteriocin were examined Vidhya Prakash et al., which L. fermentum and L. casei 335 framed demonstrated compelling against anti-toxin safe E.coli and tranquillize safe Salmonella typhi microorganisms (Prakash et al. 2018). A notable, microscopic organism of the Bacillus sort has solid flexibility to various conditions and that few species produce profoundly safe spores; they have been segregated from fish (Aly et al. 2008; Ghosh et al. 2002; Sugita et al. 1998), delicate corals and wipes (Ivanova et al. 1999) just as other marine spineless creatures (Khandeparker and Anil 2003; Miyanishi et al. 2003). One benefit of utilizing Bacillus spp. as probiotics in aquaculture is that they will probably not use qualities from Gram-negative microscopic organisms (e.g., Vibrio) that may present anti-microbial opposition (Rabinowitz and Roberts 1986). Bacillus cereus is around 60 delegates of the broadly fluctuated Bacillus variety. It incorporates the supposed Bacillus cereus gathering from the start with fundamentally the same as species B. mycoides, B. thuringiensis and B. anthracis. Bacillus is a large and ubiquitous bacteria family that is frequently isolated from soil and waste environment. There are tiny differences between these four species. B. cereus is commonly found as a saprophyte in soil, water, plants, and air, from which it is efficiently transported to nourishment, either from novel crude material or during nourishment preparations. At that point, they are associations with the Bacillus bunch are all around considered the great producer of antimicrobial substances, including peptide and lipopeptide anti-toxins, and bacteriocins (Stein 2005). Therefore, the current research mainly focuses on developing an antibiofilm agent from Bacillus cereus to eradicate MRSA disease (Jiang et al. 2020).
In the present study, we explore bacteriocin delivering Bacillus cereus microorganisms separated from many regions in Salem. We attempt to identify and characterize bacteriocin producing bacteria from food waste soil. We assessed the antibacterial capacity and molecular mass of a bacteriocin from isolated bacteria. Those bacteria are growing an alternative source of the bacteriocin to control many important antibiotic-resistant pathogenic bacteria in the future.
Methodology
Sample Collection
A soil sample collected in the kitchen waste disposal area, K.R. Thoppur, Salem District, Tamil Nadu, India (11°40′46.9″N 78°00′28.9″E). 5 g of soil sample was collected by using a clean and dry sterile spatula in a clean polythene bag.
Spread Plate Method
1 g of soil was dissolved in 1 ml of 0.8% NaCl to make soil suspension. Serial dilution was carried out. To get a concentration range from 10–1 to 10–3. A volume of 0.1 ml of each dilution was transferred aseptically to nutrient agar plates. The sample was spread uniformly. The plates were incubated overnight at 37 °C (Prashad 2014).
Screening of Bacteriocin Producing Bacteria
The single colonies obtained from each sample were screened for their antimicrobial activity against the methicillin-resistant Staphylococcus aureus (Rosado and Seldin 1993). Nutrient agar plates are preparing to overlay with 20 ml of soft agar (0.75% agar) inoculated with MRSA Concentration of 106 CFU/ml. To patch the single colonies and plates were incubated at 37 °C for 16–18 h and the appearance of clear zones showing the antagonistic activity was observed.
Genomic DNA Extraction of Bacteria
The genomic DNA was extracted from the isolated bacteria colonies were by using the slightly modified protocol described (Govindarajan et al. 2016). The 12 h cultures of bacterial isolates were taken in the microcentrifuge tube. The tube was centrifuged at 10000 rpm for 10 min. The pellet was collected and 90 μl of 10% SDS was added. The tubes were incubated at 37 °C for 1 h(Govindarajan et al. 2016). After incubation, an additional 150 μl of 5 M NaCl was added before the addition of 100 μl of 10% Cetyl trimethyl ammonium bromide (CTAB). The sample was mixed thoroughly and incubated at 65 °C kept in a water bath for 30 min, after incubation, add phenol, chloroform and Isoamylalcohol in the ration of 25:24:1 (Vol/Vol/Vol) (Govindarajan et al. 2016). The tube was centrifuged at 13000 rpm for 15 min and the aqueous layer was separated into a fresh tube. Then it was precipitated with 70% ethanol and centrifuged at 7000 rpm for 5 min. Pellets were suspended in 30 μl of TE buffer(Govindarajan et al. 2016). The DNA sample was separated according to their molecular weights under the electrophoresis system. Finally, the DNA band was visualized under a gel documentation system (Lark, Germany). The DNA concentration was determined by measuring the absorbance at the ratio 260/280 nm and the DNA suspension was stored at − 20 °C is used for further analysis.
