Abstract
Familial cardiomyopathy is an inherited disease that affects the structure and function of heart muscle and has an extreme range of phenotypes. Among the millions of affected individuals, patients with hypertrophic (HCM), dilated (DCM), or left ventricular non-compaction (LVNC) cardiomyopathy can experience morphologic changes of the heart which lead to sudden death in the most detrimental cases. TNNC1, the gene that codes for cardiac troponin C (cTnC), is a sarcomere gene associated with cardiomyopathies in which probands exhibit young age of presentation and high death, transplant or ventricular fibrillation events relative to TNNT2 and TNNI3 probands. Using GnomAD, ClinVar, UniProt and PhosphoSitePlus databases and published literature, an extensive list to date of identified genetic variants in TNNC1 and post-translational modifications (PTMs) in cTnC was compiled. Additionally, a recent cryo–EM structure of the cardiac thin filament regulatory unit was used to localize each functionally studied amino acid variant and each PTM (acetylation, glycation, s-nitrosylation, phosphorylation) in the structure of cTnC. TNNC1 has a large number of variants (> 100) relative to other genes of the same transcript size. Surprisingly, the mapped variant amino acids and PTMs are distributed throughout the cTnC structure. While many cardiomyopathy-associated variants are localized in α-helical regions of cTnC, this was not statistically significant χ2 (p = 0.72). Exploring the variants in TNNC1 and PTMs of cTnC in the contexts of cardiomyopathy association, physiological modulation and potential non-canonical roles provides insights into the normal function of cTnC along with the many facets of TNNC1 as a cardiomyopathic gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cardiomyopathy is a group of acquired or hereditary diseases characterized by structural and functional abnormalities of heart muscle. These anomalies can be associated with a large number of variants located in sarcomeric protein-encoding genes (Garfinkel et al. 2018; Landstrom et al. 2008; Tadros et al. 2020; van der Velden and Stienen 2019; Yotti et al. 2019). Through genetic testing, the presence of these variants can be discovered in patients who have a suspected inherited cardiovascular disease and/or patients who have family members presenting with a pathogenic variant (Musunuru et al. 2020). Severity of clinical outcomes varies widely, and common effects of these cardiomyopathies include heart failure and/or fatal arrhythmia (Semsarian et al. 2015). Generally, incidence of cardiomyopathy in the general population has been estimated to be at least 1:500 (Maron and Maron 2013) and possibly as high as 1:200 (Semsarian et al. 2015), and the percentage of cardiomyopathy increased by almost 27% during 10 years from 2005 to 2015 (Vos et al. 2016).
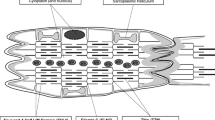
There are three major types of cardiomyopathy: dilated cardiomyopathy (DCM), hypertrophic cardiomyopathy (HCM), and restrictive cardiomyopathy (RCM). Characterized by dilated left ventricle and depressed contractility, with normal or decreased ventricular wall thickness (Fig. 1), DCM is considered the most common type, with an estimated prevalence of 1 per 2500 up to 1 per 250–400 individuals, but the vast majority is ischemic and not genetic (Hershberger et al. 2013; Jefferies and Towbin 2010; Towbin et al. 2006; Weintraub et al. 2017). Although DCM-acquired causes include infectious, environmental, and systematic factors, 25–35% of DCM cases are due to familial genetic variants (Luk et al. 2009; Maron et al. 2006). The seriousness of manifestations may vary from being asymptomatic to having acute heart failure (Choudhry et al. 2019). Ventricular dilation elevates end-diastolic pressure, which in turn contributes to progressive ventricular dilation, responsible for systolic dysfunction (Reichart et al. 2019). Severity of structural and functional abnormalities exhibited along with DCM progression further results in heart failure and, subsequently, may eventually necessitate heart transplant (Hershberger et al. 2013; Kirk et al. 2009; Weintraub et al. 2017).
Coronal section diagrams of the human heart in different pathological conditions. Hearts with: normal morphology (left); DCM (middle) with dilated ventricles; and HCM (right) with hypertrophic myocardium, interventricular septal thickening, and decreased left ventricular volume. Images generated with BioRender.com
HCM is the most common inherited heart disease (Maron et al. 2012), of which the estimated prevalence may be as high as 1:200 (Semsarian et al. 2015). According to both European Society of Cardiology (ESC) and American Heart Association (AHA) guidelines, HCM is typically characterized by changes in the shape of the heart such as abnormally increased wall thickness (Fig. 1) (Elliott et al. 2014; Gersh et al. 2011a, 2011b), which causes decrease of ventricular volume as well as end-diastolic pressure, yet systolic function is normal or even increased. Similar to DCM, individuals presenting with HCM can be asymptomatic (Brieler et al. 2017). It is reported as the most common cause of sudden death in the young, including athletes (Maron et al. 2006). During the past decades, multiple variants of genes expressing sarcomere proteins were detected, proving that HCM is a genetically heterogeneous disease (Wolf 2019). Thus, family history is always important for the diagnosis of HCM.
In contrast to DCM and HCM, RCM is much more rare and accounts for 5% of all cardiomyopathy cases (Brown et al. 2020). RCM can be characterized by increased myocardial stiffness which causes reduced ventricular filling and leads to diastolic dysfunction (Muchtar et al. 2017). Patients with RCM may exhibit clinical manifestations ranging from mild to severe, where the patient may need an implantable cardioverter defibrillator and ultimately a heart transplant due to eminent risk of sudden death (Webber et al. 2012; Wilkinson et al. 2010; Wittekind et al. 2019). The causes of RCM can be inherited or acquired, but most cases are acquired (Muchtar et al. 2017). Most RCM cases are due to infiltration or storage of abnormal substances in myocardium or fibrotic injury, and three leading causes include amyloidosis, cardiac sarcoidosis, and hemochromatosis (Costabel et al. 2017).
In addition to the three major types of cardiomyopathy described above, there are some uncommon types of cardiomyopathy. One example is left ventricular non-compaction cardiomyopathy (LVNC), which is recognized as a primary cardiomyopathy (Maron et al. 2006). It is characterized by left ventricular trabeculation, deep intertrabecular recesses and non-compacted myocardium (Jenni et al. 2007). LVNC results from failure of compaction of loose myocardial meshwork during fetal development (Samsa et al. 2013). Singh and Patel (2020) estimated the prevalence of LVNC to be between 0.05% and 0.24%, with 22% to 38% of patients showing biventricular involvement. Patients with LVNC can present with a variety of manifestations including arrhythmias, thromboembolism and even heart failure (Ikeda et al. 2015).
TNNC1 is among the genes associated with sarcomeric cardiomyopathy (Marques and de Oliveira 2016; Tadros et al. 2020; Willott et al. 2010; Yotti et al. 2019). TNNC1 variants are found to be enriched in DCM (Mazzarotto et al. 2020) and also have association with HCM (Ingles et al. 2019; Pua et al. 2020). According to a comprehensive statistical analysis by Tadros et al. (2020), TNNC1-positive cardiomyopathy patients have a relatively severe prognosis and early onset in comparison with the other two subunits of Tn. However, as a rare-variants associated gene, TNNC1 has limited diagnostic value, so patients do not have access to such genomic diagnosis. Additionally, some patients with genetic variants are now more likely to pass those variants on to the next generation because of elongated lifespan, since mortality has been reduced due to improvements in disease management (Richards and Garg 2010). Thus, TNNC1 variants may not be as rare as initially believed. Because of these advancements and multi-generational access, it is suggested that genetic testing be done on the family member with the highest severity of phenotype in order to confidently identify pathogenic variant(s) (Musunuru et al. 2020). Studying rare variants in TNNC1 helps to understand the underlying molecular/cellular mechanisms, and ultimately should pave the way for developing novel therapies to target the contractile apparatus, which may provide opportunities to broadly treat cardiac dysfunction irrespective of etiology. Companies such as Myokardia and Cytokinetics are developing treatments that target the sarcomere, implicating such therapies as feasible. Tadros et al. (2020) analyzed variants of the three troponin subunits to identify disease-associated hotspots. Their analysis showed that TNNC1-positive probands had younger ages of diagnosis and poorer clinical outcomes compared to genes for other cardiac troponin subunits, but no portion of the cardiac troponin C (cTnC) sequence reached statistical significance to be identified as a distinct hotspot.
