Abstract
Microorganisms secrete antimicrobial peptides (AMPs) which are part of the innate immune system and rapidly increase in concentration in the host upon challenge by pathogens, which they produce themselves. Kimchi, a traditional Korean food fermented by Bacillus organisms, is found to be ideal for AMP production. Our aim was to investigate the therapeutic potential of antimicrobial substances produced by Bacillus species. Peptide K1R was subjected to fermentation in a culture media containing carbon and nitrogen sources and metal ions. A protein band around 4.6 kDa was detected in tricine-SDS-PAGE and confirmed by in situ inhibitory activity of the gel. Peptide K1R was stable over a broad range of pH (6.5–9), thermo tolerant up to 60 °C and showed unaltered activity at low temperatures (0–4 °C). The complete amino acid sequence of peptide K1R was AVQGTLEDALNLSKGALNQVQKAIQNGDXLTVXGXLGTIXLAVSX. The antagonistic effect of peptide K1R against multiple drug resistant (MDR) pathogens such as Salmonella Typhimurium and Enterococcus sp. verified its potential application in treating MDR cases. The antioxidant activity of peptide K1R was also comparable to that of standard ascorbic acid.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antimicrobial peptides (AMP) constitute a distinct class of naturally existing antimicrobial molecules that have activity against a wide range of pathogenic microorganisms. Currently, conventional antibiotics are under intense pressure from developing resistance. AMPs are exciting leads in the development of novel biocidal agents. They also are alternatives to chemical food preservatives.
Despite continuing efforts, the increasing prevalence of resistance to common antibiotics among pathogenic bacteria has become one of the most significant concerns in modern medicine. With significantly reduced investments in antimicrobial research and development among major pharmaceutical companies, novel alternatives to the existing treatment strategies are not being produced at a sufficient rate to keep pace with the emergence of resistance, and the supply pipeline runs perilously close to drying up (Falagas et al. 2006).
AMPs serve as important defensive weapons against a broad spectrum of bacterial and fungal pathogens throughout the animal and plant kingdoms (Andreu and Rivas 1998). The sources range from single-celled microorganisms, such as bacteria themselves (bacteriocins) (Joerger 2003), to invertebrates (Brown and Hancock 2006). AMPs have a direct effect on microorganisms. In addition they have been proven to promote the accumulation of immune cells including macrophages, neutrophils and lymphocytes (Bowdish et al. 2005), neutralize lipopolysaccharides (endotoxin) derived from gram-negative bacteria (Rosenfeld et al. 2008), aid in wound healing, stimulate angiogenesis (Elsbach 2003), and control the actions of the innate and adaptive immune response with little or no resistance development. AMPs exert their microbicidal effect via disruption of the microbial cell membrane together with intracellular action and some of them exhibit both properties which is concentration dependent (Yeaman and Yount 2003; Brogden 2005).
AMPs are considered as one of the major candidates for the development of novel therapeutic agent alternatives to conventional antibiotic therapy. They are being extensively evaluated as novel antimicrobial drugs AMPs because they damage the membrane integrity of microbes and thus are thought to be less likely to induce resistance (Sang and Blecha 2008). Collectively, these peptides demonstrate a broad range of antiviral and antibacterial activities and modes of action, and it is important to distinguish between the direct microbicidal and indirect activities against pathogens. The structural requirements of peptides for antiviral and antibacterial activities are evaluated in light of the diverse set of primary and secondary structures described for host-defense peptides (Jenssen et al. 2006).
The production of AMPs by Bacillus strains has been increasingly characterized in the recent past and many peptides produced by this group of bacteria have been extensively employed in fermentation technology to manufacture antibiotics. Bacillus and related species have been the object of particular interest because of their safety, widespread distribution in diverse habitats, and remarkable ability to survive adverse conditions owing to the development of endospores. The AMPs from Bacillus spp., includes various classes of bacteriocins (Klaenhammer 1993), antimicrobial surface-active biosurfactants such as lipopeptides, glycopeptides and nonribosomally synthesized cyclic peptides (Mukherjee et al. 2006; Rodrigues et al. 2006).