PCR Amplification of 16S rDNA Gene
A reaction mixture containing approximately 50 ng of template DNA, l0X PCR buffer, a 20 pmol concentration of each PCR primer (27F AGA GTT TGATCMTGG CTC AG and 1492 R TAC GGY TAC CTT GTT ACG ACTT), a 2.5 mM concentration of dNTPs and 2.5U of Taq DNA polymerase in a total volume of 50 µl was prepared. After a 10 min denaturation at 94 °C, the reaction mixture was run through 35 cycles of denaturation for 20 s at 94 °C, annealing at 58 °C, and extension for 1 min at 72 °C, followed by a final extension for 10 min at 72 °C. 10 µl of PCR product was electrophoresed on 1% agarose gel to determine the size of the product. Both negative and reagent controls were included in PCR amplification (Lu et al. 2000).
Phylogenetic Analysis
The 16S rDNA amplicon of these four isolates was used to decide the sequence of the first 500 nucleotides. A blast search assumed to find out the similarity of these two sequences with the most similar 16S rDNA sequences of the GenBank database. The sequence of 16S rDNA was aligned with related sequences retrieved from the database to construct a neighbour-joining tree showing the nearest phylogenetic tree (Chowdhury et al. 2004).
Production of Bacteriocin
The isolated strain was grown in Luria broth at 37 °C for 12 h. After incubation, the broth was centrifuged at 6000 rpm for 10 min and the cells were separated. To collect the supernatant and pellet, then they are used to determine the antimicrobial activity (salasiah kormin et al. 2001).
Determination of Inhibitory Spectrum
An agar well diffusion method was used to access the production of antimicrobial compounds by the selected isolates from bacteria against MRSA. Duplicate plates were used for each target organism (Anita et al. 2014). 100 µl of bacterial culture supernatant be added to the wells in the plates. The finding of clear inhibition zones around the wells on the inoculated plates is an indication of antimicrobial activities (Anita et al. 2014).
SDS-PAGE and Zymogram Analysis
SDS-PAGE was performed according to the method of (Laemmli 1970) using Proviga (India) electrophoretic apparatus. 15% separating and 4% stacking gel were used to separate the proteins. Per well, 50 μl samples were loaded. SDS-PAGE was run until the bromophenol blue front reached the end of the gel (30 V for stacking gel and 50 V for separating gel was applied) (Selvam et al. 2016). While one half of the gel was stained with Coomassie brilliant blue R250, the other half was washed 3times with sterile ddH2O, overlaid with cultures of MRSA and incubated at 37 °C for 24 h.
Time Kill Assay
In time kill studies, an inoculum of MRSA was prepared from test organisms subcultured until the exponential phase and Cultured in Luria Britani broth were transferred in a separate flask, and crude bacteriocin extract was added from an MRSA culture flask. A growth control flask with tested strain without bacteriocin was included in the experiment. The cultures were incubated at 37 °C, and samples were collected obtained at every 3 h interval. Every sample to make a duplicate. Growth rates and bacterial concentrations were determined by measuring OD at 600 nm at different time points (Selvam et al. 2016).