Several advances have occurred since the study period included in the analysis by Tadros et al. (2020). First, initial structures of the vertebrate cardiac thin filament with and without Ca2+ have been determined by cryo–EM (Oda et al. 2020; Yamada et al. 2020). Second, additional pathogenic variants in TNNC1 have been identified and characterized (e.g., Johnston et al. 2019; Landim-Vieira et al. 2020a). And third, additional variants in TNNC1 have appeared in genomic databases. The purpose of this review is to summarize variants in TNNC1 including those that were identified after the cutoff date for the dataset examined by Tadros et al. (2020), and to suggest structural regions of cTnC that should be considered for further analysis. Using the Yamada et al. (2020) cryo–EM structure (Fig. 2), we sought to compile and visualize amino acids affected by TNNC1 variants, and also cTnC residues that can be altered by post-translational modifications in the context of the thin filament, and discuss the underlying molecular and cellular mechanisms.
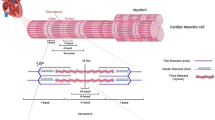
Representation of a regulatory unit from the cardiac thin filament in the Ca2+-free state, based on cryo–EM structure PDB 6KN7 (Yamada et al. 2020). The troponin complex (spacefill) is composed of three subunits: troponin C (cTnC, red), troponin I (cTnI, blue) and troponin T (cTnT, yellow). Tropomyosin and actin are exhibited as ribbons (gray). The top of the structure is oriented toward the pointed (minus) end, and the bottom is oriented toward the barbed (plus) end and Z-disk. Troponin C (red) is enlarged on the right. (Color figure online)
Structure and function of cTnC
Troponin C (TnC) is the expression product of either of two genes in the human genome, TNNC1 and TNNC2, which are located on chromosomes 3 and 20, respectively, and that code for cTnC and fast skeletal troponin C (sTnC), respectively. TNNC1 consists of six exons and five introns, and encodes TnC that is expressed not only in cardiomyocytes (cTnC—the primary focus of this review) but also in slow skeletal muscle (Katrukha 2013). cTnC is comprised of 161 amino acids (Roher et al. 1986) that fold into a dumbbell-shape with two globular domains. The two domains of TnC are connected by a central segment (D–E linker) (Sia et al. 1997; Slupsky and Sykes 1995). In crystal structures of the troponin core domain, cTnC displays a flexible, intrinsically disordered D–E linker, while sTnC presents a rigid, well-ordered α-helical linker (Takeda et al. 2003; Vinogradova et al. 2005). The two globular domains of TnC are referred to by their NH2- and COOH-terminal locations in the primary sequence. Beyond location, the two domains are distinguished by their structural and functional roles in the thin filament. cTnC exhibits predominantly α-helical content. In addition to the first α-helix (N–helix), TnC has two α-helix-containing EF-hand divalent cation-binding sites in each of the two globular domains, with different affinities and selectivity for Ca2+ versus Mg2+.
Vertebrate striated muscle contraction is triggered by Ca2+ binding to the NH2-terminal domain of TnC (Gordon et al. 2000). Both NH2-terminal domain EF-hands (sites I and II) in sTnC bind Ca2+ during a cytoplasmic Ca2+-transient to activate contraction. In contrast, site I of cTnC is inactive as a result of evolutionary selection. Therefore, cTnC has only site II for Ca2+ regulation of contraction. In both cTnC and sTnC, Ca2+ binding to the regulatory NH2-terminal domain leads to conformational changes—initially in TnC, followed by conformational changes within troponin, and then the thin filament—that are ultimately responsible for removal of inhibition of actomyosin interactions (Oda et al. 2020; Yamada et al. 2020). Specifically, as cytosolic Ca2+ levels increase, Ca2+ binding to sites I and II of the NH2-terminal domain of sTnC reveals a hydrophobic pocket in the NH2-terminal domain (Slupsky and Sykes 1995) while in contrast, Ca2+ binding to site II of cTnC’s NH2-terminal domain primes the NH2-terminal domain for opening, but without fully exposing the corresponding hydrophobic pocket (Sia et al. 1997). In both sTnC and cTnC, the α-helical switch peptide of TnI’s COOH-terminal domain ultimately binds the hydrophobic pocket of TnC’s NH2-terminal domain in the presence of Ca2+. Consequently, the C-terminal mobile domain of TnI is pulled away from actin, allowing tropomyosin to shift across the actin surface (Yamada et al. 2020), exposing myosin-binding sites on actin subunits and enabling the formation of cross-bridges when myosin motor domains bind.
TnC sites III and IV are located in the COOH-terminal domain and are generally thought not to participate directly in activation of contraction. Compared with NH2-terminal site II in cTnC and sites I and II in sTnC, sites III and IV in both cTnC and sTnC bind Ca2+ with higher affinity but less selectivity because Mg2+ can also bind under physiological conditions. Divalent cation binding at sites III and IV stabilizes binding of an α-helical segment within TnI’s NH2-terminal portion at TnC’s COOH-terminal domain; in other words, in both cardiac and skeletal troponin, TnI and TnC bind each other in an antiparallel, or head-to-tail fashion (Takeda et al. 2003; Vinogradova et al. 2005). In the relaxed state (e.g., low Ca2+ levels during cardiac diastole), sites III and IV are thought to be occupied primarily by Mg2+ (Potter and Gergely 1975) although it is not certain what fraction of these COOH-terminal sites might be occupied by Ca2+, especially in cTnC when heart rate changes (Badr et al. 2016; Fuchs and Grabarek 2011).
As described above, cTnC plays a critical role in the conformation changes that occur on the thin filament in order to regulate the transition between on and off states of the regulatory units and, consequently, cardiac muscle contraction. A very useful model of Ca2+-regulation of striated muscle contraction is the three-state model proposed by McKillop and Geeves (1993). According to this model, the thin filament is not simply switched on and off. Instead, there is a dynamic equilibrium of three states: blocked (B), closed (C), and open (M) states. During diastole, the cardiac thin filament is thought to be in mainly in the blocked state. Upon Ca2+ binding to site II, a series of thin filament conformational changes result in the formation of a weak contact between thin and thick filament (closed state). This interaction moves tropomyosin towards the groove of the F-actin and fully allows myosin to strongly interact with actin and generate force (open state). Once cytosolic Ca2+ levels decrease as Ca2+ is taken up by the sarcoplasmic reticulum, a new series of conformational changes happen at the thin filament returning it back to a closed and blocked state. While the McKillop and Geeves (1993) model was formulated from biochemical data, its significance was enhanced by being generally consistent with structural data (Pirani et al. 2005; Poole et al. 2006; Risi et al. 2017). Variants in cTnC can alter the dynamics of troponin subunit interactions. For example, cTnC-G159D has been reported to disrupt cTnI-cTnC interaction, preventing a normal shift in the open/closed cTnC conformation and resulting in blunted phopho-cTnI desensitization (Biesiadecki et al. 2007).
Since the recognition of TNNC1 as a cardiomyopathy-associated gene, numerous functional studies have been published exhibiting consistent patterns of Ca2+ sensitization and desensitization for HCM and DCM, respectively (summarized in Tadros et al. 2020). However, the mechanism(s) by which the variants throughout cTnC can disturb Ca2+ binding to site II is unknown. That is because there are disparate cases, where some variants located at or near site II could directly affect Ca2+ binding (e.g., Miszalski-Jamka et al. 2017), while other variants located in the COOH-terminal domain, i.e., distant from site II, can also disturb Ca2+ binding to the NH2-terminal domain allosterically (e.g., Landim-Vieira et al. 2020a; Miszalski-Jamka et al. 2017; Pinto et al. 2009, 2011; Ploski et al. 2016). Such altered NH2-domain response observed in the presence of D145E has been shown to prevent Ca2+ binding to site IV and to increase Ca2+ binding to site II allosterically (Swindle and Tikunova 2010). Interestingly, even though D141A has also been reported to abolish Ca2+ binding to site IV, no significant effect on myofilament Ca2+ sensitivity was observed (Negele et al. 1992).
Influencing myofilament Ca2+ sensitivity is not only a consequence of amino acid variants. TnC-targeting small molecules which can bind to either the NH2- or COOH-terminal domains have been reported to disturb Ca2+ binding to site II. In these cases, Ca2+ sensitization can be achieved by direct binding of the molecule to the NH2-terminal domain or by COOH-terminal binding which can cause an allosteric NH2-terminal response. As an example of a Ca2+ sensitizer molecule, bepridil has been reported to act similarly to variants located in the NH2-terminal domain of cTnC. This NH2-terminal domain-binding molecule (Wang et al. 2002) was shown to influence myofilament Ca2+ sensitivity in addition to affecting both the number and kinetics of acto-myosin cross-bridges (Gonzalez-Martinez et al. 2018). On the other hand, a Ca2+ sensitizer drug MCI-154 that binds to the COOH-terminal domain of cTnC elicits an allosteric NH2-domain response by increasing Ca2+ binding to site II (Li et al. 2018). The COOH-terminal domain of cTnC has been also described as a target for desensitizing molecules. For example, (-)-epigallocatechin‐3‐gallate (EGCg) has been reported to bind to the COOH-terminal domain as a Ca2+ desensitizer with positive results on reversing diastolic dysfunction detected in mouse models of cardiomyopathy (Friedrich et al. 2016; Warren et al. 2015).