The main objective of this work was to determine the effect and activity of AMPs which may contribute in the arena of pharmaceuticals, microbiology, and biotechnology for the welfare of mankind. We have been working to find out alternatives to the available antibiotic therapy to eliminate multi-drug-resistant bacteria. AMPs are naturally synthesized compound that exert antagonistic effect on other species. The bactericidal effects produced by microorganisms allow them to exist in their surroundings as a means of survival. These naturally produced compounds can be alternatives to the available antibiotic therapy. The therapeutic application of AMP hindered by problems such as low stability, toxicity, renal clearance and high manufacturing cost. In this study, we put our effort in utilizing our newly isolated AMP produced by Bacillus sp. K1R. K1R was isolated from fermented food, kimchi (a popular traditional Korean food). It shows stability towards a wide range of tested conditions.
Materials and Methods
Materials
Sephadex G-50 and Sephadex G-25 were purchased from Pharmacia (Uppsala, Sweden). All the other reagents were of analytical grades.
Isolation and Screening
The bacterial strain K1R was isolated from the traditional Korean food, kimchi. For isolation, 1 g of kimchi was mixed with 9 mL of 0.85% NaCl and kept in incubator at 37 °C for 24 h. After diluting up to 107 times with distilled water, the diluted sample was inoculated in Mueller–Hinton agar plates. After growth of bacteria, the plates were subjected for screening and identification. This was done according to the Bergey’s manual of systematic bacteriology. Many researchers have demonstrated kimchi as a source of many bacterial strains capable of producing AMPs (Lee et al. 2011, 1999; Yamanaka et al. 2007).
Sequence Similarities
The BLAST program (http://www.ncbi.nlm.nih.gov/blast) was employed to assess the degree of DNA similarity.
Optimization of Culture Media
Optimization of culture media has great significance in the improved production of AMP. Different researchers suggest different types of media for the enhanced production of bacteriocins. Media optimization was performed with 1% of different carbon sources (Lactose, Sorbitol, Sucrose, Glucose, Mannitol, Starch, Fructose, and Maltose), 1% of different nitrogen sources (Tryptone, Yeast extract, Peptone, Oat Meal, Beef extract, Malt, Soymeal) and 0.01% of different metal ions (KH2PO4, FeSO4, NaCl, Na2HPO4, CaCl2, NaH2PO4, MgSO4, ZnSO4, and MgCl2) till final optimization (Fig. 2d). de Man, Rogosa and Sharpe bacterial growth media (commonly termed as MRS media) was taken as a control media and the amount of the three sources was varied. Finally, 2% beef extract and 2% maltose were established as the optimized media and fermentation was carried out in 250 mL Erlenmeyer flasks containing 20% of the media (50 mL) with constant shaking at 160 rpm at 37 °C. No effect on metal ions was observed.
Peptide Production and Purification
Seed culture prepared on MRS media, was incubated for 16 h at 37 °C at 160 rpm. This was transferred to mass culture containing optimized medium and incubated for 30 h, followed by harvesting by cold centrifugation at 10,000×g for 35 min. Ammonium sulfate precipitation method was used to retrieve peptides from the supernatant of harvested broth (Bollag et al. 1996), followed by centrifugation at 10,000×g at 4 °C for 50 min. The obtained precipitate was dialyzed and solubilized against 10 mM Tris/HCl (pH 7.5). Amicon ultrafiltration technique was used after ammonium sulfate precipitation method to purify the suspended pellets in buffer and 30 and 10 kDa filter papers (Millipore Co.) were used in this process respectively for desalting procedure. Following dialysis and filtration, the precipitate of peptide extract was purified with a Sephadex G-50 column (1.5 × 65 cm) using the same buffer. The active fractions were pooled, concentrated, and further purified with a Sephadex G-25 column (1.4 × 25 cm) using the same buffer system. The fractions were monitored for antimicrobial activity against indicator bacteria Mycobacterium smegmatis ATCC 9341. The fractions positive for antimicrobial activity were pooled and stored at 4 °C. The protein content was estimated using the Bradford Method (Bradford 1976).