Antibiofilm Activity
The culture of MRSA was incubated into the 96 well flat-bottomed tissue culture plates and it’s incubated at 37 °C for 2 days. After 2 days, the different concentration (10–40%) of bacteriocin was added and incubated at 37 °C for 24 h using the blank group as a control. After incubation, the spent media was aspirated gently and the wells were washed with 200 µl of PBS and then air-dried. 200 µl of 99% ethanol was added and incubated for 15 min for fixation, and aspirated and allowed to dry later (Selvam et al. 2016). The wells were stained with 200 µl of 0.1% crystal violet for 5 min. The excess stain was gently rinsed with distilled water and the plate was air-dried (Mataraci and Dosler 2012). The stain was resolubilized in 200 µl of 33% acetic acid and the cell concentration was measured at OD 595 nm and check the morphological changes in antibiofilm activity (Rukmanikrishnan et al. 2020).
Result
Screening of Bacterial Isolates for Bacteriocin Production
As described in the materials and methods section, Four bacterial isolates were obtained from soil samples and tested for the bacteriocin activity toward MRSA as an indicator organism. Among them, one bacterial (PU3) isolates showed the antagonistic activity against MRSA. The antagonistic bacterial strain reported to be effective in supressing selected as found to the zone of inhibiting the growth of MRSA (Fig. 1). In our bacteriocin efficacy was compared with three isolates, PU3 produced maximum activity against MRSA.
Antibacterial Activity
The agar well diffusion method was used to determine the bacteriocin minimum inhibitory activity. The findings revealed that the bacterial strain PU3 produced a high level of bacteriocin (100 AU/ml) when compared to other strains (Fig. 2).
To assess their morphological and biochemical characterization, the isolated strain PU3 was subjected to the microscopic examination, the findings revealed that the PU3 strain exhibited rod in shape, motile, spore formed and can cultivate at 37 °C. In addition, the various biochemical and physiological tests showed that PU3 is catalase, urease, citrate, gelatin, oxidase and lipase positive; however, starch hydrolysis, methyl red, Voges-Proskauer and indole negative, as depicted in Table 1. Based on the characterization and the PU3 strain was phenotypically identified as Bacillus sp.
Isolation of Genomic DNA From Bacteria
The genomic DNA isolation from soil bacteria by alkaline lysis method. The isolation DNA was l 1% agarose gel, after the run finish of gel, the genomic DNA band was observed under the UV eliminator.
Identification of Bacteriocin Producing Bacteria Gene Sequence
The ribosomal RNA gene sequences are relatively conserved among the bacteria. Therefore, the 16S rDNA sequence was analyzed to confirm the species level identity of the bacterial isolate. The 16S rDNA region (1500 bp) was amplified from the genomic DNA of the bacterial isolate (Fig. 3). The sequences were aligned (1419 bp) and searched for sequence homology in Ribosomal Database Project II (RDP). The results revealed that the sequence had a high score bit of homology with the 16S rDNA sequence of Bacillus cereus. The selected bacterial isolate (PU3) was conformed as Bacillus cereus and the nucleotide sequence of the 16S rDNA of the PU3 strain was deposited in GenBank under the accession number KX548263.
Phylogenetic Tree Analysis
Based on BLAST and phylogenetic results, strain PU3 was identified as Bacillus cereus. The 16S rDNA sequence of isolates determined direct analysis of the genetic distance. The phylogenetic tree and related type strain were used to assemble the phylogram using the neighbor-joining method and displayed in tree view (Fig. 4).
SDS-PAGE and Zymogram Analysis
To estimate the molecular weight of the bacteriocin, the protein in the active fraction was resolved on 15% SDS-PAGE. Based on the electrophoretic mobility, the molecular weight of the single protein band that exhibited antimicrobial activity was estimated to be ~ 3 kDa as indicated by the complete clear zone of inhibition for MRSA, respectively (Fig. 5).
Time Kill Assay
The bactericidal activity was assessed by time-kill assay in the result showed that the Bacillus cereus producing bacteriocin had good antibacterial activity against MRSA at different time duration (Fig. 6).