Genetic variation in human TNNC1
Cardiomyopathy associated variants in Ca2+-regulatory proteins of the cardiac thin filament—including all three subunits of cardiac troponin—have been identified but are rare in comparison with the number of variants in thick filament proteins (Tardiff 2011; Willott et al. 2010; Yotti et al. 2019). This relative rarity of pathogenic variants in cardiac troponin is generally thought to be related to the high degree of evolutionary conservation of sequences, particularly for cTnC, and may derive from the severe outcomes at an early age (i.e., before reproduction) resulting in genetic variants not being passed on to new generations.
Sites that produce genetic variations in human TNNC1 can be easily mapped using a cryo–EM structure of the cardiac thin filament (PDB 6KN7; Yamada et al. 2020) in which troponin is based on an X-ray crystal structure PDB 4Y99 (Takeda et al. 2003). The functionally identified variants here are A8V, L29Q, A31S, C84Y, Q122AfsX30, E134D, N144D, D145E (HCM/RCM); I4M, Y5H, G34S, Q50R, E59D, D75Y, E94V, M103I, D132N, I144D, D145E, I48V, G159D (DCM); and D62N, M81I, E94A, R102C (LVNC) (Table 1, Figs. 3, 4 and 5). Regardless of the type of cardiomyopathy, there is no distinct pattern as to where these variants are located in cTnC considering that the variants identified are found in either the NH2-terminal or COOH-terminal. Beyond these variants, there are many others which have not been characterized as having cardiomyopathy associations (Table 1). With this being said, there is a distinct possibility of false negatives in Table 1 with regard to disease casualty, as the majority of variants have not been experimentally studied in detail. Using the American College of Medical Genetics and Genomics (ACMG) criteria, these variants are able to be categorized as pathogenic or putative (Richards et al. 2015). The pathogenic variants included in this analysis are A8V, A31S, A31T, M47I, P54H, C84Y, D88N, L97Q, D149G, G159D (Table 1). Variants that are pathogenic but have no clinically classified cardiomyopathies within this list are A31T, M47I, P54H, L97Q, D149G (Table 1).
(Top) PyMOL 3D structure of cTnC (red) showing amino acid variants (spheres) in cTnC (ribbon) that have been published in peer-reviewed journals and been associated with either HCM/RCM (green) or HCM/DCM (yellow) cardiomyopathies. From PDB 6KN7 (Yamada et al. 2020) as illustrated in Fig. 1. (Bottom) Representation of cTnC primary sequence and secondary structure regions with variants highlighted with their respective colors
(Top) PyMOL 3D structure of cTnC (red) showing amino acid variants (spheres) in cTnC (ribbon) that have been published in peer-reviewed journals and been associated with either DCM (blue), HCM/DCM (yellow), or LVNC/DCM (violet) cardiomyopathies. From PDB 6KN7 (Yamada et al. 2020) as illustrated in Fig. 1. (Bottom) Representation of cTnC primary sequence and secondary structure regions with variants highlighted with their respective colors
(Top) PyMOL 3D structure of cTnC showing amino acid variants (spheres) in cTnC (ribbon) that have been published in peer-reviewed journals and been associated with either LVNC (cyan) or LVNC/DCM (violet) cardiomyopathies. From PDB 6KN7 (Yamada et al. 2020) as illustrated in Fig. 1. (Bottom) Representation of cTnC primary sequence and secondary structure regions with variants highlighted in cyan
TNNC1 is a gene that has relatively many variants (115) compared to other genes with its same transcript size (483 bases that code for amino acids) (Watkins et al. 2019). Multiple variants associated with one amino acid were counted as distinct. For example, the Aspartic acid on the second residue has two variants, D2N, and D2G, so this is counted as two variants not one (Table 1). Of the 161 amino acids in the primary sequence of cTnC, there are 77 (47.8%) with at least one identified variant (Table 1). Among these 77 affected residues, variants at 42 residues (26.1%) have been identified as significantly impacting the normal physiology (structure and function) of the heart, culminating in cardiomyopathy-related phenotypes. Not all of these variants have been functionally studied although there is evidence of disease association in patients (Table 1). Further categorization of variants associated with HCM/RCM, DCM, HCM/DCM, and LVNC are 3.7%, 7.5%, 15.5%, 2.5%, respectively. This analysis does not take into account the variant’s level of pathogenicity. A conclusion can be drawn that there are more HCM/DCM-inducing variants in cTnC than other cardiomyopathy phenotypes. By structurally observing the location of the functionally studied variants in cTnC, 15 variants are located within α-helical regions and 10 are located on the linkers, within both NH2- and COOH-terminal domains (Figs. 3, 4 and 5). Microsoft Excel’s χ2 function was used to statistically examine this observation. Even though this trend suggests that cardiomyopathy inducing variants might be more likely to be located within α-helical regions, this is a clear reflection of the proportion of cTnC structure comprised of α-helices χ2 (1, N = 27) = 0.124, p = 0.72.
Mechanisms of TNNC1 cardiomyopathies
Post-translational modifications (PTMs) in cTnC
A post-translational modification (PTM) in the context of proteins occurs when functional groups get temporarily or permanently incorporated into consensus regions via covalent mechanisms as enzymatic and non-enzymatic processes that are fundamental to protein functionality (Harmel and Fiedler 2018; Müller 2018). Through biological systems, PTMs have been reported to be extensively important in maturation of synthesized proteins, signal transduction, and metabolism (Duan and Walther 2015). PTMs are known to cause a variety of outcomes in vitro and in vivo on a physiological level (Wang et al. 2014).
The PTMs included in this study include acetylation, glycation, S-nitrosylation, and phosphorylation (Table 2, Fig. 6). Acetylation of residues in cTnC is important for striated muscle contraction, as over 80% of proteins in striated muscle that are part of the contractile machinery are acetylated (Lundby et al. 2012). This lysine-targeted PTM is carried out by histone deacetylases (HDACs) and acetyltransferases (HATs), and typically involves the addition of an acetyl group onto specific sidechains within specific sequences (Table 2) (Gupta et al. 2008). This modification is responsible for increasing Ca2+ affinity to cTnC at low-affinity sites and high-affinity sites, by 3.5-fold, and twofold, respectively (Grabarek et al. 1995). The acetylated M1 residue in native protein is not included in Fig. 5 as the sequence of cTnC in the Yamada et al. (2020) structure begins at D2; this does not eliminate the possibility of the existence of a modification in vivo. Residues K17, K43, and K92 in cTnC are also reported as acetylated (Table 2, Fig. 6). The irreversible glycation PTM, also known as advanced glycation end-product (AGE) modification, is thought to primarily affect cardiac proteins that are located within cells that have a high turnover rate and is observed in vitro when exposed to excess hexose sugar (Janssens et al. 2018). Physiological effects include altering the rapid cyclic dynamic movements of the troponin complex which in turn affect the association/dissociation kinetics of Ca2+ in cTnC. Thus the presence of a glycated amino acid residue can contribute to changes in myofilament Ca2+ sensitivity and underlie heart failure and/or diabetic cardiomyopathy (Janssens et al. 2018). Residues K6 and K39 in cTnC are found to be glycated (Table 2, Fig. 6). The S-nitrosylation is completed when nitric oxide (NO) adds to thiols, and in turn alters myofilament contractility within the muscle cell. This process is accelerated under normal conditions when stress is present (Figueiredo-Freitas et al. 2015). Under physiological conditions, S-nitrosylation of cTnC desensitizes myofilaments to Ca2+ and reduces cross-bridge turnover (Figueiredo-Freitas et al. 2015). This modification therefore leads to reduced cardiac muscle contractility, posing physiological threats to affected individuals. The two Cys residues in cTnC, C35 and C84, are found to undergo S-nitrosylation PTM (Table 2, Fig. 6).