Protein Estimation and Molecular Weight Determination
Protein concentration was estimated by the Bradford method (Bradford 1976), using bovine serum albumin as a standard. The molecular weight determined by Tricine SDS–PAGE (Schägger 2006).
In-situ Analysis
In-situ analysis was done against an indicator organism Mycobacterium smegmatis ATCC 9341 (1.5 × 108 CFU/mL) by overlaying the processed gel from Tricine SDS–PAGE which was washed with 50 mM Tris–HCl buffer (pH 7.5) containing 2.5% Triton X-100 several times to 0.6% agar on Mueller–Hinton media and incubated at 37 °C.
Effects of Temperature and pH on Antimicrobial Activity
The thermal stability of peptide K1R was determined by exposing to 0, 10, 20, 37, 50, 60, 70, and 80 °C for 30 min and at standard autoclave condition before analysing the residual activity. Similarly, pH stability was analyzed at various pH values (4.0–13.5) using 100 mM pH buffers: citric acid/phosphate (4–5.5), tris/HCl acid (6.5–9.5), and potassium chloride/sodium hydroxide (10–13.5).
Effect of Chemicals on Antimicrobial Activity
The effect of different chemicals such as oxidizing agents (hydrogen peroxide and sodium perborate), reducing agent (β-mercaptoethanol), chelating agents (EDTA and EGTA), detergents (SDS, CHAPS, Triton X-100, Tween 20, Tween 80, and deoxycholic acid), metal ions (Ca++, Mg++, Co++, Cu++, Ni++, Zn++, Mn++, and Ba++) and solvents (acetone, chloroform, dimethyl sulfoxide, methanol, ethanol, 2-propanol, 1-butanol, toluene, diethyl ether, TCA, sodium chloride, and potassium chloride) on antimicrobial activity of AMP was observed.
Antimicrobial Activity of Peptide K1R
Minimum Inhibitory Concentration Test
Minimal Inhibitory Concentration (MIC) test was based on agar dilution method that was used to determine the lowest concentration of the assayed antimicrobial peptide which inhibits the growth of the bacteria being examined. For this test, agar solutions with specified numbers of bacterial counts (1.5 × 108 CFU/mL) were permitted promptly onto nutrient agar plates containing reference standard antibiotic and antimicrobial peptide concentrations and a plate without antibiotics as control and incubated at 37 °C (Wiegand et al. 2008). After 12 h of incubation, the presence of bacterial colonies indicated the growth of the organism and compared with control plate. Bacitracin and vancomycin were used as reference standard antibiotics.
Indicator organisms selected for the test are as follows:
-
1.
Gram negative bacteria: Escherichia coli KCTC 1923, Salmonella typhimurium KCTC 1925, Pseudomonas aeruginosa KCTC 1637, Alcaligenes faecalis ATCC 1004.
-
2.
Gram positive bacteria: Enterococcus faecalis ATCC 29,212, Bacillus subtilis ATCC 6633, Mycobacterium smegmatis ATCC 9341, VRE 4, MRSA 5 − 3, MRSA 4–5, VRSA, VRE 98, VRE89, VRE 82, Staphylococcus aureus KCTC 1928, Micrococcus luteus ATCC 9341.
Agar Disk Diffusion Assay
The antimicrobial activity of peptide produced by Bacillus sp. K1R was detected by agar disk diffusion assay (Jorgensen and Ferraro 2009) and tested against all indicators explained in minimum inhibitory concentration test. A 50 μL aliquot of K1R was applied on disks (8 mm) on the agar plates previously inoculated with individual indicator strain.
Amino Acid Sequence Analysis
The amino acid sequence of AMP from Bacillus sp. K1R was determined by Edman degradation using a Procise® Model 491 HT Protein Sequencer (Applied Biosystems, USA). The molecular weight of the peptide was also verified using the same sequence (Gasteiger et al. 2005).
