Abstract
Endophytic actinomycetes associated with medicinal plants of Chhattisgarh are rich source of novel antimicrobial compounds. The aim of this study was to evaluate the production of antibiotics by endophytic Streptomyces sp. K-R1 associated with root of Abutilon indicum against multidrug resistant human pathogens. The 16S rRNA sequence of the isolate K-R1 has been identified as Streptomyces parvulus NBRC 13193T (AB184326) with the GenBank database (accession number KY777591). Streptomyces parvulus K-R1 grown under submerged fermentation condition optimized by central composite design (glucose—11.25 g/l, soybean meal 12.17 g/l, calcium carbonate—1.47 g/l, sodium chloride—11.25 g/l; pH 7.18 at 30.51 °C with 4% seed inoculum for 10 days of incubation) exhibited highest activity against multidrug resistant Staphylococcus aureus JNMC-3, Staphylococcus epidermidis JNMC-4, Klebsiella pneumoniae MTCC-3384, Klebsiella pneumoniae JNMC-6, Pseudomonas aeruginosa MTCC-741, Proteus vulgaris JNMC-7, Candida albicans MTCC-183 and Aspergillus niger MTCC-872. The structures of the active compounds were elucidated by UV–Vis spectroscopy, 1H and 13C NMR, FTIR and ESIMS. Actinomycin D, actinomycin X0β and fengycin were detected in crude extracts and major components were eluted by HPLC at 10.63, 10.88 and 6.3 min respectively. In this case, a high yield of antibiotics was achieved with Streptomyces parvulus K-R1 fermented in glucose soybean meal broth media with a production of 240 mg/l after 5 days, 290 mg/l after 8 days and 340 mg/l after 11 days, which can be used in industrial fermentation process to obtain high yields. To the author’s best knowledge, it is the first report of fengycin production by Streptomyces parvulus.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Medicinal plants are known to be rich source of bioactive compounds with therapeutic potential. Several bioactive molecules from the plants have been commercially exploited. Since the endophytic actinomycetes symbiotically associate with their plant host and have genetically evolved together over a long period of time, they benefit each other by producing metabolites of biological and physiological significance complementary to each other and influence the metabolite production efficiency of endophytic Streptomyces species (Golinska et al. 2015). Hence, the possibility of the products from endophytes may be used for biocontrol of the related plant disease. Ultimately, the compounds from endophytic Streptomyces sp. once isolated and characterized, may also have potential uses in modern medicine, agriculture and in various industries.
Endophytic actinomycetes especially from the largest genus in the group, Streptomyces have been found to produce novel antibiotics like, munumbicin A–D from Streptomyces sp. NRRL 30562 from Kennedia nigriscans (Castillo et al. 2002); kakadumycin from Streptomyces sp. NRRL 30566 from Grevillea pteridifolia (Castillo et al. 2003) and coronamycin from Streptomyces sp. MCU-2110 from Monstera sp. (Ezra et al. 2004); p-amino acetophenonic acids from Streptomyces griseus from Kandelia candel (Guan et al. 2005); 6-alkalylsalicylic acid from Streptomyces laceyi MS53 from Ricinus communis (Kim et al. 2006) and p-amino acetophenonic acid from Streptomyces sp. HK10552 from Aegiceras coeniculatum (Wang et al. 2010); maklamicin from Micromonospora sp., an endophyte of Abrus pulchellus (Igarashi et al. 2011); linfuranone A from Microbispora sp. GMKU 363 from Clinacanthus siamensis (Indanada et al. 2013). Two new flavonoids 7-methoxy-3,3′,4′,6-tetrahydroxyflavone and 2′,7-dihydroxy-4′,5′-dimethoxyisoflavone together with four known compounds such as fisetin, naringenin, 3′-hydroxydaidzein and xenognosin B isolated from endophytic Streptomyces sp. BT01 from root of Boesenbergia rotunda inhibited the growth of Staphylococcus aureus ATCC25932, Bacillus cereus ATCC 064 and Bacillus subtilis ATCC 6633 (Taechowisan et al. 2014). Granaticinic acid from Streptomyces thermoviolaceus NT1, an endophyte of Catharanthus roseus has ability to inhibit several Gram-positive, Gram-negative and fungal pathogens (Roy and Banerjee 2015).
Since, Chhattisgarh state has a rich diversity of medicinal plants and the endophytic actinomycetes are relatively unexplored in this area, the present study was planned to isolate endophytic actinomycetes from selected medicinal plants. To the best of our knowledge, this is the first report on the isolation, characterization and antibiotic production ability study of endophytic actinomycetes from medicinal plant (Abutilon indicum) of Chhattisgarh. In the present study, Streptomyces sp. K-R1 has been isolated from root of Abutilon indicum and identified as Streptomyces parvulus by 16S rRNA analysis. The endophytic Streptomyces parvulus K-R1 we examined had broad spectrum antimicrobial activity against multidrug resistant clinical human pathogens commonly present in the environment of Chhattisgarh. Hence the drug can be employed for curing acute and chronic human diseases caused by these pathogens. Response surface methodology has been employed to optimize the optimum concentration of nutrients and the fermentation conditions for enhanced production of antimicrobial compounds in submerged fermentation. Three metabolites were isolated from fermented broth of Streptomyces sp. K-R1 and characterized as actinomycin-D, actinomycin X0β and fengycin. However, Production and optimization of Actinomycins by endophytic Streptomyces sp. associated with medicinal plants has not been reported in literature. In addition, there are still no reports that Streptomyces parvulus can produce fengycin. Although, Streptomyces parvulus isolated from soil and plants has been reported to produce actinomycin D and actinomycin X0β. Hence, endophytic Streptomyces parvulus K-R1 may be exploited as a potential source for the commercial production of Actinomycins and fengycin. Further investigation on the strain for commercial application will be of immense pharmaceutical importance.
Materials and Methods
Endophytic Actinomycetes K-R1
The investigated endophytic actinomycetes were isolated from medicinal shrub Abutilon indicum (L.) Sweet growing in the garden of School of Studies in Life Science, Pt. Ravishankar Shukla University, Raipur, Chhattisgarh, India (latitude: 21.1797oN, longitude: 81.7787oE). For the isolation of endophytic actinomycetes plant parts were surface sterilized with 0.1% tween 20 for 5 min followed by 70% ethanol for 5 min and 4% sodium hypochlorite for 10 min, washed again with 70% ethanol for 5 min and finally rinsed in sterile distilled water. The outer tissues were removed from the samples with a sterile knife blade and the inner tissues were excised, macerated with sterile distilled water by using mortar and pestle. Macerated samples (0.2 ml) were spread on starch casein nitrate agar medium w/v (g/l) (soluble starch—10 g, K2HPO4—2 g, KNO3—2 g, NaCl—2 g, Casein—0.3 g, MgSO4—0.05 g, CaCO3—0.02 g, FeSO4—0.01 g; pH 7.2) supplemented with nalidixic acid (50 mg/l) and nystatin (50 mg/l) to suppress the growth of bacteria and fungi respectively (Chandrakar and Gupta 2017). The plates were incubated at 28 ± 2 °C for 21 days. After attaining visible powdery growth, colonies were transferred on starch casein agar slants for storage and preservation. Several isolated colonies were selected for antimicrobial testing against multidrug resistant Gram-positive and Gram-negative pathogens. On culture, showing broad spectrum antimicrobial activity, designated K-R1 was taxonomically characterized and used for the production of antimicrobial compound.
Characterization and Identification of Endophytic Actinomycetes K-R1
Cultural, biochemical and physiological characteristics of endophytic isolate K-R1 was examined as per International guidelines (Shirling and Gottlieb 1966). Morphological identification of the isolate was done by cover slip culture method (Kawato and Shinobu 1959). The ability of isolate for utilization of different carbon and nitrogen sources was carried out by growing the isolate in basal mineral salt agar medium (g/l) [(NH4)2SO4—2.64 g, KH2PO4—2.38 g, K2HPO4. 2H2O—5.65 g, MgSO4·7H2O—1 g; agar—15 g; trace salt solution—1 ml ; pH 6.8–7.0] at 1.0% (w/v) concentration and results were recorded after 7, 14 and 21 days using glucose and proline as positive control and carbon source and nitrogen source free medium as negative control (Pridham and Gottlieb 1948). For molecular identification of the potent antibiotic producing endophytic actinobacterial isolate K-R1 from root of Abutilon indicum, 16S rRNA gene was amplified using the universal primers of 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′). The EzTaxon database (http://www.eztaxon.org/) was employed in order to assess the degree of gene sequence similarity. The most similar sequences in the EzTaxon database are selected using the whole (full-length) query sequence as a query in the BLAST search. Multiple alignments with sequences of most closely related Streptomyces and calculations of level of sequence similarity were carried out using EzEditor software (Jeon et al. 2014) (http://www.ezbiocloud.net/sw/ezeditor). The phylogenetic tree for multiple sequence alignments stored in EzEditor program was constructed using the neighbor-joining method by MEGA 5.0 software (Tamura et al. 2011). The topology of phylogenetic tree was evaluated by bootstrap resampling method with 1000 replicates. In this study, the 16S rRNA sequence of the isolate K-R1 has been submitted in the GenBank database of National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov).