(Top) PyMOL 3D structure of cTnC (red) showing post-translationally modified amino acids (spheres) in cTnC (ribbon). Acetylation is magenta, S-nitrosylation is green, phosphorylation is orange, and glycation is teal. From PDB 6KN7 (Yamada et al. 2020) as illustrated in Fig. 1. (Bottom) Representation of post-translationally modified amino acids in cTnC primary sequence and secondary structure regions with amino acids highlighted with their respective colors. PTMs were identified using UniProt, PhosphoSitePlus, and published literature (Table 1)
Another PTM with an important role in Ca2+ homeostasis through coordination with other PTMs is phosphorylation (Irie et al. 2015). Phosphorylation involves the introduction of a phosphate (PO4) group to an amino acid sidechain (typically Ser, Thr or Tyr), in turn affecting the protein’s conformational properties, function and interactions with other molecules (Ardito et al. 2017). Phosphorylation sites are often located in disordered regions of proteins throughout biological systems. Of the 15 Ser, Thr and Tyr residues in human cTnC, two Ser residues—S89 and S98—have been identified as targets for phosphorylation (Table 2, Fig. 6) (Huttlin et al. 2010). Decreased phosphopeptide count in heart tissue was shown to be correlated with differences in signaling and tissue heterogeneity (Huttlin et al. 2010). While relatively little is known about the physiological effects of cTnC phosphorylation, cTnI phosphorylation has received much more attention, initially because of its relevance to autonomic regulation of cardiac function (Solaro et al. 1976). Phosphorylation of cTnI at Ser-42 and Ser-44 may affect its interaction with cTnC and alter Ca2+ activation (Kobayashi and Solaro 2005). This alteration takes place because the modified reside in cTnI interacts with Glu-10 in cTnC, which is in the N–helix of the regulatory NH2-terminal domain of cTnC (Kobayashi and Solaro 2005). Similarly, phosphorylation of cTnI residues Ser-23 and Ser-24 cTnI is important in regulating cardiac muscle contractility, largely through modulation of Ca2+ binding by cTnC (Biesiadecki et al. 2007). Although phosphorylation has been extensively studied, Ca2+-dependent allosteric changes in the thin filament by phosphorylation of TnI are not fully understood (Dong et al. 2007). Furthermore, PTMs directly involving residues in cTnC can impact the regulation of Ca2+ binding (Grabarek et al. 1995). It will be important to continue to establish under what physiological conditions the residues in cTnC identified in Table 2 become modified and to what extent (i.e., what fraction of cTnC molecules are modified at that residue), and also what other PTMs may occur in cTnC.
Co-localization of amino acid variants and PTMs
The S-nitrosylated residue C84 in cTnC (Table 2) may also be altered by an HCM-inducing C84Y variant (Table 1) (Figueiredo-Freitas et al. 2015; Landstrom et al. 2008). The C84Y variant involves replacing a cysteine at position 84 with a tyrosine. Not only does the removal of Cys preclude the possibility of S-nitrosylation at that site, but the introduction of Tyr to the amino acid sequence introduces a new possibility for PTM. Tyrosine is commonly modified by sulfation (Yang et al. 2015) and phosphorylation (Harney et al. 2005). Considering that S-nitrosylation is known to decrease myofilament Ca2+ sensitivity, the replacement of C84 with Tyr increases Ca2+ affinity of isolated cTnC as well as Ca2+ sensitivity of myofilament function in the intact sarcomere (Pinto et al. 2009).
The significance of Cys residues in TnC extends beyond the naturally occurring phenomena listed in Tables 1 and 2. Grabarek et al. (1990) introduced a disulfide into sTnC via site-directed mutagenesis that inactivated the Ca2+-regulatory nature of TnC. This disulfide bridge was located between the B–C linker and helix D of the central helix (Grabarek et al. 1990). In contrast, disulfide bridge formation between the native Cys residues (C35 and C84) in cTnC results in a constitutively activated cTnC (referred to as aTnC) that does not require Ca2+ for activation of force generation, unloaded shortening, or rapid kinetics of isometric tension redevelopment (Chase et al. 1994a, 1994b; Hannon et al. 1993; Martyn et al. 1994). This intermolecular disulfide bridge is thought to produce conformational changes that mimic some aspects of Ca2+-binding; distortion of the structure of the NH2-terminal domain would likely reduce dynamics of cTnC, could alter site II in a manner that is similar to the structure cTnC in the Ca2+-bound state, and probably induces opening of the hydrophobic patch for cTnI-binding. Site-specific variants to the Cys-84 residue in cTnC (Table 1) would decrease or eliminate the likelihood of PTMs at that and nearby residues, and in turn lead to changes in the structure or function of cTnC. Characterizing PTMs in the pathogenic, HCM-inducing variant C84Y has provided, and will certainly continue to provide insights into how PTMs act on a molecular level to influence phenotypes.
The acetylated residue K92 (Table 2) also experiences a K92R variant (Table 1). The introduction of an Arg amino acid to the sequence is conservative in the sense that both Lys and Arg have basic sidechains, but it brings forward the possibility of new PTMs. Commonly, PTMs associated with Arg are classified as non-enzymatic and enzymatic. Non-enzymatic PTMs are carbonylation and advanced glycation end products (AGEs) (Slade et al. 2014). Enzymatic PTM’s of Arg residues are citrullination, methylation, phosphorylation, ADP-ribosylation (Slade et al. 2014). K92 lies in the middle of the flexible DE-linker that connects the NH2- and COOH-terminal domains of cTnC, not far from residue C84 (Takeda et al. 2003; Yamada et al. 2020). This region is important for Ca2+ dependent regulation of cardiac troponin (Manning et al. 2012). Knowing this, removal or modification of a residue in this region may alter its regulatory properties. Additionally, the introduction of amino acid variants into the sequence of cTnC as discussed in this review may have the possibility to alter the likelihood of PTMs at the affected amino acid or nearby residues. This does not remove the possibility of PTMs, as PTMs are necessary for function and species complexity (Darling and Uversky 2018). At the time of writing, there are no reports on the pathogenicity of the K92R variant (Table 1). Therefore, this variant could be further clinically studied to further investigate the physiological significance of PTM at K92 (Table 2) and to determine the disease relevance of both PTMs and the variant.
Haploinsufficiency vs. dominant negative mechanism
All of the pathogenic variants in TNNC1 identified to-date encode single or double amino acid substitutions. Therefore, mutant cTnC is expected to incorporate normally into the sarcomere, thereby causing disease via a dominant negative (‘poison peptide’) mechanism (Gomes and Potter 2004). This contrasts with what is observed for cardiomyopathic variants in the genes encoding myosin-binding protein C and titin, which typically produce truncating variants and elicit disease via haploinsufficiency (Yotti et al. 2019). Upon incorporation into the contractile apparatus, cTnC mutants trigger contractile dysfunction due to altered interactions with cTnT, cTnI or Ca2+ binding affinity, or a combination thereof. Although numerous TNNC1 variants have been reported to be associated with cardiomyopathy (Table 1), only several have been extensively characterized structurally and functionally. The DCM-associated cTnC-G159D variant has been suggested to impair interactions with full-length cTnT and enhance interactions with full-length cTnI, as determined indirectly by a mammalian two-hybrid assay (Mogensen et al. 2004). The cTnC-G159D variant was also previously shown to decrease myofilament Ca2+ sensitivity and blunt the lusitropic effect of protein kinase A-mediated phosphorylation of cTnI at the N-terminal serine residues (Biesiadecki et al. 2007). The first study to directly demonstrate that cTnT binds to cTnC showed that the intrinsically disordered carboxy-terminus of cTnT directly participates in Ca2+-dependent interactions with the regulatory NH2-domain of cTnC, and that the binding affinity between these two subunits is increased in the presence of a DCM-associated variant in cTnC (I4M; Table 1) (Johnston et al. 2019). Additional DCM-associated cTnC variants (Y5H, M103I, and I148V) have been shown to decrease Ca2+ sensitivity of isometric tension generation (Pinto et al. 2011). It should be noted that the proband carrying the Y5H variant also had a genetic variant encoding a R1045C substitution in the β-myosin heavy chain (Pinto et al. 2011). Therefore, the relative contributions of these variants to the pathophysiology observed in this proband remain uncertain, and this lessons applies to other probands, some of which are compound heterozygous (Landim-Vieira et al. 2020a; Lim et al. 2008; Ploski et al. 2016), with variants in both cTnC and another cardiomyopathy gene (Table 1).