Anti-oxidant Activity of Peptide K1R
2,2-diphenyl-1-picrylhydrazyl (DPPH) method was employed to evaluate the anti-oxidant activity using the process described by (Zhang et al. 2006) with some modification. DPPH is a stable free radical containing an odd electron in its structure and usually utilized for the detection of radical scavenging activity in chemical analysis. Hundred microliters of various concentrations of peptide solutions was added to 100 µL of DPPH (0.3 mM) in methanol solution. The mixture was kept in dark at room temperature for 30 min and the absorbance at 517 nm measured on spectrophotometer. Methanol was used as the blank whereas distilled water was used as the negative control. Ascorbic acid was used as the positive control.
DPPH radical scavenging ability was calculated by the following equation:
where, Abssample and Absblank are the absorbance of sample and blank respectively.
Results
Identification of Bacillus Strain
For molecular phylogeny, the 16S rRNA sequence of the local isolate was compared to the sequence of 10 Bacillus species. To determine the relation of the local isolate with these Bacillus strains, multiple sequence alignment was performed between the 16S rRNA gene sequences of various Bacillus species and local isolate. Similarly, computer assisted RNA searches against the bacterial database similarly revealed that the 16S rRNA sequence was over 99% identical with the 10 Bacillus strains. The 16S rRNA sequence of Bacillus sp. K1R was deposited in GenBank under the Accession No. AYTO01000043. The phylogenic tree is shown in Fig. 1.
Phylogenetic tree based on 16S rRNA gene sequences, showing the phylogenetic position of strain CSB40 to some other related members of the genus Bacillus. Reference sequences were retrieved from GenBank under the AccessionNumbers indicated in parentheses after the strain designation. Numbers at nodes are percentage bootstrap values based on 1000 replications; only values greater than 77% are shown. Bar 0.005 substitutions per nucleotide position
Optimization of the Culture Media
Many bacteria can be grown in the laboratory in culture media that are designed to provide all the essential nutrients in solution for bacterial growth: however for the maximum production of antimicrobial compounds, nutrients and growth factors should be optimized (Jack et al. 1995).
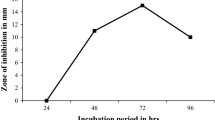
In case of carbon source, Bacillus strain K1R was inoculated in culture media containing different carbon sources and incubated up to 108 h. A disk diffusion assay was performed to evaluate the activity of AMP produced by Bacillus strain K1R against the indicator organism. Incubation time versus zone of inhibition graph was plotted and it was observed that maltose was the best carbon source for this strain. Similarly, this process was repeated to choose the best nitrogen source and metal ion source. In case of nitrogen source, beef extract turned out to be the best but in case of metal ions, none of them had a favourable effect on antimicrobial activity. Therefore, maltose and beef extract were chosen as the best carbon and nitrogen source. The final optimization was done by varying the amount of beef extract and maltose, and it was clear that peptide K1R shows its maximum activity at 30 h of inoculation, when 2% maltose (MAL) and 2% beef extract (BE) are present in media. In this research, Mycobacterium smegmatis ATCC 9341 was chosen as the indicator organism because it is very easy to grow, non-pathogenic in nature, and simple model to work. The final optimization of carbon, nitrogen, and metal ion source is quantitatively illustrated in Fig. 2. (In figure MH: Mueller Hinton).
Quantitative representation of optimization of culture media for Bacillus species K1R. a Effect of various carbon sources on the activity of antimicrobial peptide produced by Bacillus strain K1R. b Effect of various nitrogen sources on the activity of antimicrobial peptide produced by Bacillus strain K1R. c Effect of various metal ions on the activity of antimicrobial peptide produced by Bacillus strain K1R. d Effect of different amount of beef extract and maltose on the activity of antimicrobial peptide produced by Bacillus strain K1R
Production of Antimicrobial Peptide
The seeding of the new culture was carried out in MRS media after 16 h of growth, it was transferred to the main culture containing optimized media and conditions and allowed to grow for 30 h. The AMP secreted by the strain K1R displayed antimicrobial activity against Mycobacterium smegmatis ATCC 9341, Enterococcus faecalis ATCC 29,212 and Micrococcus luteus ATCC 9341.