Selection of Suitable Medium for Antimicrobial Compound Production by K-R1
The selection of suitable medium for antimicrobial compound production by isolate K-R1 was done by growing the isolate in ten different culture medium w/v (Hi-media, Mumbai, India) viz., czapek-dox broth (CZB), glycerol asparagines broth (ISP-5), glucose soybean meal broth (GSB), inorganic salt starch broth (ISP-4), nutrient broth (NB), potato dextrose broth (PDB), sabouraud broth (SB), soybean meal broth (X-media), starch casein nitrate broth (SCNB) and yeast extract malt extract broth (ISP-2). 50 ml of above broth culture medium in 150 ml of conical flask was inoculated with 1 ml of Streptomyces sp. K-R1 seed culture and incubated at 28 ± 2 °C for 14 days. After incubation, the filtrate was separated from the culture broth by Whatman no. 1 filter paper and assessed against multidrug resistant Gram-positive and Gram-negative human pathogens for antimicrobial activity. The inoculums of the test pathogens were standardized by growing the organisms in Muller Hinton broth (Hi-media, Mumbai, India) and adjusting the optical density of the culture broth to a turbidity corresponding to 0.08 at 620 nm using a spectrophotometer, which was equivalent to 108 CFU/ml. The medium in which the endophytic isolate K-R1 exhibited maximum zone of inhibition was used for further study.
Experimental Design and Optimization by RSM
Response surface methodology (RSM) with central composite design (CCD) was employed in order to optimize the optimum environmental conditions viz., temperature, pH, incubation time, inoculum volume and nutritional parameters viz., glucose, soybean meal, calcium carbonate and sodium chloride for enhanced production of antimicrobial compound by endophytic Streptomyces sp. K-R1. The model was studied within a range of five coded levels designated as (− α), (− 1), (0), (+ 1) and (+ α) with 30 experimental runs (Online Resource 1). Model was validated by growing the endophytic Streptomyces sp. K-R1 under CCD optimized media (glucose soybean meal broth) and un-optimized media (starch casein nitrate broth). The broths were centrifuged at 4000 rpm for 10 min and the supernatant was tested against Staphylococcus aureus MTCC-96 by agar well diffusion method. Antimicrobial activity was expressed in terms of inhibition zone in diameter (mm).
Submerged Fermentation and Production of Antimicrobial Compound
A stock culture of endophytic Streptomyces sp. K-R1 was grown and maintained on glucose soybean meal agar (GSB) slant. The stock culture was prepared by transferring 5 ml of sterile water into 14 days old well sporulated slant of isolate K-R1; the surface of the medium was scrapped with sterile inoculating needle; transferred into 45 ml of seed medium with the same components as the agar slant. The seed culture medium was incubated at 28 °C for 48 h (Balagurunathan and Subramanian 1993). Four milliliters (4%) of the inoculum from seed culture medium in each flaks after incubation was aseptically transferred into a lot of 1000 ml Erlenmeyer flasks (15 nos) containing 300 ml of production medium optimized by central composite design: glucose 11.25 g/l, soybean meal 12.17 g/l, calcium carbonate 1.47 g/l and sodium chloride 11.25 g/l; pH 7.18 in each flask. The flasks were incubated at 30.51 °C for 10 days in static condition. Aliquots (2 ml) of cultures were collected after every 4 days of incubation and checked for antibiotic production by evaporating in rotary evaporator at 40 °C. After fermentation, the medium was centrifuged at 4000 rpm for 10 min and the supernatant was extracted with ethyl acetate in the ratio of 1:1 (v/v). Gentle mixing was done for 15 min using separating funnel and allowed to stand for 30 min to separate the organic phase (3400 ml) from the aqueous phase. Both the phases were checked for antimicrobial activity against Staphylococcus aureus MTCC-96 by agar well diffusion method and concentrated in rotary vacuum evaporator at 40 °C. A total of 2.25 g dark reddish orange color active crude was collected from 5 liters of fermentation broth.
Statistical Analysis
Optimization for the production of antimicrobial compound by endophytic Streptomyces sp. K-R1 in different media was analyzed by SPSS software package (version 16.0). Means of the eight measurements of inhibition zone diameter for each experiment was compared using Duncans Multiple Range Test (DMRT) at (p ≤ 0.05). Central composite design (CCD) and statistical analysis of multiple factors was carried out with the Design Expert software package (version 9.0.1, State-Ease Inc., USA). Model was analyzed by ANOVA and the quality of polynomial model equation was assessed by the coefficient of determination R2. The significance of regression coefficient was determined by the p value.
Purification and Bioactivity of Antimicrobial Compound
To check the antimicrobial compound clearly by bioautography, the crude antibiotic was partially purified by thin layer chromatography (TLC) by using three different solvents viz., n-butanol: ethyl acetate: water v/v (9:9:1), n-hexane: ethyl acetate v/v (9:1), chloroform: methanol v/v (6:4) as a mobile phase on pre coated silica gel aluminium plate (Merck 60 F-254; 0.5 mm thick, Germany). Sample was loaded using a capillary tube 1.5 cm above from the bottom of TLC plate in a row along a line and spot was left to dry. The TLC plate was placed vertically in a glass jar containing the solvents and incubated at room temperature for 30 min. When the solvent reached near the top, the plate was taken out and dried. Separated component was visualized under visible and ultraviolet light (254 and 366 nm) and their Rf value was calculated. For bioautography analysis, developed TLC plate was dried overnight and the bands were scraped out separately and dissolved in ethyl acetate (500 µl), then centrifuged at 4000 rpm for 10 min. Supernatant was subjected to antimicrobial activity assay against multidrug resistant pathogens by agar well diffusion method. Further, purity of antimicrobial compound was analyzed by HPLC (Shimadzu). Antimicrobial compound was dissolved in ethyl acetate (1 mg/ml) and was subjected to chromatography on silica gel column Hypersil BDS C18 (150 × 4.6 mm, 5 µ pore size), injection volume was 20 µl. The mobile phase was solvent A: acetonitrile and solvent B: 5 mM ammonium acetate in water by gradient preparation of 95–10% solvent B for 0.01–10.0 min and 10–10% solvent B for 10–30 min; with a flow rate of 1 ml/min and total run time of 30 min. Purified compound was evaluated for its minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) against multidrug resistant human pathogens using Mueller–Hinton broth (Standard NCCL method 2000). Dilution range of the antimicrobial compound taken was from the 1 to 0.0315 mg/ml respectively. Bacterial and fungal pathogens were procured from Institute of Microbial Technology (IMTECH), Chandigarh, India. They included, Bacillus cereus MTCC-430, Staphylococcus aureus MTCC-96, Bacillus subtilis MTCC-441, Staphylococcus epidermidis MTCC-435, Escherichia coli MTCC-1687, Klebsiella pneumoniae MTCC-3384, Proteus vulgaris MTCC-744, Pseudomonas aeruginosa MTCC-741, Candida albicans MTCC-183, Aspergillus niger MTCC-872. Clinical human pathogenic bacteria were procured from Pandit Jawaharlal Nehru Medical College (JNMC), Raipur; Chhattisgarh, India. They included Bacillus cereus JNMC-1, Bacillus subtilis JNMC-2, Staphylococcus aureus JNMC-3 and Staphylococcus epidermidis JNMC-4, Escherichia coli JNMC-5, Klebsiella pneumoniae JNMC-6 and Proteus vulgaris JNMC-7. Antibiotic susceptibility of pathogenic organisms was tested by disc diffusion method. Antibiotic disc (Hi-media, Mumbai, India) used in the study at 10 mcg concentration. The antibiotic discs were placed on Muller–Hinton agar plates previously seeded with 16–18 h old bacterial cultures (108 CFU/ml) grown in Muller–Hinton broth. Plates were incubated at 37 °C for 24 h. The zones of inhibition were measured to the nearest millimeter.
Structure Elucidation and Identification of Active Compound
The structure of the active compounds isolated from HPLC was elucidated by UV–Vis spectroscopy, 1H and 13C NMR, FT-IR and ESIMS. The ultra violet and visible spectrum of the active compounds from Streptomyces sp. K-R1 was determined in ethyl acetate (0.01 mg/ml) with a JASCO, UV–Visible spectrophotometer at 200–800 nm. The spectrum was plotted as wavelength along X-axis verses absorbance along Y-axis. FT-IR spectrum of the antimicrobial compound was recorded using JASCO FT-IR model no. 4100 at 400–4000 cm−1 using KBr pellet technique. The 1H and 13C NMR spectrum of the antimicrobial compound was determined in JEOL 400-MHz using CDCl3 as a solvent. Mass spectrum of the antimicrobial compound was recorded using Shimadzu LCMS with APCI and ESI probes (model LC-2010EV). Electrospray ionization was operated in the positive and negative ion mode and mass spectra were recorded over a range of 100–1300 m/z.