Antibiofilm Activity
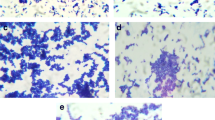
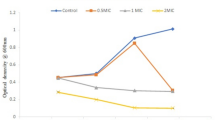
The different concentrations of bacteriocin were incubated with MRSA for 4 h at 37 °C for adherence to the wells of microtiter plates; it inhibited biofilm attachment in a concentration-dependent manner (Fig. 7). About 0.020 ± 0.001 attachment of biofilm was observed in the presence of 30–40% of bacteriocin and also examined morphological changes of bacteriocin treated with biofilm. Bacteriocin showed significant inhibitory activity against MRSA biofilm formation at 24 h, about its concentration (Fig. 8).
Discussion
Bacteriocins are promising antimicrobial operators, ribosomally integrated peptides and delivered by Gram-positive and Gram-negative microbes for the evident motivation behind wrecking their rivals (Cotter et al. 2005). The quantity of MRSA disease has been expanding and turns into a difficult issue in general well-being worldwide. Noval antimicrobial operators are quickly expected to battle this safe medication issue. The utilization of bacteriocin as an elective specialist to beat the issue is promising (Papagianni and Anastasiadou 2009). In this way, the microorganisms enduring such a domain are relied upon to deliver a wide range of antimicrobial movement. In my investigation, a few bacterial strains separated from the dirt examples indicated antimicrobial movement against MRSA. The inhibitory range of these strains was assessed and found that they showed antimicrobial action against Gram-positive microscopic organisms MRSA. The bacteriocins show a tight range towards the related strains (Kim et al. 2001). The 16S rDNA sequencing of these strain was completed to learn the species-level recognizable proof locale was exceptionally preserved, for assurance that the species were distinguished as Bacillus cereus. The phylogenetic tree was the manufacture and microscopic organisms are ordered. They are now detailed for this investigation 16S rDNA quality sequencing uncovered that all the strains have a place with Bacillus licheniformis yet neglected to show huge variety in the arrangement of these strains. By and large, the 16S rDNA arrangement is utilized in Bacillus order as a structure for animal varieties portrayal (Stackebrandt and Swiderski 2002). The utilization of 16S rDNA quality sequencing is used for the ID of bacterial soil test disconnects, which showed vague biochemical profiles (Woo et al. 2000, 2001). A few specialists have likewise revealed the B. licheniformis is known to deliver bioactive proteins with various atomic loads from 2.0 to 150 kDa that was purged from the way of life supernatant. Cerein 7, an antibacterial peptide delivered by B. cereus (Oscariz and Pisabarro 2001), bacillocin 490 (Martirani et al. 2002) and lichenin (Cladera-Olivera et al. 2004) are hydrophobic proteins with low atomic mass (< 2 kDa). In my present examination, the bacteriocin movement protein band was complied with in SDS PAGE investigation and afterwards, Zymogram action indicated an away from the restraint of ~ 3 kDa. The comparative aftereffect of some report that the innovation of type I, II thermostable bacteriocins and phenol-dissolvable modulins (PSMδ; MW 3 kDa) by S. epidermidis can specifically eliminate bacterial pathogens, for example, S. aureus (Cogen et al. 2010; Claudel et al. 2019). A period slaughter test was performed with bacteriocin to affirm the bactericidal impact of B. cereus. Development hindrance and adequacy of the bacteriocin were time-subordinate, creating particular time-kill profiles for the tried microscopic organisms. The unrefined concentrate of bacteriocin licheniocin 50.2 demonstrated noteworthy bactericidal actions against L. monocytogenes beginning with the primary hour of utilization (Beric et al. 2013). The advancement of biofilm is a serious issue in the clinical environment and nourishment ventures. In specific reports of biofilm, the cloned and over-communicated bacteriocin enterocin-B displayed antibiofilm movement against L. monocytogenes, A. baumanii and S. aureus. This anti-biofilm action was expanded essentially with a blend of enterocin A (Ankaiaha et al. 2018). In my outcome, a significant decrease in biofilm improvement ability to MRSA with bacteriocin was seen at various fixations.