Purification of Antimicrobial Peptide
Ammonium Sulfate Precipitation Method
After 30 h of growth, the broth of strain K1R was harvested and tested for antimicrobial activity against indicator organism (data not shown). Cell-free culture supernatant was precipitated using ammonium sulfate (80% w/v) and stored at 4 °C overnight with continuous stirring. The pellet was collected, suspended and dialyzed in Tris–HCl buffer (pH 7.5).
Column Chromatography
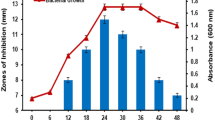
The crude extracts were applied to Sephadex G-50 column and eluted with 10 mM Tris–HCl buffer (pH 7.5) as a mobile phase followed by Sephadex-G25 column and 2 mL fraction were collected. Figure 3 shows the comprehensible concept of chromatographic steps involved during purification, and the selection of the active fractions by observing the antimicrobial activity vs. protein content plot i.e., zone of inhibition versus absorbance at 595 nm.
Determination of Molecular Weight
Active fractions (9–14) obtained from Sephadex G-25 were pooled and concentrated and were separated by Tricine SDS–PAGE. Ultra-low molecular weight marker proteins (1.7–42 kDa) were used to determine the approximate molecular size of the active protein band. After electrophoresis, one gel was stained with Coomassie brilliant blue and another was kept for the identification of AMPs band on an unstained gel for in-situ examination. The antimicrobial compound was confirmed by the presence of single band in first gel having molecular weight around 4.6 kDa and in situ examination result showed a distinct zone of inhibition spot against the indicator organism (Figs. 4, 5).
Effects of Temperature and pH on Antimicrobial Activity
The influence of temperature and pH on the antimicrobial activity of peptide is represented in Figs. 6 and 7. Peptide K1R was found to be stable around 30–60 °C whereas it lost about 25% of its activity at 70 °C. The autoclaved sample lost its activity completely. On analysing the residual activity, it was found to be highly stable around pH 6.5–9. It lost 50% of its activity when the pH was increased up to 12. Finally, it showed 35% of residual activity at pH 13.5.
Effect of Chemicals on Antimicrobial Activity
The effect of various chemicals in terms of residual activity is represented in Table 1. From the table, it can be seen that the intact activity of peptide K1R remained stable in the presence of oxidizing agents such as hydrogen peroxide and sodium perborate and detergents such as deoxycholic acid and SDS. Triton X-100 and toluene had similar effects. Metal ions such as calcium and magnesium inhibited activity.
Antimicrobial Activity of Peptide K1R
Minimum Inhibitory Concentration Test
The antimicrobial activity of peptide K1R in terms of minimum inhibitory concentration (MIC) is shown in Table 2. The emergence of MDR has created a challenge to pharmaceutical researches. Here, we carried out antimicrobial activity study to understand the efficacy of our newly extracted AMP in comparison to that of already known drugs such as vancomycin and bacitracin. Peptide K1R displayed antagonistic effect against different MDR pathogens. It showed a strong effect on pathogens such as vancomycin resistant Enterococci 4, vancomycin-resistant Enterococci 82, vancomycin-resistant Enterococci 89, and vancomycin-resistant Enterococci 98 compared with vancomycin and bacitracin. The effect of peptide K1R was similar to the effect of vancomycin against Enterococcus faecalis ATCC 29,212 and better than that of bacitracin against the same pathogen. The MIC value of peptide K1R against Micrococcus luteus ATCC 9341 was found to be effective than that of bacitracin. The resultant effect of peptide K1R vancomycin-resistant gram-positive organisms was better than others. Strains of staphylococcus resistant to vancomycin was not susceptible to it and showed no promising results. K1R was found to be more effective than other strains in our previous study (Choi et al. 2012).
Agar Disk Diffusion Assay
The inhibitory spectrum of peptide K1R based on agar disk diffusion assay is shown in Table 3. The pattern of antimicrobial activity determined via agar disk diffusion assay supports the results of MIC test however MIC >128 µg/mL corresponded to the 14 mm zone of inhibition in agar diffusion assay.