Stability of Antimicrobial Compound in the Presence of Enzymes, Detergents and Temperatures
The sensitivity of the purified antimicrobial compound to denaturation by enzymes w/v viz., trypsin, proteinase K and lysozyme (Hi-media, Mumbai, India) was tested (Munimbazi and Bullerman 1998). All the enzymes were dissolved in distilled water at concentration 1 mg/ml. 100 µl of the antimicrobial solution (1 mg/ml) was mixed with 100 µl enzyme and incubated at 37 °C for 1 h. The antibiotic solution without any enzymes was used as control and antibacterial activity of the mixture was tested against Staphylococcus aureus MTCC-96 by agar well diffusion method.
Effect on the antimicrobial compound by various detergents viz., tween-20, tween-80, triton X-100 and sodium dodecyl sulphate (SDS) was determined by mixing the detergents with the antimicrobial compounds (Munimbazi and Bullerman 1998). The detergents were dissolved in distilled water at a concentration of 0.01 g/ml. 100 µl of the antimicrobial solution (1 mg/ml) was mixed with 100 µl of detergents and incubated at 37 °C for 1 h. Antimicrobial solution without any detergent was used as control. The effect of temperature on stability of the antimicrobial compound, screw capped ampoules, each with 1 mg/ml of antibiotic in DMSO were kept at 30–100 °C for 30 min in water bath. The antimicrobial solutions were cooled to room temperature and volumes were brought to the original concentration and the residual antimicrobial activity was determined against Staphylococcus aureus MTCC-96.
In order to determine the effect of antimicrobial compound from endophytic Streptomyces sp. K-R1 on the ergo sterol present in the fungal cell wall membrane, ergo sterol was used as the reversal agent. Sabarouds dextrose agar plates seeded with 0.5% ergo sterol was prepared along with a control plate which was lacking ergo sterol. The plates were seeded with Candida albicans MTCC-183. Wells were made with a sterile cork borer and 100 µl of the antimicrobial solution (1 mg/ml) was added to the well. The plates were incubated at 28 °C for 48 h and observed for inhibition zone in diameter (mm).
Residual activity (RA) of antimicrobial metabolites was calculated using the equation:
where RA is the residual activity, HT is mean inhibition halo of extract after treatment (mm), HC is mean inhibition halo of the control (mm), 6 is diameter of the well (mm).
Results
Characterization and Identification of Endophytic Streptomyces sp. K-R1
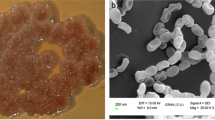
The isolate showed good growth on all types of media. It showed light grey color aerial mycelium in starch casein nitrate agar (SCA), glycerol asparagine agar (ISP-5) and tyrosine agar (ISP-7), medium grey color in yeast extract malt extract agar (ISP-2), oat meal agar (ISP-3) and inorganic salt starch agar (ISP-4). The isolate showed orange yellow color pigment on all types of media. Aerial filament showed Retinaculum Apertum (RA) type of spore chain, open loops with extended spirals of wide diameter structure. The isolate K-R1 was able to degrade starch, lecithin, lipid and xanthine degradation. The isolate was able to grow at 1–10% NaCl and in the presence of 0.01% sodium azide, 0.1% phenol and 0.001% potassium tellurite. It was able to grow at 28–37 °C and able to utilize l-valine, l-histidine and l-hydroxyproline as a nitrogen source. Similarly, the isolate was able to utilize sucrose, meso-inositol, mannitol, l-rhamnose, raffinose, d-melezitoze, adonitol, d-melibiose, dextran and xylitol as a carbon source. The nucleotide sequence of 16S rRNA and phylogenetic tree generated from representative strains of the related genera showed that endophytic Streptomyces sp. K-R1 had high levels of sequence similarity to species of Streptomyces parvulus NBRC 13193T (AB184326) (Fig. 1). The 16S rRNA analysis revealed that, endophytic Streptomyces sp. K-R1 is phylogenetically closely related to Streptomyces parvulus (the sequence similarity levels were 99.93%). Nearly, complete (1438 bp) 16S rRNA sequence of endophytic Streptomyces sp. K-R1 has been submitted in the GenBank database of National Center for Biotechnology Information (NCBI) (accession number KY777591).
Selection of Suitable Media for Antimicrobial Compound Production
Antimicrobial compound production by endophytic Streptomyces sp. K-R1 was carried out on ten different types of media in a submerged culture for 14 days (Table 1). Among them, glucose soybean meal broth media composed of w/v (g/l) (glucose—10 g, soybean meal—10 g, sodium chloride—10 g, calcium carbonate—1 g; pH 7.5) was found to be best media for biomass as well as antimicrobial compound production (p = 0.001). Hence, this media was further selected for optimization and fermentation studies. Minimum production of biomass and antimicrobial compound was achieved using ISP-2 medium. However, antimicrobial compound production was totally inhibited in the medium PDB and SB.
Optimization of Culture Condition for Antimicrobial Compound Production
The results of response surface methodology (RSM) experiment for studying the effect of independent physical variables on antibiotic production by endophytic Streptomyces parvulus K-R1 grown on glucose soybean meal broth media showed that, the maximum production of antibiotics was achieved at run no. 15 when (temperature—28 °C, pH 7.25, incubation time—8 days, inoculum size—3%). While the minimum production of antibiotic was observed in run no. 4 at: (temperature—37 °C, pH 7.25, incubation time—8 days, inoculum size—3%). The goodness of the model was checked by the coefficient of determination (R2). In the present study, (R2) value of 0.92 indicated that the response model can explain 92% of the total variation. The response surface graphs were employed in order to determine the degree of interaction between environmental factors that have the most significant effect on antibiotic production by Streptomyces parvulus K-R1 (Fig. 2).
Response surface graph shows: (1) significant interaction between temperature and pH (p = 0.0140) (2) temperature and incubation time (p = 0.0002) (3) temperature and inoculum size (p = 0.0349) (4) pH and incubation time (p = 0.0223) (5) non significant interaction between pH and inoculum size (p = 0.4608) (6) incubation time and inoculum size (p = 0.8369) on antibiotics production by endophytic Streptomyces parvulus K-R1
A significant interaction was found between the temperature and pH (p = 0.0140), temperature and incubation time (p = 0.0002), temperature and inoculum size (p = 0.0349), pH and incubation time (p = 0.0223). Maximum production of antibiotic was observed when decrease in temperature (between: 32.5 and 30 °C) and increases in pH (between: 7.0 and 7.5), increases in incubation time (between 5 and 10 days) and inoculum size (between: 2 and 4%). It was observed that, higher temperature (> 31 °C) gradually decreased the production of antibiotics. The interaction effect between pH and incubation time revealed maximum production of antibiotic when the pH was 7.25 and incubation time was 8 days. Further increase or decrease in their values gradually decreased in the production. The final optimized environmental conditions obtained after CCD for maximum antibiotic production by Streptomyces parvulus K-R1 were: (temperature—30.51 °C, pH—7.18, incubation time—9.16 days, inoculum size—3.91%).
The optimized environmental conditions were further maintained at constant level for the optimization of nutritional conditions for maximum production of antibiotics by Streptomyces parvulus K-R1. The maximum production of antibiotics was obtained in the run no. 23 with the production medium containing glucose—10.0 g/l, soybean meal—12.50 g/l, calcium carbonate—1.50 g/l, sodium chloride—12.50 g/l. While the minimum production of antibiotic was obtained in the run no. 10 with the production medium containing glucose—12.50 g/l, soybean meal—15.0 g/l, calcium carbonate—1.50 g/l, sodium chloride—12.50 g/l. In the present study, the coefficient of determination (R2) was found to be 0.93, which indicated that the response model can explain 93% of the total variation. A significant interaction was found between the glucose and calcium carbonate (p = 0.0062), glucose and sodium chloride (p = 0.0001), soybean meal and calcium carbonate (p = 0.0061), soybean meal and sodium chloride (p = 0.0044), calcium carbonate and sodium chloride (p = 0.0037) (Fig. 3).