Altered interactions with TnI and/or TnT, or perhaps tropomyosin/actin
In contrast to DCM variants, HCM variants in cTnC tend to increase myofilament Ca2+ sensitivity. For example, the HCM-associated cTnC-A8V variant significantly increases Ca2+ sensitivity of isometric tension via direct enhancement of Ca2+-binding to site II and increased binding to full-length cTnI (Gonzalez-Martinez et al. 2018; Kawai et al. 2017; Martins et al. 2015; Zot et al. 2016). Another HCM-associated variant in cTnC that leads to markedly increased myofilament Ca2+ sensitivity is C84Y (Landstrom et al. 2008). The double, compound heterozygous cTnC variant (E59D/D75Y) associated with DCM (Lim et al. 2008) has also been shown to decrease myofilament Ca2+ sensitivity and reduce strong actin-myosin binding (Dweck et al. 2010). Interestingly, the HCM-associated cTnC-D145E variant has been shown to alter Ca2+ binding in the C-domain of cTnC (Marques et al. 2017; Swindle and Tikunova 2010), which could alter interactions with the N-terminal region of cTnI and/or C-terminal region of cTnT. Some cTnC variants can also impact actomyosin adenosine triphosphatase (ATPase) activity (e.g., Veltri et al. 2017). For example, it has been reported that the HCM-associated cTnC-A31S variant increases Ca2+-activated actomyosin ATPase activity in addition to increasing Ca2+ sensitivity of isometric force (Parvatiyar et al. 2012). Taken together, TNNC1-encoded cardiomyopathic variants generally cause myofilament dysfunction via disruption of Ca2+-binding and/or altered interactions with other troponin subunits.
It is now evident, however, that the pathogenicity of TNNC1 variants is not always explained by defects in sarcomere contractility. For example, the functional consequences of TNNC1-encoded compound heterozygous missense variants (D132N and D145E) that were identified in two siblings with early-onset DCM (Landim-Vieira et al. 2020a). These variants were inherited, one from each parent. Strikingly, we found no significant differences in the contractile parameters of permeabilized cardiac muscle preparations reconstituted with 50% D145E/50% D132N cTnC variants compared with WT (Landim-Vieira et al. 2020a). This observation raised the intriguing possibility that pathogenic variants in the TNNC1 alleles may cause cardiomyopathy through mechanisms beyond myofilament dysfunction.
Potential non-canonical (mitochondrial/nuclear) roles of cTnC
In a seminal study in 2009 on cardiomyocyte renewal in humans by Bergmann and colleagues from the Frisén lab, TNNT2 and TNNI3 were coincidentally identified for the first time as being directly associated with isolated cardiomyocyte nuclei (Bergmann et al. 2009). By immunofluorescence microscopy and immunoblotting, both subunits were unambiguously shown to be associated with human cardiomyocyte nuclei that were purified from human cardiac ventricular tissue and then sorted by immunolabeling and flow-cytometry (Bergmann et al. 2009). In a follow up study, Asumda and Chase (2012) identified all three subunits of cardiac troponin (cTnI, cTnT and cTnC) along with Tm and nuclear actin within the nuclei of cultured neonatal rat ventricular cardiomyocytes by immunofluorescence confocal microscopy. Evidence from this report also showed that nuclear expression of the myofilament proteins occurs early during cardiomyogenic differentiation of rat, multipotent, bone marrow-derived mesenchymal stem cells (Asumda and Chase 2012). Further, bioinformatics predictions were used to show that cTnI and cTnT contain putative nuclear localization signals (NLS) in both of the aforementioned studies (Asumda and Chase 2012; Bergmann et al. 2009). Some pathogenic variants may introduce or nullify a NLS, therefore potentially altering nucleocytoplasmic shuttling (Chase et al. 2013). Interestingly, using the approach previously applied to cTnC WT (Chase et al. 2013), cTnC variant V64A (Table 1) is predicted to localize to the cardiomyocyte nucleus to a much greater extent than WT or any other cTnC variant that affects a single amino acid. It remains to be tested experimentally, however, whether cTnC-V64A is in fact concentrated in cardiomyocyte nuclei.
Although the precise roles of nuclear-localized myofilament proteins in cardiomyocytes remain largely unclear, cancer cells might be able to provide some insight. Certain cancer cell lines and human tumor tissues display over-expression of troponin at the gene and protein levels (Johnston et al. 2018). Bioinformatic pathway analyses suggest that troponin subunits may play a role in the organization of chromatin, DNA repair processes, RNA metabolism, and gene transcription (Johnston et al. 2018). These results point to the intriguing possibility that myofilament proteins might have dual roles in cardiomyocytes: to regulate muscle contraction and participation in Ca2+-dependent signaling in the nucleus, which could indeed have profound implications for understanding the molecular pathophysiological mechanisms of cardiomyopathies caused by thin filament variants.
In 2015, Wu and colleagues provided compelling evidence for the possibility that a cardiomyopathic variant in a troponin subunit has a regulatory role in the nuclear compartment of cardiomyocytes (Wu et al. 2015). Using induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) derived from human patients harboring a DCM-linked sarcomeric variant (cTnT-Arg173Trp), they found increased nuclear localization of cTnTArg173Trp compared to iPSC-CMs derived from non-DCM relatives (Wu et al. 2015). To ascertain the biological significance of nuclear-localized TNNT2, co-immunoprecipitation followed by mass-spectrometry was used to identify potential interacting proteins (Wu et al. 2015). Interestingly, they found TNNT2 was bound, either directly or indirectly, to histone H3 and histone demethylases, among other interacting partners (Wu et al. 2015). After further correlating these observations with changes in chromatin marks, the authors proposed that TNNT2 participates in the epigenetic regulation of phosphodiesterase (PDE) gene expression, which was altered as a result of the cTnT-Arg173Trp DCM variant (Wu et al. 2015). A more recent study also revealed that a RCM variant in cTnI might directly interact with histone deacetylase 1 and repress gene expression of phosphodiesterase 4D in mutant cardiomyocytes (Zhao et al. 2020a; Zhao et al. 2020b). These results are particularly captivating because they not only transform the way we view the pathogenesis of inherited cardiomyopathies, but also how we view myofilament proteins from a fundamental, basic science perspective. Because other thin myofilament proteins have been identified within cardiomyocyte nuclei, it would seem likely that this phenomenon is not exclusive to these specific variants in TNNT2 and TNNI3 or even the WT sequences. In addition to thick filament components MYH7 and MYBPC3, pathogenic genetic variants may also occur in thin filament genes for TPM1, TNNC1, TNNI3 and TNNT2 which are linked to cardiomyopathy. The two variants demonstrated to alter gene expression raise the possibility that, in addition to disrupting the biomechanical properties of the sarcomere, they and other variants could also perturb the architecture of chromatin, the architecture of the entire nucleus, or even the not-well-understood elevation of ploidy and nuclearity of cardiomyocytes (Landim-Vieira et al. 2020b).
Future directions
Cardiac TnC is a highly conserved protein from an evolutionary viewpoint. Thus investigating the locations of the surprisingly large number of variants in the cardiomyopathic gene TNNC1 and PTMs of cTnC is crucial to further understand the underlying mechanisms of heart disease progression. Through extensive analyses, this review categorizes all of the currently documented variants in cTnC which may or may not cause cardiomyopathies. Additionally, compiling residues altered by amino acid variants and/or PTMs in cTnC allows us to localize the affected residues in the 3D structure. It will be important to assess the functional significance of cTnC PTMs, as their functional roles are relatively undiscussed in the literature. Further work should be done to investigate variants by characterizing human iPSC-CMs engineered with established pathogenic variants in TNNC1 HCM/RCM (A8V) and DCM (I4M). There are pathogenic variants A31T, M47I, P54H, L97Q, and D149G that do not have any known cardiomyopathy associated (Table 1). These variants need to be investigated to further warrant a possible cardiomyopathy association. Studying structural dynamics (HDX) and protein–protein interactions (e.g., by crosslinking) of cTnC in the thin filament (native or reconstituted) should also be completed along with high-resolution structural analyses (NMR and cryo–EM). Finally, further probing non-canonical roles of cTnC from a basic science perspective and also from a translational perspective would provide a better understanding of these sarcomeric proteins beyond muscle contraction.
Data availability
Data are already available in public databases.
Code availability
Not applicable.