Amino Acid Sequence Analysis
The amino acid sequence of the AMP produced by peptide K1R was found to be AVQGTLEDALNLSKGALNQVQK AIQNGDXLTVXGXLGTIXLAVSX. This sequence did not show significant homology with the reported peptides of similar type. However, it showed similarity with the hypothetical proteins sequences (Table 4) from Bacillus sp. in the National Centre for Biotechnology Information (NCBI) protein database using BLAST (basic local alignment search tool; http://blast.ncbi.nlm.nih.gov/Blast.cgi). Bold amino acid sequences of hypothetical proteins in Table 4 show the similarity with AMP produced from organism K1R. Based on the amino acid sequence and by the use of protein identification and analysis tools on the ExPASy Server, the molecular weight of the peptide was found to be ~4578.23 dalton which verifies the result from Tricine SDS–PAGE and in-situ analysis.
Anti-oxidant Activity of Peptide K1R
To obtain information about the mechanisms underlaying the antioxidative effects of AMP, the radical scavenging effect was examined by measuring changes in the absorbance of DPPH radical at 517 nm. Ascorbic acid and peptide K1R both showed concentration-dependent scavenging of DPPH radicals. The effect of peptide K1R was similar and comparable to that of ascorbic acid at all concentrations tested (1–1000 µg/mL). The results of free radical scavenging activity as % inhibition are shown in Fig. 8. Peptide K1R showed a percentage inhibition of 70.97 at 1000 µg/mL whereas standard ascorbic acid exhibited 75.36% inhibition at the same concentration. In both cases, graded increase in percentage of inhibition was observed for the increase in the concentration of ascorbic acid. The antioxidant activity of peptide K1R was found to be comparable to that of other peptides (Borquaye et al. 2015).
Discussion
The morphology of colonies, biochemical characteristics and the utilization of nutrients suggested the selected strain to be a Bacillus species. Purified products from fermented food alter the pre-consumption of dietary items which in turn, alter the ways by which fermentation-enriched chemicals act on our own intestinal microbiota profile (Selhub et al. 2014). The functional activities of the purified products include antimutagenic/anticancer (Kong et al. 2010), anti-obesity (Kim et al. 2011), antiatherosclerotic, and immunomodulatory effects (Noh et al. 2012).
In this study, a potent AMP was derived from microbial strain K1R, identified as Bacillus, from 16S rRNA gene sequences analysis. This study was designed to characterize and determine the antimicrobial activity of K1R. Beef extract and maltose were found to be the suitable nutrient sources for maximum peptide production. Media optimization was based upon our previous research on antimicrobial peptides (Choi et al. 2012; Regmi et al. 2016). Bacteriocins are usually produced in complex media (Lejeune et al. 1998; Parente and Ricciardi 1994). Physicochemical factors have a dramatic effect on the production of bacteriocins (Biswas et al. 1991). Standard culture media as well as enriched media are not always good producer of peptides (Jack et al. 1995; Tomás et al. 2002). AMP was purified using Sephadex G-50 and Sephadex G-25 gel filtration column chromatography. It was found to be stable over wide range of temperature (30–60 °C) and pH (6.5–9) conditions which was comparable and even wider range than other similar studies (Alkotaini et al. 2013; Bizani and Brandelli 2002; Cheigh et al. 2002; Kamoun et al. 2005; Shin et al. 2008). The molecular weight of the AMP K1R was found to be ~4600 Da which was also verified by the result from the amino acid sequence using ExPASy server. N-terminal amino acid sequence analyses of peptide K1R were performed by automated Edman degradation and five of them were unknown residues that could not be identified are shown as X. The reaction was not blocked in the sequencing cycles of the analysis. The presence of unknown residues (X) also may be due to uncleaved amino acids like proline during the procedure. Peptide K1R remained stable in the presence of oxidizing agents such as hydrogen peroxide and sodium perborate and detergents such as deoxycholic acid and SDS. Antimicrobial effects, in terms of MIC values of the purified peptide were obtained, and two well-known reference antibiotics, bacitracin and vancomycin were used. It is remarkable that peptide K1R displayed antagonistic effect against different MDR pathogens such as VRE 4, VRE 82, VRE 89, VRE 98 including Enterococcus faecalis and Salmonella typhimurium where MIC was in the range of 4–32 µg/mL. Disk diffusion assay also verified the MIC result. The antioxidant properties of peptide K1R were tested using free radical DPPH method and it was comparable to the standard ascorbic acid, which signifies its activity in terms of hydrogen atom donating capacity/electron transfer capability.