Response surface graph shows: (1) non significant interaction between glucose and soybean meal (p = 0.7506) (2) significant interaction between glucose and calcium carbonate (p = 0.0062) (3) glucose and sodium chloride (p = 0.0001) (4) soybean meal and calcium carbonate (p = 0.0061) (5) soybean meal and sodium chloride (p = 0.0044) (6) calcium carbonate and sodium chloride (p = 0.0037) on antibiotic production by endophytic Streptomyces parvulus K-R1
It is clear from the figure that, lower concentration of glucose (between: 12.5 g and 11.25 g/l) and higher concentration of calcium carbonate (between: 1.25 g and 1.50 g/l) increases the production of antibiotics. The higher concentrations (> 11.5 g/l) of glucose and sodium chloride revealed gradual decrease in antibiotic production. It was observed that, lower concentration of soybean meal (between: 12.50 g and 12.0 g/l) and higher concentration of calcium carbonate (between: 1.25 g and 1.50 g/l) significantly increase the production of antibiotics. The maximum production of antibiotics was observed when the concentration of soybean meal is (between: 11.25 g and 12.25 g/l) and sodium chloride is (11.0–11.50 g/l). In addition, higher concentration of calcium carbonate (between: 1.25 g and 1.60 g/l) and lower concentration of sodium chloride (between: 12.5 g and 11.5 g/l) significantly increases the antibiotic production by Streptomyces parvulus K-R1. The final optimized nutritional conditions by CCD for maximum production of antibiotic by Streptomyces parvulus K-R1 were as: (glucose—11.25 g/l, soybean meal—12.17 g/l, calcium carbonate—1.47 g/l, sodium chloride—11.25 g/l).
Validation of Optimization Studies
The model was validated by growing the Streptomyces parvulus K-R1 under CCD optimized physical and nutritional parameters predicted by RSM (Fig. 4). The average antibacterial activity against Staphylococcus aureus MTCC-96 obtained experimentally by CCD was found to be 28.62 mm which was in close to the predicted value of 27.74 mm. Similarly, in the study of nutritional parameters, the average antibacterial activity against Staphylococcus aureus MTCC-96 obtained experimentally by CCD was found to be 26.12 mm which was in close to the predicted value of 25.73 mm. Optimization by CCD in the glucose soybean meal broth medium w/v (g/l) containing : glucose—11.25 g, soybean meal—12.17 g, calcium carbonate—1.47 g, sodium chloride—11.25 g; pH 7.18, inoculated with 4% seed inoculum incubated at 30.51 °C for 10 day enhanced the 1.23 fold increase of antibiotic production as compared from un-optimized starch casein nitrate broth medium.
Fermentation and Production of Antimicrobial Compound
The endophytic Streptomyces sp. K-R1 had highest biomass and antibiotic production in glucose soybean meal broth having medium composition w/v (g/l): glucose 11.25 g, soybean meal 12.17 g, calcium carbonate 1.47 g, sodium chloride 11.25 g; pH 7.18 at 30.51 °C with 4% seed inoculum for 10 days of incubation. Antimicrobial compound production and biomass production by Streptomyces sp. K-R1 was observed up to 14 days of incubation. Antibiotic production was detected in culture broth after 2 days of incubation and reached maximum on 10th day of incubation. Further increase in the incubation time gradually found to decrease the activity. However, mycelium growth gradually increased up to 14 days of incubation. These results showed that the antibiotic production by endophytic Streptomyces sp. K-R1 started at the early logarithmic phase of growth and reached the maximum at the mid exponential phase of growth. From 5 l fermentation broth, ethyl acetate extracted reddish orange color active compound (2.25 g) was obtained after concentration in rotary evaporator at 40 °C. Furthermore, different concentrations of ethyl acetate extracted antimicrobial compound 5, 10, 15 and 20 mg was tested against pathogenic bacteria viz., Klebsiella pneumoniae MTCC-3384, Klebsiella pneumoniae JNMC-6 and Proteus vulgaris MTCC-744, Proteus vulgaris JNMC-7 in order to determine the minimum inhibitory activity range of antimicrobial compound against multi drug resistant bacteria (Fig. 5). The growths of pathogens were continuously declined at 5–20 mg/ml concentration of antimicrobial compound. Furthermore, the MIC ranges of antimicrobial compound against pathogens were tested between 1 and 5 mg/ml respectively. The compound exhibited MIC values below 1 mg/ml and MBC values at 1 mg/ml against pathogens.
Purification and Bioefficacy of Antimicrobial Compound from Streptomyces sp. K-R1
Purification and isolation of antibiotics from endophytic Streptomyces parvulus K-R1 associated with root of Abutilon indicum shown in Fig. 6. Separation of active compound from the crude extract was carried out by thin layer chromatography. The crude antibiotic was dissolved in ethyl acetate and spotted on TLC plate. Different solvents were tried to separate the antibiotic, finally the best solvent system n-butanol: ethyl acetate: water (v/v 9:9:1) was used. A single separated yellow color band was observed with the Rf value 0.66 (UV active). The active spot is confirmed by testing the compound (1 mg/ml) against multi drug resistant pathogens. Further purity of the antimicrobial compound was checked by reverse phase HPLC (Shimadzu). Three compounds with antimicrobial activity were separated by the HPLC with retention time of 10.63, 10.88 and 6.3 min respectively (Online Resource 2). The purity of the first compound was found as 68.925%, second compound was 1.2% and third compound was 6.783% respectively. The first compound was found to be present in major quantity in the extract as compared from other two compounds. The purified antibiotic (1 mg/ml) was found to be active against multidrug resistant Gram-positive, Gram-negative and fungal pathogens (Table 2).
The purified antibiotic exhibited activity against multi drug resistant pathogens (resistant to at least three different classes of antibiotics) viz., Staphylococcus aureus JNMC-3, Staphylococcus epidermidis JNMC-4, Klebsiella pneumoniae MTCC-3384, Klebsiella pneumoniae JNMC-6, Pseudomonas aeruginosa MTCC-741, Proteus vulgaris JNMC-7 and Escherichia coli JNMC-5. The compound also showed activity against Bacillus cereus MTCC-430 and Staphylococcus aureus MTCC-96 (resistant to amoxicillin), Bacillus cereus JNMC-1 (resistant to gentamycin and tetracycline) and Escherichia coli MTCC-1687 (resistant to penicillin). In addition, the compound also showed antifungal activity against Candida albicans MTCC-183 and Aspergillus niger MTCC-872 (resistant to fuconazole and ketoconazole). The minimum inhibitory concentration (MIC) of the purified antimicrobial compound from Streptomyces parvulus K-R1 revealed the value of 0.12 mg/ml against MDR pathogen Pseudomonas aeruginosa MTCC-741; 0.25 mg/ml against Staphylococcus aureus MTCC-96 and Bacillus subtilis JNMC-2; 0.5 mg/ml against Bacillus subtilis MTCC-441, Proteus vulgaris MTCC-744, and Staphylococcus epidermidis JNMC-4, Candida albicans MTCC-183 and Bacillus cereus JNMC-1; 1 mg/ml against Bacillus cereus MTCC-430, Staphylococcus epidermidis MTCC-435, Escherichia coli MTCC-1687, Klebsiella pneumoniae JNMC-6, Proteus vulgaris JNMC-7 and Aspergillus niger MTCC-872. The minimum bactericidal concentration (MBC) was also determined by sub culturing the contents of the tube of MIC showing no growth on to antibiotic free liquid medium and examining for bacterial growth. The MBC values of the compounds against all the tested multidrug resistant human pathogens were 1 mg/ml.
Structure Elucidation and Identification of Antibiotic from Streptomyces sp. K-R1
Three bioactive compounds were isolated from glucose soybean meal broth medium and their structures were elucidated by UV–Vis, 1H NMR, 13C NMR, FT-IR and ESI-MS analysis (Online Resource 3). The compounds showed typical UV–Vis spectra with maximum absorbance at 443.1, 423.1 and 254.7 nm, similar to those of known Actinomycins and surfactin. ESI-MS of isolated compounds revealed molecular ion peaks at m/z 1255.40 [M + H]+ for compound 1; m/z 1271.63 [M + H] + for compound 2 and m/z 1096.5 [M − H]−; m/z 1464.2 [M − H]−, m/z 1113.6 [M + H] + for compound 3. The molecular weights of the compounds were identical to those of actinomycin D, actinomycin X0β and fengycin respectively (Fig. 7). The spectrum of the compound actinomycin D and actinomycin X0β was further confirmed by comparing the spectra with an authentic literature (Solanki et al. 2015; Zhang et al. 2016) (Tables 3, 4, 5). Compound 3: Fengycin was found to be present in the minor quantity in the extract of endophytic Streptomyces sp. K-R1. The 13C NMR spectrum exhibits carbonyl resonances between 173 and 179 ppm, which indicates the carbon signals of various amino acids. The carbon signals between 115 and 140 ppm correspond to C=C of alkenes and the resonances of various fatty acid chains are found between 10 and 40 ppm. Some unsaturated carbon atoms showed resonances at 122 and 131 ppm can be attributed to olefinic fatty acid residue. The FTIR spectrum of the compound showed bands characteristics of peptides at 1630.5 cm−1 (stretching mode of the CO–N bond), bands at 2952–2924.5, 2854.1 cm−1 and at 1466.6, 1376 cm−1 showed aliphatic chains (–CH3, –CH2–). The ESIMS, NMR and FTIR spectrum of the compound showed similarity to cyclic lipopeptides like surfactin produced by Bacilli, as reported by other researchers (Wei et al. 2010; Jha et al. 2016).