References
Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L (2017) The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int J Mol Med 40:271–280. https://doi.org/10.3892/ijmm.2017.3036
Asumda FZ, Chase PB (2012) Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation 83:106–115. https://doi.org/10.1016/j.diff.2011.10.002
Badr MA, Pinto JR, Davidson MW, Chase PB (2016) Fluorescent protein-based Ca2+ sensor reveals global, divalent cation-dependent conformational changes in cardiac troponin C. PLoS ONE 11:e0164222. https://doi.org/10.1371/journal.pone.0164222
Bergmann O et al (2009) Evidence for cardiomyocyte renewal in humans. Science 324:98–102. https://doi.org/10.1126/science.1164680
Biesiadecki BJ, Kobayashi T, Walker JS, John Solaro R, de Tombe PP (2007) The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ Res 100:1486–1493. https://doi.org/10.1161/01.RES.0000267744.92677.7f
Brieler J, Breeden MA, Tucker J (2017) Cardiomyopathy: an overview. Am Fam Physician 96:640–646
KN Brown VS Pendela RR Diaz 2020 Restrictive (infiltrative) cardiomyopathy. In: StatPearls. Treasure Island (FL)
Budde H et al (2019) The newly detected myosin binding protein C/troponin interaction, function and thin filaments integrity affected by cTn mutants. J Muscle Res Cell Motil 40:240. https://doi.org/10.1007/s10974-019-09534-w
Carnevale A et al (2020) Genomic study of dilated cardiomyopathy in a group of Mexican patients using site-directed next generation sequencing. Mol Genet Genomic Med. https://doi.org/10.1002/mgg3.1504
Chase PB, Martyn DA, Hannon JD (1994a) Activation dependence and kinetics of force and stiffness inhibition by aluminiofluoride, a slowly dissociating analogue of inorganic phosphate, in chemically skinned fibres from rabbit psoas muscle. JMRCM 15:119–129
Chase PB, Martyn DA, Hannon JD (1994b) Isometric force redevelopment of skinned muscle fibers from rabbit with and without Ca2+. Biophys J 67:1994–2001. https://doi.org/10.1016/S0006-3495(94)80682-4
Chase PB, Szczypinski MP, Soto EP (2013) Nuclear tropomyosin and troponin in striated muscle: new roles in a new locale? J Muscle Res Cell Motil 34:275–284. https://doi.org/10.1007/s10974-013-9356-7
Choudhry S, Puri K, Denfield SW (2019) An update on pediatric cardiomyopathy. Curr Treat Options Cardiovasc Med 21:36. https://doi.org/10.1007/s11936-019-0739-y
Chung WK, Kitner C, Maron BJ (2011) Novel frameshift mutation in Troponin C (TNNC1) associated with hypertrophic cardiomyopathy and sudden death. Cardiol Young 21:345–348. https://doi.org/10.1017/S1047951110001927
Costabel U, Wessendorf TE, Bonella F (2017) Epidemiology and clinical presentation of sarcoidosis. Klin Monbl Augenheilkd 234:790–795. https://doi.org/10.1055/s-0042-105569
Darling AL, Uversky VN (2018) Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front Genet 9:158. https://doi.org/10.3389/fgene.2018.00158
Dong WJ, Jayasundar JJ, An J, Xing J, Cheung HC (2007) Effects of PKA phosphorylation of cardiac troponin I and strong crossbridge on conformational transitions of the N-domain of cardiac troponin C in regulated thin filaments. Biochemistry 46:9752–9761. https://doi.org/10.1021/bi700574n
Duan G, Walther D (2015) The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1004049
Dweck D, Hus N, Potter JD (2008) Challenging current paradigms related to cardiomyopathies. Are changes in the Ca2+ sensitivity of myofilaments containing cardiac troponin C mutations (G159D and L29Q) good predictors of the phenotypic outcomes? J Biol Chem 283:33119–33128. https://doi.org/10.1074/jbc.M804070200
Dweck D, Reynaldo DP, Pinto JR, Potter JD (2010) A dilated cardiomyopathy troponin C mutation lowers contractile force by reducing strong myosin-actin binding. J Biol Chem 285:17371–17379. https://doi.org/10.1074/jbc.M109.064105
Elliott PM et al (2014) 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 35:2733–2779. https://doi.org/10.1093/eurheartj/ehu284
Figueiredo-Freitas C et al (2015) S-Nitrosylation of sarcomeric proteins depresses myofilament Ca2+ sensitivity in intact cardiomyocytes. Antioxid Redox Signal 23:1017–1034. https://doi.org/10.1089/ars.2015.6275
Franaszczyk M et al (2020) Analysis of de novo mutations in sporadic cardiomyopathies emphasizes their clinical relevance and points to novel candidate genes. J Clin Med. https://doi.org/10.3390/jcm9020370
Friedrich FW, Flenner F, Nasib M, Eschenhagen T, Carrier L (2016) Epigallocatechin-3-gallate accelerates relaxation and Ca2+ transient decay and desensitizes myofilaments in healthy and Mybpc3-targeted knock-in cardiomyopathic mice. Front Physiol 7:607. https://doi.org/10.3389/fphys.2016.00607
Fuchs F, Grabarek Z (2011) The Ca2+/Mg2+ sites of troponin C modulate crossbridge-mediated thin filament activation in cardiac myofibrils. Biochem Biophys Res Commun 408:697–700. https://doi.org/10.1016/j.bbrc.2011.04.092
Garfinkel AC, Seidman JG, Seidman CE (2018) Genetic pathogenesis of hypertrophic and dilated cardiomyopathy. Heart Fail Clin 14:139–146. https://doi.org/10.1016/j.hfc.2017.12.004
Gersh BJ et al (2011a) 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 124:e783-831. https://doi.org/10.1161/CIR.0b013e318223e2bd
Gersh BJ et al (2011b) 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 124:2761–2796. https://doi.org/10.1161/CIR.0b013e318223e230
Gomes AV, Potter JD (2004) Molecular and cellular aspects of troponin cardiomyopathies. Ann N Y Acad Sci 1015:214–224
Gonzalez-Martinez D et al (2018) Structural and functional impact of troponin C-mediated Ca2+ sensitization on myofilament lattice spacing and cross-bridge mechanics in mouse cardiac muscle. J Mol Cell Cardiol 123:26–37. https://doi.org/10.1016/j.yjmcc.2018.08.015
Gordon AM, Homsher E, Regnier M (2000) Regulation of contraction in striated muscle. Physiol Rev 80:853–924. https://doi.org/10.1152/physrev.2000.80.2.853
Grabarek Z, Mabuchi Y, Gergely J (1995) Properties of troponin C acetylated at lysine residues. Biochemistry 34:11872–11881. https://doi.org/10.1021/bi00037a027
Grabarek Z, Tan R-Y, Wang J, Tao T, Gergely J (1990) Inhibition of mutant troponin C activity by an intra-domain disulphide bond. Nature 345:132–135. https://doi.org/10.1038/345132a0
Gupta MP, Samant SA, Smith SH, Shroff SG (2008) HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J Biol Chem 283:10135–10146. https://doi.org/10.1074/jbc.M710277200
Hannon JD, Chase PB, Martyn DA, Huntsman LL, Kushmerick MJ, Gordon AM (1993) Calcium-independent activation of skeletal muscle fibers by a modified form of cardiac troponin C. Biophys J 64:1632–1637
Harmel R, Fiedler D (2018) Features and regulation of non-enzymatic post-translational modifications. Nat Chem Biol 14:244–252. https://doi.org/10.1038/nchembio.2575
Harney DF, Butler RK, Edwards RJ (2005) Tyrosine phosphorylation of myosin heavy chain during skeletal muscle differentiation: an integrated bioinformatics approach. Theor Biol Med Model 2:12. https://doi.org/10.1186/1742-4682-2-12
Hershberger RE, Hedges DJ, Morales A (2013) Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol 10:531–547. https://doi.org/10.1038/nrcardio.2013.105
Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Geßner R (2001) First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat 17:524. https://doi.org/10.1002/humu.1143
Huttlin EL et al (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189. https://doi.org/10.1016/j.cell.2010.12.001
Ikeda U, Minamisawa M, Koyama J (2015) Isolated left ventricular non-compaction cardiomyopathy in adults. J Cardiol 65:91–97. https://doi.org/10.1016/j.jjcc.2014.10.005
Ingles J et al (2019) Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med 12:e002460. https://doi.org/10.1161/CIRCGEN.119.002460
Irie T et al (2015) S-nitrosylation of calcium-handling proteins in cardiac adrenergic signaling and hypertrophy. Circ Res 117:793–803. https://doi.org/10.1161/CIRCRESAHA.115.307157
Janssens JV, Ma B, Brimble MA, Van Eyk JE, Delbridge LMD, Mellor KM (2018) Cardiac troponins may be irreversibly modified by glycation: novel potential mechanisms of cardiac performance modulation. Sci Rep 8:16084. https://doi.org/10.1038/s41598-018-33886-x
Jefferies JL, Towbin JA (2010) Dilated cardiomyopathy. Lancet 375:752–762. https://doi.org/10.1016/S0140-6736(09)62023-7