Conclusion
Bacteriocins are produced by many bacteria that have a lot of antagonistic activity. They can suppress MRSA, which has become a significant concern in hospitals around the world. These non-pathogenic bacterial strains are found in nature. In this work, the isolated PU3 bacterial strains from soil samples have an antagonistic impact against MRSA. In the future, they could exploit the bacteriocin produced by these microorganisms as a source of peptide antibiotics against harmful bacteria and for the control of drug-resistant MRSA infection. While resistance to these peptides in target strains has yet to be observed under laboratory circumstances, resistance mechanisms are common in producer strains. To avoid the spread of resistance and loss of efficacy, caution should be given while using bacteriocins and related compounds in clinical settings.
References
Alejandro B, María T, Germán B (2013) Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26(2):185–230
Aly SM, Mohamed MF, John G (2008) Effect of probiotics on the survival, growth and challenge infection in Tilapia nilotica (Oreochromis niloticus). Aqua Res 39:647–656. https://doi.org/10.1111/j.1365-2109.2008.01932.x
Anita M, Hari Sing L, Balawant Singh R (2014) Isolation of antimicrobial producing bacteria from soil samples collected from Bhopal Region of Madhya Pradesh, India. Int J Curr Microbiol App Sci 3(12):563–569
Ankaiaha D, Palanichamya E, Antonyrajb Repally CB, Ayyannaa SIB, Arul V (2018) Cloning, overexpression, purification of bacteriocin enterocin-B and structural analysis, interaction determination of enterocin-A, B against pathogenic bacteria and human cancer cells. Int J Biol Macromole 116:502–512
Beric T, Stankovic S, Draganic V, Kojic M, Lozol J, Fira D (2013) Novel antilisterial bacteriocin licheniocin 50.2 from Bacillus licheniformis VPS50.2 isolated from soil sample. J Appl Microbiol 116:502–510. https://doi.org/10.1111/jam.12393
Chowdhury SP, Khanna S, Verma SC, Tripathi AK (2004) Molecular diversity of tannic acid degrading bacteria isolated from tannery soil. J Appl Microbiol 97(6):1210–1219
Cladera-Olivera F, Caron GR, Brandelli A (2004) Bacteriocin production by Bacillus licheniformis P40 in cheese whey using response surface methodology. Biochem Engin J 21:53–58. https://doi.org/10.1016/j.bej.2004.05.002
Claudel JP, Auffret N, Leccia MT, Poli F, Corvec S, Dréno B (2019) Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatol 235:287–294. https://doi.org/10.1159/000499858
Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, Torpey JW, Otto M, Nizet V, Kim JE (2010) Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol 130:192–200. https://doi.org/10.1038/jid2009.243
Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. https://doi.org/10.1038/nrmicro1273
David L, Ashwini C, Olaya R (2013) Christophe beloin from in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2:288–356
Eladawy M, El-Mowafy M, Adel El-Sokkary MM, Barwa R (2019) Effects of lysozyme, proteinase k, and cephalosporins on biofilm formation by clinical isolates of Pseudomonas aeruginosa. Interd Pers Infect Diseases 2020:9. https://doi.org/10.1155/2020/6156720
Ghosh K, Sen SK, Ray AK (2002) Characterization of bacilli isolated from the gut of rohu Labeo rohita fingerlings and its significance indigestion. J Appl Aqua 12:33–42. https://doi.org/10.1300/J028v12n03-04
Gomes F, Martins N, Isabel CFR, Ferreira MH (2019) Anti-biofilm activity of hydromethanolic plant extracts against Staphylococcus aureus isolates from bovine mastitis. Heliyon 5(5):e01728. https://doi.org/10.1016/j.heligen.2019.eo1728