Production of Antibiotics (Actinomycin D, actinomycin X0β and fengycin) by Streptomyces sp. K-R1
Three antibiotics (actinomycin D, actinomycin X0β and fengycin) were detected in crude extracts and were eluted by HPLC at 10.63 min, 10.88 min and 6.3 min respectively. Production of antibiotics was observed in glucose soybean meal broth medium (under CCD optimized culture conditions) at every three days of incubation (Fig. 8). The yields of antibiotics from glucose soybean meal broth media were 240 mg/l after 5 days of incubation, 290 mg/l after 8 days and 340 mg/l after 11 days of incubation. The biomass of endophytic Streptomyces sp. K-R1 was continuously increased from 5th day to 16th day of incubation but the production of antibiotics was found to be maximum at 11th day of incubation and constant after 11 day of incubation.
Stability of Antibiotics in the Presence of Enzymes, Detergents and Temperature
The effect of enzymes, detergents and temperature on activity of antimicrobial compound from Streptomyces parvulus K-R1 was checked by mixing the antimicrobial compound and enzymes w/v (Table 6). The statistical analysis revealed that treatments with detergents differed significantly (p = 0.001). The antimicrobial compound retained higher residual activity in the presence of SDS (96.20%) and Tween-80 (98.73%). However, residual activity was significantly dropped to 93.67 and 89.53% in the presence of triton X-100 and tween-20 respectively. Similarly, antimicrobial activity of the compound was differed significantly (p = 0.011) when treated with enzymes. The compound retained higher residual activity in the presence of lysozyme (92.40%) and proteinase K (91.14%) suggesting that the active compound has stable nature in the presence of these enzymes when incubated for 1 h at 37 °C. However, residual activity significantly dropped to 89.87% in the presence of trypsin, suggesting that the active metabolite has peptide bonds, that when hydrolyzed leads to reduction of activity. In addition, the antimicrobial compound was found to be thermo tolerant in nature; the compound did not completely loose its activity even at 100 °C. However, significant decline in the residual activity was observed after 50 °C.
Effect of ergo sterol on antifungal activity of the antimicrobial compound from Streptomyces parvulus K-R1 has been evaluated. The control plate without ergo sterol showed an inhibition zone diameter of 17.75 mm against Candida albicans MTCC-183, whereas the plate containing reversal agent ergo sterol, showed a reduced inhibition zone diameter of 16.25 mm against Candida albicans MTCC-183. Thus, the antimicrobial compound from Streptomyces parvulus K-R1 probably binds to the ergo sterol present in the fungal cell membrane resulting in the leakage of intracellular material and eventually death of the cell.
Discussion
Endophytic Streptomyces genus has been widely used as an important biological tool for producing novel antibiotics (Golinska et al. 2015). Therefore, isolation and identification of antibiotics from Streptomyces sp. associated with medicinal plants is of great significance. In the present study, Streptomyces parvulus NBRC 13193T (AB184326) has been isolated from root of Abutilon indicum by surface sterilization method on starch casein agar medium. Previously this organism as an endophyte has also been reported from stems and root part of medicinal plant Dracaena cochinchinensis (Khieu et al. 2015) and root of Aloe vera (Chandrakar and Gupta 2017). Recently, two strains of Streptomyces parvulus NBRC 13193T and Streptomyces parvulus 12811T has been reported from stem and root part of medicinal plant Dracaena cochinchinensis (Salam et al. 2017). On the basis of the presence of Streptomyces parvulus in different host plants we can say that this organism is not a host specific. In this study, Streptomyces parvulus K-R1 exhibited broad spectrum antimicrobial activity against multidrug resistant Gram-positive bacteria viz. Bacillus cereus MTCC-430, Staphylococcus aureus MTCC-96, Bacillus subtilis MTCC-441, Staphylococcus epidermidis MTCC-435, Bacillus cereus JNMC-1, Bacillus subtilis JNMC-2, Staphylococcus aureus JNMC-3, Staphylococcus epidermidis JNMC-4; Gram-negative bacteria viz. Escherichia coli MTCC-1687, Klebsiella pneumoniae MTCC-3384, Proteus vulgaris MTCC-744, Pseudomonas aeruginosa MTCC-741, Escherichia coli JNMC-5, Klebsiella pneumoniae JNMC-6, Proteus vulgaris JNMC-7 and fungal pathogens viz. Candida albicans MTCC-183, Aspergillus niger MTCC-872. Similar findings have been reported in related studies of endophytic Streptomyces parvulus NBRC 13193T and Streptomyces parvulus 12811T associated with root of Dracaena cochinchinensis, which showed remarkable antifungal activity against plant pathogens viz., Fusarium graminearum, Aspergillus carbonarius and Aspergillus westerdijkiae (Salam et al. 2017). On the basis of broad spectrum antimicrobial activity of Streptomyces parvulus against plants and human pathogens we can say that, they have formed a symbiotic and beneficial association with plants. It is reported from the earlier studies that, actinomycin D from endophytic Streptomyces sp. Tc022 associated with root of Alpinia galanga exhibited strong antifungal activity against Colletotrichum musae and Candida albicans (Taechowisan et al. 2006). Similarly, other researchers reported actinomycin D from Streptomyces parvulus (KJ200636.1) associated with Codonopsis lanceolata showed potential anti virulence activity against Staphylococcus aureus infection which significantly inhibited biofilm formation by Staphylococcus aureus ATCC-25923 and ATCC-6538 (sensitive to methicillin) and Staphylococcus aureus ATCC-33591 (resistant to methicillin) (Lee et al. 2016). Difference in the antimicrobial activity of Streptomyces parvulus between similar species with different host tissues might be related with the chemical difference of host plants and the physiological condition of the host plant which trigger the production of antibiotics for the survival of Streptomyces parvulus against pathogens.
In the present study, Streptomyces parvulus K-R1 produced actinomycin D, actinomycin X0β and fengycin when grown in glucose soybean meal broth medium. However, there are still no reports that Streptomyces parvulus can produce fengycin. Although, Streptomyces parvulus isolated from soil and plants has been reported to produce actinomycin D and actinomycin X0β (Taechowisan et al. 2006; Lee et al. 2016). In addition, in our earlier studies, similar strain of Streptomyces parvulus Av-R5 has been isolated from root of Aloe vera, which produces similar compound actinomycin D and actinomycin X0β but not fengycin. Hence, the difference in the antibiotic production ability of endophytic Streptomyces parvulus between similar species with different host tissues might be related with the chemical difference of host plant and the physiological condition of the host plant which trigger the production of antibiotic like fengycin for the survival of endophytic Streptomyces parvulus against plant pathogenic fungal organisms. In the present study, actinomycin D and actinomycin X0β were separated by HPLC with the retention time of 10.63, 10.88 and 6.3 min along with the purity of 68.925, 1.2 and 6.873% respectively. Actinomycin D was found to be present in large quantity in the extract of endophytic Streptomyces parvulus (K-R1) possessing antimicrobial activity against human pathogens. In addition, actinomycin D can be synthesized by different species of Streptomyces as part of a mixture of several Actinomycins viz. actinomycin X2, actinomycin X0β and actinomycin D (Chen et al. 2012; Hamza et al. 2013). In our study, the antibiotic compound isolated from Streptomyces parvulus K-R1 recovered from root of Abutilon indicum also exhibited inhibitory against fungus. This antifungal property may be attributed to the presence of fengycin which was detected in the extract and identified by LCMS and NMR spectrum. Fengycin is a lipopeptides bio surfactant generally produced by Bacillus subtilis and particularly active against filamentous fungi viz. Aspergillus niger, Fusarium graminearum and Sclerotinia sclerotiorum (Wei et al. 2010). Hence, endophytic Streptomyces parvulus K-R1 associated with root of Abutilon indicum may be exploited as a potential source for the commercial production of fengycin, as this strain is not reported to produce fengycin. Further investigations on the strain for commercial production and application of fengycin will be of immense pharmaceutical importance.