Jenni R, Oechslin EN, van der Loo B (2007) Isolated ventricular non-compaction of the myocardium in adults. Heart 93:11–15. https://doi.org/10.1136/hrt.2005.082271
Johnston JR, Chase PB, Pinto JR (2018) Troponin through the looking-glass: emerging roles beyond regulation of striated muscle contraction. Oncotarget 9:1461–1482. https://doi.org/10.18632/oncotarget.22879
Johnston JR et al (2019) The intrinsically disordered C terminus of troponin T binds to troponin C to modulate myocardial force generation. J Biol Chem 294:20054–20069. https://doi.org/10.1074/jbc.RA119.011177
Katrukha IA (2013) Human cardiac troponin complex Structure and functions. Biochemistry (Mosc) 78:1447–1465. https://doi.org/10.1134/S0006297913130063
Kawai M, Johnston JR, Karam T, Wang L, Singh RK, Pinto JR (2017) Myosin rod hypophosphorylation and CB kinetics in papillary muscles from a TnC-A8V KI mouse model. Biophys J 112:1726–1736. https://doi.org/10.1016/j.bpj.2017.02.045
Kirk R et al (2009) Outcome of pediatric patients with dilated cardiomyopathy listed for transplant: a multi-institutional study. J Heart Lung Transplant 28:1322–1328. https://doi.org/10.1016/j.healun.2009.05.027
Kobayashi T, Solaro RJ (2005) Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu Rev Physiol 67:39–67. https://doi.org/10.1146/annurev.physiol.67.040403.114025
Landim-Vieira M et al (2020) Familial dilated cardiomyopathy associated with a novel combination of compound heterozygous TNNC1 variants (* equal contributions). Front Physiol 10:1612. https://doi.org/10.3389/fphys.2019.01612
Landim-Vieira M, Schipper JM, Pinto JR, Chase PB (2020) Cardiomyocyte nuclearity and ploidy: when is double trouble? J Muscle Res Cell Motil. https://doi.org/10.1007/s10974-019-09545-7
Landstrom AP et al (2008) Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J Mol Cell Cardiol 45:281–288. https://doi.org/10.1016/j.yjmcc.2008.05.003
Li MX et al (2018) The calcium sensitizer drug MCI-154 binds the structural C-terminal domain of cardiac troponin C. Biochem Biophys Rep 16:145–151. https://doi.org/10.1016/j.bbrep.2018.10.012
Lim CC et al (2008) A novel mutant cardiac troponin C disrupts molecular motions critical for calcium binding affinity and cardiomyocyte contractility. Biophys J 94:3577–3589. https://doi.org/10.1529/biophysj.107.112896
Lu C et al (2018) Molecular analysis of inherited cardiomyopathy using next generation semiconductor sequencing technologies. J Transl Med 16:241. https://doi.org/10.1186/s12967-018-1605-5
Luk A, Ahn E, Soor GS, Butany J (2009) Dilated cardiomyopathy: a review. J Clin Pathol 62:219–225. https://doi.org/10.1136/jcp.2008.060731
Lundby A et al (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep 2:419–431. https://doi.org/10.1016/j.celrep.2012.07.006
Manning EP, Guinto PJ, Tardiff JC (2012) Correlation of molecular and functional effects of mutations in cardiac troponin T linked to familial hypertrophic cardiomyopathy: an integrative in silico/in vitro approach. J Biol Chem 287:14515–14523. https://doi.org/10.1074/jbc.M111.257436
Maron BJ, Maron MS (2013) Hypertrophic cardiomyopathy. Lancet 381:242–255. https://doi.org/10.1016/S0140-6736(12)60397-3
Maron BJ, Maron MS, Semsarian C (2012) Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol 60:705–715. https://doi.org/10.1016/j.jacc.2012.02.068
Maron BJ et al (2006) Contemporary definitions and classification of the cardiomyopathies: an American Heart Association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation 113:1807–1816. https://doi.org/10.1161/CIRCULATIONAHA.106.174287
Marques MdA, de Oliveira GAP (2016) Cardiac troponin and tropomyosin: structural and cellular perspectives to unveil the hypertrophic cardiomyopathy phenotype. Front Physiol 7:429. https://doi.org/10.3389/fphys.2016.00429
Marques MdA et al (2017) Allosteric transmission along a loosely structured backbone allows a cardiac troponin C mutant to function with only one Ca2+ ion. J Biol Chem 292:2379–2394. https://doi.org/10.1074/jbc.M116.765362
Martins AS et al (2015) In vivo analysis of troponin C Knock-In (A8V) mice: evidence that TNNC1 is a hypertrophic cardiomyopathy susceptibility gene. Circ Cardiovasc Genet 8:653–664. https://doi.org/10.1161/CIRCGENETICS.114.000957
Martyn DA, Chase PB, Hannon JD, Huntsman LL, Kushmerick MJ, Gordon AM (1994) Unloaded shortening of skinned muscle fibers from rabbit activated with and without Ca2+. Biophys J 67:1984–1993. https://doi.org/10.1016/S0006-3495(94)80681-2
Mazzarotto F et al (2020) Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation 141:387–398. https://doi.org/10.1161/CIRCULATIONAHA.119.037661
McKillop DFA, Geeves MA (1993) Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J 65:693–701. https://doi.org/10.1016/S0006-3495(93)81110-X
Miszalski-Jamka K et al (2017) Novel genetic triggers and genotype-phenotype correlations in patients with left ventricular noncompaction. Circ Cardiovasc Genet. https://doi.org/10.1161/CIRCGENETICS.117.001763
Mogensen J et al (2004) Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol 44:2033–2040. https://doi.org/10.1016/j.jacc.2004.08.027
Muchtar E, Blauwet LA, Gertz MA (2017) Restrictive cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res 121:819–837. https://doi.org/10.1161/CIRCRESAHA.117.310982
Müller MM (2018) Post-translational modifications of protein backbones: unique functions, mechanisms, and challenges. Biochemistry 57:177–185. https://doi.org/10.1021/acs.biochem.7b00861
Musunuru K et al (2020) Genetic testing for inherited cardiovascular diseases: a scientific statement from the American Heart Association. Circ Genom Precis Med 13:e000067. https://doi.org/10.1161/HCG.0000000000000067
Negele JC, Dotson DG, Liu W, Sweeney HL, Putkey JA (1992) Mutation of the high affinity calcium binding sites in cardiac troponin C. J Biol Chem 267:825–831
Oda T, Yanagisawa H, Wakabayashi T (2020) Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin. J Struct Biol. https://doi.org/10.1016/j.jsb.2020.107450
Parvatiyar MS, Landstrom AP, Figueiredo-Freitas C, Potter JD, Ackerman MJ, Pinto JR (2012) A mutation in TNNC1-encoded cardiac troponin C, TNNC1-A31S, predisposes to hypertrophic cardiomyopathy and ventricular fibrillation. J Biol Chem 287:31845–31855. https://doi.org/10.1074/jbc.M112.377713
Pinto JR, Parvatiyar MS, Jones MA, Liang J, Ackerman MJ, Potter JD (2009) A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J Biol Chem 284:19090–19100. https://doi.org/10.1074/jbc.M109.007021
Pinto JR et al (2011) Functional characterization of TNNC1 rare variants identified in dilated cardiomyopathy. J Biol Chem 286:34404–34412. https://doi.org/10.1074/jbc.M111.267211
Pirani A, Xu C, Hatch V, Craig R, Tobacman LS, Lehman W (2005) Single particle analysis of relaxed and activated muscle thin filaments. J Mol Biol 346:761–772
Ploski R et al (2016) Evidence for troponin C (TNNC1) as a gene for autosomal recessive restrictive cardiomyopathy with fatal outcome in infancy. Am J Med Genet A 170:3241–3248. https://doi.org/10.1002/ajmg.a.37860
Poole KJV et al (2006) A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol 155:273–284. https://doi.org/10.1016/j.jsb.2006.02.020
Potter JD, Gergely J (1975) The calcium and magnesium binding sites on troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J Biol Chem 250:4628–4633
Pua CJ et al (2020) Genetic studies of hypertrophic cardiomyopathy in singaporeans identify variants in TNNI3 and TNNT2 that are common in chinese patients. Circ Genom Precis Med. https://doi.org/10.1161/CIRCGEN.119.002823
Reichart D, Magnussen C, Zeller T, Blankenberg S (2019) Dilated cardiomyopathy: from epidemiologic to genetic phenotypes: a translational review of current literature. J Intern Med 286:362–372. https://doi.org/10.1111/joim.12944
Richards AA, Garg V (2010) Genetics of congenital heart disease. Curr Cardiol Rev 6:91–97. https://doi.org/10.2174/157340310791162703
Richards S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Risi C, Eisner J, Belknap B, Heeley DH, White HD, Schröder GF, Galkin VE (2017) Ca2+-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A 114:6782–6787. https://doi.org/10.1073/pnas.1700868114