Govindarajan RK, Revathi S, Rameshkumar N, Krishnan M, Kayalvizhi N (2016) Isolation and characterization of tannase producing bacteria from the gut of gryllotalpa krishnanij. Microbiol Biotech Food Sci 6(2):813–817
Harsh M, Des F, Mary C, ReaPaul D, Cotter CH, Paul Ross R (2018) Fighting biofilms with lantibiotics and other groups of bacteriocins. Npj Biofilms and Microbiomes 4:9. https://doi.org/10.1038/s41522-018-0053-6
Ivanova EP, Gorshkova NM, Nedashovskaya OI, Vysotsky MV, Svetashev VI, Mikhailov VV (1999) Taxonomy of B. subtilis, B. pumilus and B. horti of marine origin. Marine Biol 25:483–487
Jiang Y, Geng M, Bai L (2020) Targeting biofilms therapy: current research strategies and development hurdles. Microorgan 8(8):1222. https://doi.org/10.3390/microorganisms8081222
Karthikeyan V, Santhosh SW (2009) Study of bacteriocin as a food preservative and the L. acidophilus strain as probiotic. PJN 8(4):335–340. https://doi.org/10.3923/pjn.2009.335.340
Khandeparker L, Anil AC, Raghukumar S (2003) Barnacle larval destination: piloting possibilities by bacteria and lectin interaction. J Exp Mar Biol Ecol 289:1–13. https://doi.org/10.1016/S0022-0981(03)00024-8
Kim SI, Chang JY, Kim IC, Lee K (2001) Characterization of bacteriocin from Bacillus subtilus ex1. Koe Appl Microbiol Biotechnol 29:50–55
Klaenhammer TR (1988) Bacteriocins of lactic acid bacteria. Biochemie 70:337–349. https://doi.org/10.1016/0300-9084(88)90206-4
Kormin S, Rusul G, Radu S, Ling FH (2001) Bacteriocin-producing lactic acid bacteria isolated from traditional fermented food malays. J Med Sci 8(1):63–68
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat 227(5259):680–685
Liu X, Wang J, Weng CX, Wang R, Cai Y (2020) Low-frequency ultrasound enhances bactericidal activity of antimicrobial agents against Klebsiella pneumoniae biofilm. BioMed Res Int. https://doi.org/10.1155/2020/5916260
Lopez S, Belloso M (2008) Use of nisin and other bacteriocins for the preservation of dairy products. Int Dairy J 18:329–343. https://doi.org/10.1016/j.idairyj.2007.11.009
Lu JJ, Perng CL, Lee SY, Wan CC (2000) Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol 38(6):2076–2080
Martirani L, Varcamonti M, Naclerio G, De Felice M (2002) Purification and partial characterization of bacillocin 490, a novel bacteriocin produced by a thermophilic strain of Bacillus licheniformis. Microb Cell Fact 1:1. https://doi.org/10.1186/1475-2859-1-1
Mataraci E, Dosler S (2012) In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant staphylococcus aureus biofilms. Antimicrob Agents Chemother 56(12):6366–6371. https://doi.org/10.1128/AAC.01180-12
Miyanishi N, Hamada N, Kabayashi T, Imada C, Watanabe E (2003) Purification and characterization of a novel extracellular beta-1-3 glucanase produced by Bacillus clussi NM-1 isolated from Ezo abalone haliotis discus hannai. J Biosci Bioeng 95:45–51. https://doi.org/10.1016/S1389-1723(03)80147-0
Montville TJ, Chen Y (1998) Mechanistic action of pediocin and nisin: recent progress and unresolved questions. Appl Microbiol Biotechnol 50:511–519. https://doi.org/10.1007/s002530051328
O’sullivan A, Garde P, Ross RP, Hill C (2003) Generation of food-grade lactococcal starters which produce the lantibiotics lacticin 3147 and lacticin 481. Appl Environ Microbiol 69:3681–3685. https://doi.org/10.1128/AEM.69.6.3681-3685.2003
Oscariz JC, Pisabarro AG (2001) Classification and mode of action of membrane-active bacteriocins produced by gram-positive bacteria. Int Microbiol 4:13–19. https://doi.org/10.1007/s101230100003
Papagianni M, Anastasiadou S (2009) Pediocins: the bacteriocins of Pediococci Sources, production, properties, and applications. Microb Cell Fact 8:3. https://doi.org/10.1186/1475-2859-8-3