Optimum culture conditions are a key factor for the growth as well as antibiotic production by microorganisms. In the present study, maximum production of antibiotics by endophytic Streptomyces parvulus K-R1 was found by using GSB medium containing glucose 11.25 g/l, soybean meal 12.17 g/l, calcium carbonate 1.47 g/l, sodium chloride 11.25 g/l at pH 7.18, incubated at 30.52 °C for 10 days with the inoculum volume 3.91%. The co-variation between cell growth and antibiotic production test, we found that production of antibiotics by Streptomyces parvulus K-R1 was started at the early logarithmic phase of growth on second day and produce maximum at mid logarithmic phase on tenth day. However, similar strain of Streptomyces parvulus AB184326 has been reported to synthesize actinomycin in Czapek-dox broth and tryptone soya broth after 10 days (Rahman et al. 2010; Hamza et al. 2013). Different investigators used different fermentation media for the production of Actinomycins by Streptomyces parvulus strains viz., Streptomyces parvulus DAUFPE 3124 began actinomycin synthesis after 6 days in chemically defined medium containing fructose 30 g/l, soy milk 30 g/l and calcium carbonate 2 g/l (Sausa et al. 2002). Similarly, actinomycin synthesis by Streptomyces parvulus RSPSN2 began after 10 days in PM-2 medium (Shetty et al. 2014). Streptomyces parvulus KUAP106 began actinomycin synthesis after 5 days in starch casein nitrate broth (Usha et al. 2010). We observed that, major as well as minor changes in the amount of the medium components could be triggering the synthesis of actinomycin by Streptomyces strains at their logarithmic growth phase.
Actinomycin synthesis by different Streptomyces parvulus strains has been strongly influenced by the carbon and nitrogen sources present in the production medium (Hamza et al. 2013). In our study we found that, antibiotic production was directly proportional to the concentration (within a certain range) of glucose and soybean meal. Response surface methodology helps in evaluation of relationship between the dependent (antibiotic production) variable and independent (media components) variables. A significant increase in the production of antibiotic was achieved when the amount of glucose increased between 10.25 to 11.50 g/l, calcium carbonate from 0.97 to 1.47 g/l, sodium chloride and soybean meal from 10.25 to 12.25 g/l. Further, increase in glucose concentration suppressed the antibiotic production by Streptomyces parvulus K-R1. In the present study, the incorporation of 2% glucose in the production medium significantly influenced the synthesis of actinomycin and also supported abundant cellular growth of endophytic Streptomyces parvulus K-R1. It was reasoned, therefore, that glucose might be consumed rapidly for the production of cell material and would be available in sufficient amount for actinomycin synthesis by the isolate K-R1. However, other researchers reported that d-fructose enhances the actinomycin synthesis by Streptomyces parvulus and complete inhibition of actinomycin synthesis by Streptomyces parvulus was observed when glucose and galactose used as a carbon source (Williams and Katz 1977; Sausa et al. 2002; Praveen et al. 2008). It was observed that, the incorporation of soybean meal as a nitrogen source was found to be good source for actinomycin production by endophytic Streptomyces parvulus K-R1. In this study, the combination of glucose and soybean meal was found to be excellent carbon and nitrogen source for cell growth as well as actinomycin synthesis by Streptomyces parvulus K-R1. In the earlier studies, glutamic acid, phosphates, sodium nitrate and l-threonine have been reported to induce the synthesis of actinomycin by different species of Streptomyces (Katz et al. 1956). The nature and the amount of carbon and nitrogen sources have been considered as a direct precursor for actinomycin D biosynthesis by Streptomyces strains. In the present study, actinomycin D biosynthesis by endophytic Streptomyces parvulus K-R1 has been achieved when the concentration of glucose was 1.12% and soybean meal 1.21% (< 2%) whereas higher concentration (> 2%) was found to be the excellent for cell growth of Streptomyces parvulus K-R1. It was reasoned therefore, that the higher concentration (> 2%) of glucose and soybean meal consumed so rapidly for the growth of cell material and little would be available as carbon and nitrogen source for actinomycin synthesis by Streptomyces parvulus K-R1. Hence, the lower concentration (< 2%) of carbon and nitrogen sources which had not been completely utilized during growth would prove to be satisfactory for actinomycin synthesis by Streptomyces parvulus K-R1. One would expect that, a carbon and nitrogen source which had been completely utilized during growth would prove unsatisfactory for subsequent antibiotic production; conversely a compound which is partly consumed during cell growth should be much more suitable for antibiotic production. Many researchers reported that higher concentration of glucose (> 1%) in the production medium repressed the synthesis of actinomycin D by Streptomyces parvulus due to the repression of the phenoxazinone synthase enzyme activity which catalyses the synthesis of phenoxazinone ring of actinomycin (Gallo and Katz 1972).
Actinomycin synthesis under optimized medium yielded much higher level of actinomycin (340 mg/l), which compared favorably to other values reported in the literature for actinomycin synthesis in the similar medium components. Although, our strain endophytic Streptomyces parvulus K-R1 yielded much higher level of actinomycin then, the medium optimized in bioreactors (Sousa et al. 2002; Praveen et al. 2008). Actinomycin biosynthesis by Streptomyces strains has been greatly influenced by carbon and nitrogen ratio. The higher yield (87.7 g/l) of actinomycin by Streptomyces parvulus DAUFPE 3124 has been achieved at fructose and l-threonine C/N ratio of 20/3.57 g/l (Sousa et al. 2001). Similarly, higher yield of actinomycin by Streptomyces thermocarboxydus 173998 has been achieved at 10 g/l carbon source viz., starch (305 mg/l), maltose (225 mg/l) and mannitol (223 mg/l); lower yield of actinomycin has been reported in the presence of arabinose (185 mg/l), fructose (192 mg/l), glucose (110 mg/l), raffinose (100 mg/l), glycerol (99 mg/l), sucrose (83 mg/l) and xylose (16 mg/l) (Hamza et al. 2013). Recently, actinomycin D synthesis by agricultural soil bacteria Streptomyces hydrogenans IB310 has been optimized through Response Surface Methodology. Highest yield of actinomycin D (18.912 mg/l) was obtained at glycerol 1.949 g/l, oat meal 2.676 g/l and tween 80 0.524 g/l (Kulkarni et al. 2017). Enhanced synthesis of Actinomycins production by Streptomyces halstedii MTCC 6817 has been obtained in the presence of fructose (410 mg/l) followed by lactose (182 mg/l), maltose (178 mg/l), xylose (92 mg/l), galactose (70 mg/l), glucose (56 mg/l) and mannose (34 mg/l) (Praveen et al. 2008). In the present study, optimum temperature between; 30.0 and 31.0 °C and pH between; 7.00 and 7.25 greatly influenced the cell growth and favor the higher yield of actinomycin synthesis by Streptomyces parvulus K-R1. In addition, Streptomyces parvulus K-R1 from root of Abutilon indicum under central composite design optimization approach resulted in 1.23 fold increase of actinomycin production as compared from un-optimized medium. So, the obtained result from optimization is very useful for improving supply of the intended actinomycin from endophytic Streptomyces parvulus K-R1 for its industrial application in future.
According to the literature Streptomyces parvulus has the advantage of producing predominantly actinomycin D (Meienhofer and Atherton 1973; Kulkarni et al. 2017). In the present study, Streptomyces parvulus K-R1 from root of Abutilon indicum also predominantly produced actinomycin D and actinomycin X0β. In addition, thin layer chromatography of the antimicrobial compound from Streptomyces parvulus K-R1 showed single band when using different solvents. Other researchers also reported that Streptomyces parvulus had only a single band for actinomycin D; while other species of Streptomyces shows multiple bands in TLC and produce different unknown compounds (Sousa et al. 2002). It was observed that, metabolites treated with proteolytic enzymes slightly inhibited antibiotic production by 89–92%. The significant reduction in the activity of active metabolite in the presence of trypsin reveals that, the active metabolite has peptide bond that when hydrolyze leads to the gradual decline in the activity and the active metabolite has polypeptide nature. Detergents increase the permeability of cell membrane for antibiotics. However some detergents have an ability to degrade the antibiotics therefore antibiotics could not be able to enter into the bacterial cell. In the present study, non significant decline in the activity of antimicrobial compound was observed in the presence of SDS, triton X-100, tween-20 and tween-80. This results suggested that, antimicrobial activity of the compound did not completely lose its activity in the presence of detergents, hence the antibiotic can easily be enter into the bacterial cell and cause death of the pathogen. Thermo stability studies exhibited that antimicrobial compound retained antimicrobial activity after treatment with different temperatures up to 30 to 100 °C. The gradual decline in the antibacterial activity of the antibiotic after treatment at 50 °C and above for 1 h of incubation was observed. This result indicates that the chemical structure of active compounds present in crude extracts does not tolerate higher temperatures for long periods of time. Actinomycins are polypeptide natures of antibiotic which has been reported to produced by various species of Streptomyces viz., Streptomyces griseoruber (Praveen and Tripathi 2009), Streptomyces parvulus (Williams and Katz 1977), Streptomyces sindenensis (Praveen et al. 2008), Streptomyces avermititis (Chen et al. 2012) and Streptomyces flavogriseus (Wei et al. 2017). In addition, only a few strains of Streptomyces parvulus have been reported to produce relatively large quantities of actinomycin D, including Streptomyces parvulus GQ451836 (180 mg/l) (Rahman et al. 2010), Streptomyces parvulus DAUFPE 3124 (133 mg/l) (Sousa et al. 2002), Streptomyces parvulus (152 mg/l) (Shetty et al. 2014) and by other species viz., Streptomyces griseoruber (210 mg/l) (Praveen and Tripathi 2009), Streptomyces thermocarbodoxydus 173998 (305 mg/l) (Hamza et al. 2013), a mutant strain of Streptomyces sindenensis (850 mg/l) (Praveen et al. 2008) and Streptomyces flavogriseus NJ-4 (960 mg/l) (Wei et al. 2017). Whereas, our endophytic Streptomyces parvulus K-R1 from root of Abutilon indicum exhibited a unique ability to produce large quantities of Actinomycins and fengycin with a production of 240 mg/l after 5 days, 290 mg/l after 8 days and 340 mg/l after 11 days of incubation in glucose soybean meal broth media; as compared to the similar species of Streptomyces parvulus reported from soil and plants (Taechowisan et al. 2006; Lee et al. 2016).