References
Alkotaini B, Anuar N, Kadhum AAH (2013) The effects of temperature, pH and carbon sources on antimicrobial peptide AN5-1 production using Paenibacillus alvei AN 5. Asian J Microbiol Biotechnol Environ Sci 15(1):195–201
Andreu D, Rivas L (1998) Animal antimicrobial peptides: an overview. Pept Sci 47(6):415–433
Biswas SR, Ray P, Johnson MC, Ray B (1991) Influence of growth conditions on the production of a bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol 57(4):1265–1267
Bizani D, Brandelli A (2002) Characterization of a bacteriocin produced by a newly isolated Bacillus sp. Strain 8 A. J Appl Microbiol 93(3):512–519
Bollag Daniel M, Rozycki Michael D, E. SJ (1996). Protein Methods. The Quarterly Review of Biology 72, no.2.
Borquaye LS, Darko G, Ocansey E, Ankomah E (2015) Antimicrobial and antioxidant properties of the crude peptide extracts of Galatea paradoxa and Patella rustica. SpringerPlus 4(1):1
Bowdish, DME, Davidson DJ, Lau YE, Lee K, Scott MG, Hancock, R. E. W. (2005) Impact of LL-37 on anti-infective immunity. J Leukocyte Biol 77(4):451–459
Bradford, MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72, 248–254
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3(3):238–250
Brown KL, Hancock, REW (2006) Cationic host defense (antimicrobial) peptides. Curr Opin Immunol 18(1):24–30
Cheigh CI, Choi HJ, Park H, Kim SB, Kook MC, Kim TS, Pyun YR (2002) Influence of growth conditions on the production of a nisin-like bacteriocin by Lactococcus lactis subsp. lactis A164 isolated from kimchi. J Biotechnol 95(3):225–235
Choi YH, Cho SS, Simkhada JR, Yoo JC (2012) A novel thermotolerant and acidotolerant peptide produced by a Bacillus strain newly isolated from a fermented food (kimchi) shows activity against multidrug-resistant bacteria. Int J Antimicrob Agents 40(1):80–83
Elsbach P (2003) What is the real role of antimicrobial polypeptides that can mediate several other inflammatory responses? J Clin Investig 111(11):1643–1645
Falagas ME, Fragoulis KN, Karydis I (2006) A comparative study on the cost of new antibiotics and drugs of other therapeutic categories. PLoS ONE 1(1):e11
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, A. B. (2005). Protein identification and analysis tools on the ExPASy server. In: John M. Walker (ed), The Proteomics Protocols Handbook, Humana Press Inc., Newyork pp. 571-607
Jack RW, Tagg JR, Ray B (1995) Bacteriocins of gram-positive bacteria. Microbiol Rev 59(2):171–200
Jenssen H, Hamill P, Hancock, R. E. W. (2006). Peptide antimicrobial agents. Clin Microbiol Rev 19 (3):491-511
Joerger RD (2003) Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poult Sci 82(4):640–647
Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49(11):1749–1755
Kamoun F, Mejdoub H, Aouissaoui H, Reinbolt J, Hammami A, Jaoua S (2005) Purification, amino acid sequence and characterization of Bacthuricin F4, a new bacteriocin produced by Bacillus thuringiensis. J Appl Microbiol 98(4):881–888
Kim EK, An SY, Lee MS, Kim TH, Lee HK, Hwang WS, Lee KW (2011) Fermented kimchi reduces body weight and improves metabolic parameters in overweight and obese patients. Nutr Res 31(6):436–443
Klaenhammer TR (1993) Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev 12(1–3):39–85
Kong C-S, Bahn Y-E, Kim B-K, Lee K-Y, Park K-Y (2010) Antiproliferative effect of chitosan-added kimchi in HT-29 human colon carcinoma cells. J Med Food 13(1):6–12