Roher A, Leiska N, Spitz W (1986) The amino acid sequence of human cardiac troponin-C. Muscle Nerve 9:73–77. https://doi.org/10.1002/mus.880090112
Samsa LA, Yang B, Liu J (2013) Embryonic cardiac chamber maturation: trabeculation, conduction, and cardiomyocyte proliferation. Am J Med Genet C Semin Med Genet 163C:157–168. https://doi.org/10.1002/ajmg.c.31366
Schlecht W, Zhou Z, Li K-L, Rieck D, Ouyang Y, Dong W-J (2014) FRET study of the structural and kinetic effects of PKC phosphomimetic cardiac troponin T mutants on thin filament regulation. Arch Biochem Biophys 550–551:1–11. https://doi.org/10.1016/j.abb.2014.03.013
Schmidtmann A et al (2005) Cardiac troponin C-L29Q, related to hypertrophic cardiomyopathy, hinders the transduction of the protein kinase A dependent phosphorylation signal from cardiac troponin I to C. FEBS J 272:6087–6097. https://doi.org/10.1111/j.1742-4658.2005.05001.x
Semsarian C, Ingles J, Maron MS, Maron BJ (2015) New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 65:1249–1254. https://doi.org/10.1016/j.jacc.2015.01.019
Sia SK, Li MX, Spyracopoulos L, Gagné SM, Liu W, Putkey JA, Sykes BD (1997) Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain. J Biol Chem 272:18216–18221
DP Singh, H Patel 2020 Left Ventricular Non-compaction (LVNC) cardiomyopathy. In: StatPearls. Treasure Island (FL)
Slade DJ, Subramanian V, Fuhrmann J, Thompson PR (2014) Chemical and biological methods to detect post-translational modifications of arginine. Biopolymers 101:133–143. https://doi.org/10.1002/bip.22256
Slupsky CM, Sykes BD (1995) NMR solution structure of calcium-saturated skeletal muscle troponin C. Biochemistry 34:15953–15964
Solaro RJ, Moir AJ, Perry SV (1976) Phosphorylation of troponin I and the inotropic effect of adrenalin in the perfused rabbit heart. Nature 262:615–617
Swindle N, Tikunova SB (2010) Hypertrophic cardiomyopathy-linked mutation D145E drastically alters calcium binding by the C-domain of cardiac troponin C. Biochemistry 49:4813–4820. https://doi.org/10.1021/bi100400h
Tadros HJ et al (2020) Meta-analysis of cardiomyopathy-associated variants in troponin genes identifies loci and intragenic hot spots that are associated with worse clinical outcomes. J Mol Cell Cardiol 142:118–125. https://doi.org/10.1016/j.yjmcc.2020.04.005
Takasaki A et al (2018) Sarcomere gene variants act as a genetic trigger underlying the development of left ventricular noncompaction. Pediatr Res. https://doi.org/10.1038/s41390-018-0162-1
Takeda S, Yamashita A, Maeda K, Maéda Y (2003) Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 424:35–41. https://doi.org/10.1038/nature01780
Tardiff JC (2011) Thin filament mutations: developing an integrative approach to a complex disorder. Circ Res 108:765–782. https://doi.org/10.1161/CIRCRESAHA.110.224170
Towbin JA et al (2006) Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA 296:1867–1876. https://doi.org/10.1001/jama.296.15.1867
van der Velden J, Stienen GJM (2019) Cardiac disorders and pathophysiology of sarcomeric proteins. Physiol Rev 99:381–426. https://doi.org/10.1152/physrev.00040.2017
van Spaendonck-Zwarts KY et al (2010) Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation 121:2169–2175. https://doi.org/10.1161/CIRCULATIONAHA.109.929646
Vasilescu C et al (2018) Genetic basis of severe childhood-onset cardiomyopathies. J Am Coll Cardiol 72:2324–2338. https://doi.org/10.1016/j.jacc.2018.08.2171
Veltri T et al (2017) Hypertrophic cardiomyopathy cardiac troponin C mutations differentially affect slow skeletal and cardiac muscle regulation. Front Physiol 8:221. https://doi.org/10.3389/fphys.2017.00221
Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ (2005) Ca2+-regulated structural changes in troponin. Proc Natl Acad Sci U S A 102:5038–5043
Vos T et al (2016) Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1545–1602. https://doi.org/10.1016/S0140-6736(16)31678-6
Wang X, Li MX, Sykes BD (2002) Structure of the regulatory N-domain of human cardiac troponin C in complex with human cardiac troponin I147–163 and bepridil. J Biol Chem 277:31124–31133. https://doi.org/10.1074/jbc.M203896200
Wang Y-C, Peterson SE, Loring JF (2014) Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res 24:143–160. https://doi.org/10.1038/cr.2013.151
Warren CM et al (2015) Green tea catechin normalizes the enhanced Ca2+ sensitivity of myofilaments regulated by a hypertrophic cardiomyopathy-associated mutation in human cardiac troponin I (K206I). Circ Cardiovasc Genet 8:765–773. https://doi.org/10.1161/CIRCGENETICS.115.001234
Watkins WS et al (2019) De novo and recessive forms of congenital heart disease have distinct genetic and phenotypic landscapes. Nat Commun 10:4722. https://doi.org/10.1038/s41467-019-12582-y
Webber SA et al (2012) Outcomes of restrictive cardiomyopathy in childhood and the influence of phenotype: a report from the Pediatric Cardiomyopathy Registry. Circulation 126:1237–1244. https://doi.org/10.1161/CIRCULATIONAHA.112.104638
Weintraub RG, Semsarian C, Macdonald P (2017) Dilated cardiomyopathy. Lancet 390:400–414. https://doi.org/10.1016/S0140-6736(16)31713-5
Wilkinson JD et al (2010) The pediatric cardiomyopathy registry and heart failure: key results from the first 15 years. Heart Fail Clin 6(401–413):vii. https://doi.org/10.1016/j.hfc.2010.05.002
Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD (2010) Mutations in Troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J Mol Cell Cardiol 48:882–892. https://doi.org/10.1016/j.yjmcc.2009.10.031
Wittekind SG, Ryan TD, Gao Z, Zafar F, Czosek RJ, Chin CW, Jefferies JL (2019) Contemporary outcomes of pediatric restrictive cardiomyopathy: a single-center experience. Pediatr Cardiol 40:694–704. https://doi.org/10.1007/s00246-018-2043-0
Wolf CM (2019) Hypertrophic cardiomyopathy: genetics and clinical perspectives. Cardiovasc Diagn Ther 9:S388–S415. https://doi.org/10.21037/cdt.2019.02.01
Wu H et al (2015) Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an iPSC model of dilated cardiomyopathy. Cell Stem Cell 17:89–100. https://doi.org/10.1016/j.stem.2015.04.020
Yamada Y, Namba K, Fujii T (2020) Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat Commun 11:153. https://doi.org/10.1038/s41467-019-14008-1
Yang Y-S, Wang C-C, Chen B-H, Hou Y-H, Hung K-S, Mao Y-C (2015) Tyrosine sulfation as a protein post-translational modification. Molecules 20:2138–2164. https://doi.org/10.3390/molecules20022138
Yotti R, Seidman CE, Seidman JG (2019) Advances in the genetic basis and pathogenesis of sarcomere cardiomyopathies. Annu Rev Genomics Hum Genet 20:129–153. https://doi.org/10.1146/annurev-genom-083118-015306
Zhaolu WQ, Luo J, Pan B, Liu L-J, Tian J (2020) Cardiac troponin I R193H mutant interacts with HDAC1 to repress phosphodiesterase 4D expression in cardiomyocytes. Genes Dev. https://doi.org/10.1016/j.gendis.2020.01.004
Zhao W et al (2020) Epigenetic regulation of phosphodiesterase 4d in restrictive cardiomyopathy mice with cTnI mutations. Sci China Life Sci 63:563–570. https://doi.org/10.1007/s11427-018-9463-9
Zot HG, Hasbun JE, Michell CA, Landim-Vieira M, Pinto JR (2016) Enhanced troponin I binding explains the functional changes produced by the hypertrophic cardiomyopathy mutation A8V of cardiac troponin C. Arch Biochem Biophys 601:97–104. https://doi.org/10.1016/j.abb.2016.03.011
Acknowledgments
We thank Mark P. Szczypinski and Elliott P. Soto for excellent assistance in examining the predicted effects of single amino acid changes in cTnC on cellular localization.
Funding
This work was supported by U.S. American Heart Association Grant 19PRE34380628 (to J.R.J.), Florida State University’s Undergraduate Research Opportunity Program (UROP) (to P.B.C.), American Heart Association Scientist Development Grant 16SDG29120002 (to M.S.P.), Duke University School of Medicine (to A.P.L.) and U.S. National Institutes of Health National Heart Lung and Blood Institute grant HL128683 (to J.R.P.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reinoso, T.R., Landim-Vieira, M., Shi, Y. et al. A comprehensive guide to genetic variants and post-translational modifications of cardiac troponin C. J Muscle Res Cell Motil 42, 323–342 (2021). https://doi.org/10.1007/s10974-020-09592-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10974-020-09592-5