Actinomycin production by various species and strains of Streptomyces has been reported earlier and studied extensively (Chen et al. 2012; Praveen et al. 2008). Commercially, actinomycin D is produced by Streptomyces parvulus (Kurosawa et al. 2005). However, production and optimization of actinomycin by endophytic Streptomyces parvulus associated with medicinal plants has not been reported in literature. Actinomycins are chromophoric peptide antibiotic produced through an oxidative condensation of two molecules of the 3-hydroxy-4-methyl-anthranilic peptide and difference found in the components of an actinomycin mixtures are due to the number, arrangements and kinds of amino acids present in the peptides (Chen et al. 2012; Katz et al. 1956; Praveen et al. 2008). Hence, the searching of actinomycin from Streptomyces species present inside the unique habitat like plants has a great deal of research into the new analogous of actinomycin with unique chemical, physical and biological characteristics. Antibiotic from Streptomyces parvulus K-R1 associated with root of Abutilon indicum was found to be most active against Pseudomonas aeruginosa MTCC-741 with the MIC of 0.12 mg/ml; Staphylococcus aureus MTCC-96 and Bacillus subtilis JNMC-2 were sensitive at 0.25 mg/ml; Bacillus subtilis MTCC-441, Proteus vulgaris MTCC-744, Bacillus cereus JNMC-1 and Staphylococcus epidermidis JNMC-4 and Candida albicans MTCC-183 was sensitive at 0.5 mg/ml; Bacillus cereus MTCC-430, Staphylococcus epidermidis MTCC-435, Escherichia coli MTCC-1687, Klebsiella pneumoniae MTCC-3384, Staphylococcus aureus JNMC-3, Escherichia coli JNMC-5, Klebsiella pneumoniae JNMC-6, Proteus vulgaris JNMC-7 and Aspergillus niger MTCC-872 was sensitive at 1 mg/ml. However, the MBC values were found to be higher than the MIC against multidrug resistant Pseudomonas aeruginosa MTCC-741, Proteus vulgaris MTCC-744, Staphylococcus epidermidis JNMC-4; Staphylococcus aureus MTCC-96, Bacillus subtilis JNMC-2, Bacillus subtilis MTCC-441 (resistant to penicillin), Candida albicans MTCC-183 (resistant to fuconazole and ketoconazole), Bacillus cereus JNMC-1 (resistant to gentamycin and tetracycline). As the MBC was higher than the MIC against particular organisms, it is inferred that the compound has bacteriostatic nature against these organisms and not bactericidal. The present results indicated that the Actinomycins and fengycin from endophytic Streptomyces parvulus K-R1 from root of Abutilon indicum has been showed extensive inhibition to the growth of Aspergillus niger MTCC-872 and Candida albicans MTCC-183. Antifungal activities of actinomycin D biosynthesized by endophytic Streptomyces parvulus against plant pathogens have been reported earlier (Salam et al. 2017; Taechowisan et al. 2006; Lee et al. 2016). Recently, actinomycin D produced by agricultural soil bacteria Streptomyces hydrogenans IB310 has been showed antagonistic activity against bacterial and fungal phytopathogens viz., Agrobacterium tumefaciens, Pseudomonas syringae, Xanthomonas campestris, Botrytis allii, Fusarium oxysporum and Ustilago maydis (Kulkarni et al. 2017). As stated in earlier reports, endophytic Streptomyces species present inside the host plants protects the plants against pathogenic microorganisms by the production of antibiotics; in return plant provides the chemical nutrients for the growth and survival of endophytic Streptomyces and influences the metabolite production efficiency of endophytic Streptomyces species. Hence, the possibility of the products from endophytes may be used for biocontrol of the related plant diseases. Ergo sterol present in the fungal cell membrane is a useful target for antifungal drugs, which causes ions and other molecules to leak out from the cell membrane and kill the pathogen (Nafis et al. 2018). Hence, the incorporation of exogenous ergo sterol in the culture medium interferes with the activity of the potential antifungal substances produced by the isolates and reduces the inhibition diameter in the assay. In our study, antibiotics from Streptomyces parvulus K-R1 showed reduced zone of inhibition after exogenous ergo sterol addition against Candida albicans MTCC-183. This could be explained by the production of antifungal compound by the same strain. Hence the antibiotic may binds to the ergo sterol present in the fungal cell membrane resulting in the leakage of intracellular materials and cause death of the pathogen. The isolation of Actinomycins from Streptomyces parvulus is not a new discovery but our present study proved that the medicinal plant Abutilon indicum has rich source of endophytic actinomycetes and producing a large amount of Actinomycins and fengycin against multidrug resistant bacterial and fungal pathogens.
Conclusion
It is concluded from the present study that endophytic Streptomyces sp. K-R1 associated with root of Abutilon indicum has antibiotic potential. Cultural characteristics and nucleotide sequence of the isolate suggested that the isolate was Streptomyces parvulus NBRC 13193T (AB184326). In this case, a high yield (340 mg/l) of actinomycin D, actinomycin X0β and a new compound fengycin is achieved with Streptomyces parvulus K-R1 fermented in glucose soybean meal broth media, which can be used in industrial fermentation process to obtain high yields. Also, glucose and soybean meal is cheaper and easily available substrates by organic wastes. Therefore, our result is encouraging as a value addition of waste and bioprocess economy. In addition, ethyl acetate extract of Actinomycins and fengycin showed broad spectrum antimicrobial activity against multidrug resistant Gram-positive, Gram-negative bacteria and fungi. Different fermentation conditions for the production of Actinomycins by Streptomyces parvulus has been reported from soil sources. To the best of our knowledge, isolation of endophytic actinomycetes for their antibiotic potentiality from medicinal plants of Chhattisgarh has not to be explored yet; hence, this is the first report on production and optimization of Actinomycins and fengycin from endophytic Streptomyces parvulus K-R1 associated with Abutilon indicum from Chhattisgarh. In addition, there are still no reports that Streptomyces parvulus can produce fengycin. Although, Streptomyces parvulus isolated from soil and plants has been reported to produce actinomycin D and actinomycin X0β. Hence, this is the first report for the synthesis of fengycin by Streptomyces parvulus. The Actinomycins and fengycin produced by isolate K-R1 showed extensive inhibition to the growth of fungal pathogens which are reported to be found in the agricultural soil and become as phytopathogens, hence the Actinomycins and fengycin from our endophytic Streptomyces parvulus K-R1 may be used for biocontrol of the related plant diseases.