Pinto M, Langer TM, Hüffer T, Hofmann T, Herndl GJ (2019) The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 14(6):e0217165. https://doi.org/10.1371/journal.pone.0217165
Prakash V, Sreetha H, Poornima KH, Lakshmimol KN, Regma R, Fathima H, Vishnu TV, Venu S, Bipin G, Pal S (2018) Antagonistic effects of bacteriocins from Lactobacillus spp. against enteric pathogens. Poll Res 37:128–134
Prashad MP (2014) Characterization of amylase gene in Bacillus species isolated from different soil samples. Int J Curr Micro Biol App Sci 3(9):891–896
Rabinowitz JC, Roberts M (1986) Translational barriers limiting expression of E. coli genes in Bacillus and other gram-positive organisms. In: Levy SB, Novick RP (eds) Banbury report 24: antibiotic resistance genes: ecology Transfer and Expression. Cold Spring Harbour Laboratory, New York
Rosado AS, Seldin L (1993) Production of a potentially novel antimicrobial substance by Bacillus polymyxa. World J Microbiol Biotechnol 9(5):521–528. https://doi.org/10.1007/BF00386287
Rukmanikrishnan B, Rajasekharan SK, Lee J, Ramalingam S, Lee J (2020) K-Carrageenan/lignin composite films: Biofilm inhibition, antioxidant activity, cytocompatibility, UV and water barrier properties. Mater Today Commun. https://doi.org/10.1016/j.mtcomm.2020.101346
Selvam D, Thangarasu A, Neelamegam R, Muthukalingan K, Nagarajan K (2016) Antimicrobial potential of earthworm wegeneriona sp. against Human Pathogens. J Anal Pharm Res 3(4):00060. https://doi.org/10.15406/japlr.2016.03.00060
She P, Wang Y, Liu Y (2019) Effects of exogenous glucose on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. Microbiol 8:933. https://doi.org/10.1002/mbo3.933
Stackebrandt E, Swiderski J (2002) From phylogeny to systematics: the dissection of the genus Bacillus. In: Applications and systematicss of Bacillus and relatives. Blackwell, Oxford. https://doi.org/10.1002/9780470696743.ch2
Stein T (2005) Bacillus subtilis antibiotics: structures, syntheses, and specific functions. Molecul Microbiol 56:845–857. https://doi.org/10.1111/j.13652958.2005.04587.x
Strempel N, Strehmel J, Overhage J (2014) Potential application of antimicrobial peptides in the treatment of bacterial biofilm infections. Curr Pharm Des 21(1):67–84. https://doi.org/10.2174/1381612820666140905124312
Sugita H, Hirose Y, Matsuo N, Deguchi Y (1998) Production of the antibacterial substance by bacillus species strain NM12, an intestinal bacterium of the Japanese coast. Aqua 165:269–280. https://doi.org/10.1023/A:1009246007208
Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG (2015) Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28(3):603–661
Woo PCY, Leung PKL, Leung KW, Yuen KY (2000) Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mole Pathol 53:211–215. https://doi.org/10.1136/mp.53.4.211
Woo MH, Vance JR, Bjornsti MA (2001) Studying DNA topoisomerase I-targeted drugs in the yeast Saccharomyces cerevisiae. Methods Mole Biol 95:303–313. https://doi.org/10.1385/1-59259-057-8:303
Funding
This research did not receive any specific grant from funding agencies in public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
SD, TA—experimental design analysis and text manuscript. SR—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dhanam, S., Arumugam, T. & Rajasekar, S. Biofilm Effects of the Soil Bacillus cereus Metabolites: Isolation, Characterization and Antimicrobial Activity Against Methicillin-Resistant Staphylococcus aureus. Int J Pept Res Ther 27, 2371–2379 (2021). https://doi.org/10.1007/s10989-021-10258-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-021-10258-0