Lee HJ, Joo YJ, Park CS, Kim SH, Hwang IK, Ahn JS, Mheen TI (1999) Purification and characterization of a bacteriocin produced by Lactococcus lactis subsp, lactis H-559 isolated from kimchi. J Biosci Bioeng 88(2):153–159
Lee H, Yoon H, Ji Y, Kim H, Park H, Lee J, Holzapfel W (2011) Functional properties of Lactobacillus strains isolated from kimchi. Int J Food Microbiol 145(1):155–161
Lejeune R, Callewaert R, Crabbé K, De Vuyst L (1998) Modelling the growth and bacteriocin production by Lactobacillus amylovorus DCE 471 in batch cultivation. J Appl Microbiol 84(2):159–168
Mukherjee S, Das P, Sen R (2006) Towards commercial production of microbial surfactants. TRENDS in Biotechnol 24(11):509–515
Noh JS, Choi YH, Song YO (2012). Beneficial effects of the active principle component of Korean cabbage kimchi via increasing nitric oxide production and suppressing inflammation in the aorta of apoE knockout mice. Br J Nutr 109 (01) 17–24
Parente E, Ricciardi A (1994) Influence of pH on the production of enterocin 1146 during batch fermentation. Lett Appl Microbiol 19(1):12–15
Regmi S, Choi YH, Choi YS, Kim MR, Yoo JC (2016). Antimicrobial peptide isolated from Bacillus amyloliquefaciens K14 revitalizes its use in combinatorial drug therapy. Folia Microbiol 1–12
Rodrigues L, Banat IM, Teixeira J, Oliveira R (2006) Biosurfactants: potential applications in medicine. J Antimicrob Chemother 57(4):609–618
Rosenfeld Y, Sahl H-G, Shai Y (2008) parameters involved in antimicrobial and endotoxin detoxification activities of antimicrobial peptides†. BioChemistry 47(24):6468–6478
Sang Y, Blecha F (2008) Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim Health Res Rev 9(2):227–235
Schägger H (2006) Tricine-SDS–PAGE. Nat Protoc 1(1):16–22
Selhub EM, Logan AC, Bested AC (2014) Fermented foods, microbiota, and mental health: ancient practice meets nutritional psychiatry. J Physiol Anthropol 33:2
Shin MS, Han SK, Ryu JS, Kim KS, Lee WK (2008) Isolation and partial characterization of a bacteriocin produced by Pediococcus pentosaceus K23-2 isolated from Kimchi. J Appl Microbiol 105(2):331–339
Tomás J, Bru E, Wiese B, de Ruiz Holgado, A. A. P., Nader-Macías ME (2002) Influence of pH, temperature and culture media on the growth and bacteriocin production by vaginal Lactobacillus salivarius CRL 1328. J Appl Microbiol 93(4):714–724
Wiegand I, Hilpert K, Hancock, R. E. W. (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3(2):163–175
Yamanaka H, Moriyoshi K, Ohmoto T, Ohe T, Sakai K (2007) Degradation of bisphenol a by Bacillus pumilus isolated from kimchi, a traditionally fermented food. Appl Biochem Biotechnol 136(1):39–51
Yeaman MR, Yount NY (2003) Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55(1):27–55
Zhang X, Koo J, Eun J (2006) Antioxidant activities of methanol extracts and phenolic compounds in Asian pear at different stages of maturity. Food Sci Biotechnol 15(1):44
Acknowledgements
This work was supported by the National Research Foundationof Korea (NRF) grant funded by the Korean government (MEST) (NRF-2015R1A2A1A15056120, NRF-2015R1D1A1A 010 59 483) and “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01128901)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Sandesh Panthi and Yun Hee Choi have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Panthi, S., Choi, Y.H., Jee, JP. et al. Antimicrobial Peptide from Bacillus Strain K1R Exhibits Ameliorative Potential Against Vancomycin-Resistant Enterococcus Group of Organisms. Int J Pept Res Ther 23, 419–430 (2017). https://doi.org/10.1007/s10989-016-9572-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-016-9572-2