References
Balagurunathan R, Subramanian A (1993) Studies on marine Streptomyces nigrifaciens taxonomy and standardization of antibiotic production. Cienc Mar 19:435–443
Castillo UF, Strobel GA, Ford EJ, Hess WM, Porter H, Jensen JB, Albert H, Robison R (2002) Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology 148:2675–2685
Castillo U, Harper JK, Strobel GA, Sears J, Alesi K, Ford E, Lin J, Hunter M, Maranta M, Ge H, Yaver D, Jensen JB, Porter H, Robison R, Miller D, Hess WM, Condron M, Teplow D (2003) Kakadumycins, novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol Lett 234:183–190
Chandrakar S, Gupta A (2017) Antibiotic potential of endophytic actinomycetes of medicinal herbs against human pathogenic bacteria. Proc Natl Acad Sci India B 87:905–915
Chen C, Song F, Wang Q, Abdel-Mageed WM, Guo H, Fu C, Hou W, Dai H, Liu X, Yang N, Xie F, Yu K, Chen R, Zhang L (2012) A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-tuberculosis activity. Appl Microbiol Biotech. https://doi.org/10.1007/s00253-012-4079-z
Ezra D, Castillo UF, Strobel GA, Hess WM, Porter H, Jensen JB, Condron MAM, Teplow DB, Sears J, Maranta M, Hunter M, Weber B, Yaver D (2004) Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU-2110) endophytic on Monstera sp. Microbiology 150:785–793
Gallo M, Katz E (1972) Regulation of secondary metabolite biosynthesis catabolic repression of phenoxazinone synthase and actinomycin formation by glucose. J Bacteriol 109:659–667
Golinska P, Wypij M, Agarkar G, Rathod D, Dahm H, Rai M (2015) Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Anton Leeuw 108:267–289
Guan S, Suttler I, Lin W, Guo D, Grabley S (2005) p-Aminoacetophenonic acids produced by a mangrove endophyte: Streptomyces griseus sub sp. J Nat Prod 68:1198–1200
Hamza AA, Ali HA, Clark BR, Murphy CD, Elobaid EA (2013) Isolation and characterization of actinomycin D producing Streptomyces sp. from Sudanese soil. Afr J Biotechnol 12:2624–2632
Igarashi Y, Ogura H, Furihata K, Oku N, Indananda C, Thamchaipenet A (2011) Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J Nat Prod 74:670–674
Indanada C, Igarashi Y, Ikeda M, Oikawa T, Thamchaipenet A (2013) Linfuranone A, a new polyketide from plant derived Microbispora sp. GMKU 363. J Antibiot 66:675–677
Jeon YS, Lee K, Park SC, Kim BS, Cho YJ, Ha SM (2014) EzEditor: a versatile sequence alignment editor for both rRNA-and protein-coding genes. Int J Syst Evol Microbiol 64:689–691
Jha SS, Joshi SJ, Geetha SJ (2016) Lipopeptide production by Bacillus subtilis R1 and its possible applications. Braz J Microbiol 47:955–964
Katz E, Pienta P, Sivak A (1956) The role of nutrition in the synthesis of actinomycin. Appl Microbiol 6:236–241
Kawato M, Shinobu R (1959) Cover slip culture of Streptomyces herbaricolour nov. sp. supplement: a simple technique for the microscopical observation. Mem Osaka Univ Lib Arts Educ 8:114–119
Khieu TN, Liu MJ, Nimaichand S, Quach NT, Ky SC, Ph QT, Vu TT, Nguyen TD, Xiong Z, Prabhu DM, Li WJ (2015) Characterization and evaluation of antimicrobial and cytotoxic effects of Streptomyces sp. HUST012 isolated from medicinal plant Dracaena cochinchinensis Lour. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00574
Kim N, Shin JC, Kim W, Hwang BY, Kim BS, Hong YS, Lee D (2006) Cytotoxic 6 alkylsalicylic acids from the endophytic Streptomyces laceyi. J Antibiot 59:797–800
Kulkarni M, Gorthi S, Banerjee G, Chattopadhyay P (2017) Production, characterization and optimization of actinomycin D from Streptomyces hydrogenans IB310, a (n antagonistic bacterium against phytopathogens). Biocatal Agric Biotechnol 10:69–74
Kurosawa K, Bui VP, Van EJL, Willis LB, Lessard PA, Ghiviriga I, Sambandan TG, Rha CK, Sinskey AJ (2005) Characterization of Streptomyces MITKK-103, a newly isolated actinomycin X2 producer. Appl Microbiol Biotechnol 72:145–154
Lee JH, Kim YG, Lee K, Kim CJ, Park DJ, Ju Y, Lee JC, Wood TK, Lee J (2016) Streptomyces-derived actinomycin D inhibits biofilm formation by Staphylococcus aureus and its hemolytic activity. Biofouling 32:45–56
Meienhofer J, Atherton E (1973) Structure activity relationship in the actinomycins. Adv Appl Microbiol 16:203–300
Munimbazi C, Bullerman LB (1998) Isolation and partial characterization of antifungal metabolites of Bacillus pumilus. J Appl Microbiol 84:959–968
Nafis A, Oubaha B, Elhidar N, Ortlieb N, Kulik A, Niedermeyer T, Hassani L, Barakate M (2018) Novel production of two new non polyenic antifungal macrolide derivatives by Streptomyces Z26 isolated from Moroccan rhizospheric soil. Online J Biol Sci. https://doi.org/10.3844/ojbsci.2018
Praveen V, Tripathi CKM (2009) Studies on the production of actinomycin-D by Streptomyces griseoruber: a novel source. Lett Appl Microbiol 49:450–455
Praveen V, Tripathi CKM, Bihari V, Srivastava SC (2008) Production of actinomycin D by the mutant of a new isolate of Streptomyces sindenensis. Braz J Microbiol 39:689–692
Pridham TG, Gottlieb D (1948) The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J Bacteriol 56:107–114
Rahman MA, Islam MZ, Khondkar P, Islam MA (2010) Characterization and antimicrobial activities of a polypeptide antibiotic isolated from a new strain of Streptomyces parvulus. Bangla Pharma J 13:14–17
Roy S, Banerjee D (2015) Broad spectrum antibacterial activity of granaticinic acid, isolated from Streptomyces thermoviolaceus NT1; an endophyte in Catharanthus roseus (L.) G. Don. J Appl Pharma Sci 5:6–11
Salam N, Khieu TN, Liu MJ, Vu TT, Ky SC, Quach NT, Phi QT, Narsing Rao MP, Fontana A, Sarter S, Li WJ (2017) Endophytic actinobacteria associated with Dracaena cochinchinensis Lour: isolation, diversity and their cytotoxic activities. BioMed Res Int. https://doi.org/10.1155/2017/1308563
Shetty PR, Buddana SK, Tatipamula VB, Naga VVV, Ahmad J (2014) Production of polypeptide antibiotic from Streptomyces parvulus and its antibacterial activity. Braz J Microbiol 45:303–312
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Solanki R, Kundu A, Das P, Khanna M (2015) Characterization of antimicrobial compounds from Streptomyces sp. World J Pharma Res 4:1626–1641
Sousa MFVQ, Lopes CE, Junior NP (2001) A chemically defined medium for production of actinomycin D by Streptomyces parvulus. Braz Arch Biol Technol 44:227–231
Sousa MFVQ, Lopes CE, Pereira NJ (2002) Development of a bioprocess for the production of actinomycin D. Braz J Chem Eng 19:277–285
Taechowisan T, Wanbanjob A, Tuntiwachwuttikul P, Taylor WC (2006) Identification of Streptomyces sp. Tc022, an endophyte in Alpinia galanga, and the isolation of actinomycin D. Ann Microbiol 56:113–117
Taechowisan T, Chanaphat S, Ruensamran W, Phutdhawong SW (2014) Antibacterial activity of new flavonoids from Streptomyces sp. BT01; an endophyte in Boesenbergia rotunda (L) Mansf. J Appl Pharma Sci 4:8–13
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Usha R, Ananthaselvi P, Venil CK, Palaniswamy M (2010) Antimicrobial and antiangiogenesis activity of Streptomyces parvulus KUAP106 from mangrove soil. Eur J Biol Sci 2:77–83
Wang F, Xu M, Li Q, Sattler I, Lin W (2010) P-amino acetophenonic acids produced by a mangrove endophyte Streptomyces sp. (strain HK10552). Molecules 15:2782–2790
Wei YH, Wang LC, Chen WC, Chen SY (2010) Production and characterization of fengycin by indigenous Bacillus subtilis F29-3 originating from a potato farm. Int J Mol Sci 11:4526–4538
Wei Z, Xu C, Wang J, Lu F, Bie X, Lu Z (2017) Identification and characterization of Streptomyces flavogriseus NJ-4 as a novel producer of actinomycin D and holomycin. Peer J. https://doi.org/10.7717/peerj.3601
Williams WK, Katz E (1977) Development of a chemically defined medium for the synthesis of actinomycin-D by Streptomyces parvulus. Antimicrob Agents Chemother 11:281–290
Zhang X, Ye X, Chai W, Lian XY, Zhang Z (2016) New metabolites and bioactive Actinomycins from marine derived Streptomyces sp. ZZ338. Marine Drugs 14:181–189
Acknowledgements
Authors are thankful to the Head, SLS, PRSU, Raipur for providing the necessary facilities for the research. One of the authors (SC) is grateful to University Grant Commission, New Delhi, India for financial support in the form of Basic Science Research (F.7-145/2007BSR), SRF. Financial support from DST under FIST program and UGC for DRS-SAP III are gratefully acknowledged. The authors also thank Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh for providing cultures and Pandit Jawaharlal Nehru Medical College, Raipur, Chhattisgarh for providing clinical cultures for research.
Funding
This study was funded by University Grant Commission, New Delhi, India (F.7-145/2007BSR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declares that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chandrakar, S., Gupta, A.K. Studies on the Production of Broad Spectrum Antimicrobial Compound Polypeptide (Actinomycins) and Lipopeptide (Fengycin) from Streptomyces sp. K-R1 Associated with Root of Abutilon indicum against Multidrug Resistant Human Pathogens. Int J Pept Res Ther 25, 779–798 (2019). https://doi.org/10.1007/s10989-018-9727-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-018-9727-4