Abstract
Glutathione S-transferases (GSTs) are a family of enzymes that function in the detoxification of variety of electrophilic substrates. In the present work, we report a novel zeta-like GST (designated as KKSG9) from the biphenyl/polychlorobiphenyl degrading organism Acidovorax sp. KKS102. KKSG9 possessed low sequence similarity but similar biochemical properties to zeta class GSTs. Functional analysis showed that the enzyme exhibits wider substrate specificity compared to most zeta class GSTs by reacting with 1-chloro-2,4-dinitrobenzene (CDNB), p-nitrobenzyl chloride (NBC), ethacrynic acid (EA), hydrogen peroxide, and cumene hydroperoxide. The enzyme also displayed dehalogenation function against dichloroacetate, permethrin, and dieldrin. The functional role of Tyr12 was also investigated by site-directed mutagenesis. The mutant (Y12C) displayed low catalytic activity and dehalogenation function against all the substrates when compared with the wild type. Kinetic analysis using NBC and GSH as substrates showed that the mutant (Y12C) displayed a higher affinity for NBC when compared with the wild type, however, no significant change in GSH affinity was observed. These findings suggest that the presence of tyrosine residue in the motif might represent an evolutionary trend toward improving the catalytic activity of the enzyme. The enzyme as well could be useful in the bioremediation of various types of organochlorine pollutants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Glutathione S-transferases (GSTs) consist of a widely distributed family of enzymes which function in the detoxification of a variety of electrophilic substrates by conjugation with reduced glutathione (GSH) [1]. The conjugation renders the electrophilic substrates more soluble which facilitates their excretion [2]. Furthermore, the protein was also found to be useful in the purification of other proteins in the form of GST-tag proteins which can be easily cleaved at the end leaving only the desired recombinant protein [3]. GSTs played a role in the detoxification of products of oxidative stress, carcinogens and in bacteria, they were often located in some degradative pathways indicating their function in the biodegradation and mineralization of several types of pollutants [4]. Furthermore, some recent studies have shown the importance of bacterial GSTs in the bioremediation of heavy metals such as mercury and arsenate [5, 6]. They are classified as cytosolic, mitochondrial, microsomal, and bacterial specific fosfomycin resistant proteins with cytosolic GSTs representing the largest class of GSTs [4].
The use of microbial enzymes in the bioremediation of different types of pollutants is gaining much attention because of its unique advantages, including low cost, environmental friendliness, and good performance [7]. Furthermore, analysis of the complete genome sequence of several organisms had continued to provide great insight into the presence of novel enzymes with potentials for bioremediation application. This can be seen in the recent discovery of novel classes of GSTs, cold active esterases and carboxylesterases [8,9,10]. GSTs offer greater advantages in the bioremediation of pollutants because of the large repertoire of substrate recognition by a single enzyme. In addition to the previously known cytosolic beta, zeta, chi, and theta GST classes, at least three different classes; eta, rho and nu were recently identified [10,11,12]. Among all these GST classes, zeta class is the one that preserved its sequence similarity across species for a considerable period of evolution [13]. They are found in both prokaryotes and eukaryotes such as in mammals, fungi, plants, and bacteria [14,15,16,17]. Zeta GSTs catalyzes isomerization reactions in tyrosine metabolism, dehalogenation of various substrates and have peroxidase activities [14, 16].
Acidovorax sp. KKS102 is a biphenyl/polychlorobiphenyl degrading organism isolated from the soil near a refinery in Japan [18]. Database suggests that Acidovorax sp. KKS102 contained at least eleven putative GSTs. While studying the GSTs of Acidovorax sp. KKS102, we came across a specific GST (WP_015016207) that contained a signature motif which is closely related to that of Zeta class but with very low sequence similarity to all zeta class GSTs. The protein is also completely lacking essential arginine residues identified in the signature motif of zeta class GSTs, it was designated as KKSG9 and selected for further studies. Furthermore, the activity of the enzyme toward some known GST substrates was investigated. The functional role of residue Tyr12 which replaces the commonly found Cys, Thr, Ser or Ala was investigated by site-directed mutagenesis. Dehalogenation function of the enzyme against dichloroacetate was also investigated. Furthermore, a molecular docking studies together with GST activity studies was carried out in order to investigate the possible interaction of the enzyme with permethrin and organochlorine pesticide dieldrin.
2 Materials and Methods
2.1 Materials
2.1.1 Organism (Acidovorax sp. KKS102)
The organism was obtained in freeze-dried form from Japan Collection of Microorganisms (JCM No. 17234) and revived using Luria–Bertani broth according to the JCM’s instructions. Putative GSTs were retrieved from NCBI database.
2.1.2 Chemicals
Unless otherwise stipulated, chemicals employed were of the highest grade obtainable. 1-chloro-2,4 dinitrobenzene (CDNB), ethacrynic acid (EA), para-nitro benzyls chloride (NBC), antibiotics, reduced glutathione (GSH), NADPH, glutathione reductase, dieldrin, dichloroacetate and all other chemicals and reagents were purchased from Sigma-Aldrich, USA. Molecular biology reagents were purchased from Invitrogen, USA. QuickChange Lightning site–directed mutagenesis kit was purchased from Agilent Technologies, USA.
2.2 Methods
2.2.1 Bioinformatic Analysis
Sequence alignment study was carried out using ClustalW2 [19]. Phylogenetic analysis was carried out using molecular evolutionary genetic analysis (MEGA) software version 6.0 [20]. Neighbor-joining method was used to trace the evolutionary history of the GSTs [21]. The Reltime method was used to calculate the divergence time for all the branch points [21]. Predicted molecular weight and isoelectric point were determined using ProtParam tool (http://us.expasy.org/tools/protparam.html).
2.2.2 PCR Amplification and Cloning of KKSG9
Genomic DNA from Acidovorax sp. KKS102 was isolated using prep ease genomic DNA isolation kit and was used for PCR amplification of KKSG9 gene. The gene was successfully amplified using primers; (forward: 5′ CACCATGCTCGCCCTCTACGGCCA3′ and reverse: 5′TCAGTCGCGGTCGGGTGCTCCT3′). PCR reaction was set using three step PCR-set up. (1) One cycle of initial denaturation at 98 °C for 10 s, (2) 30 cycles of denaturation at 98 °C for 1 s, 67.1 °C annealing for 5 s and 72 °C extension at 15 s/1 kb, (3) One cycle of final extension at 72 °C for 10 min. The amplified products were purified and cloned into pET101 D-TOPO vector. The successful clone was isolated and sequenced for confirmation.
2.2.3 Site-Directed Mutagenesis
Site-directed mutagenesis was carried out according to the method described by Liu et al., [22]. The pair of primers used during the PCR reaction for the Y12C mutation was; forward 5′-TTTCCTCCTGCACCCAGAAGGTGCTGATCGCGCTG-3′ and reverse 5′-TCTGGGTGCAGGAGGAAAAGGGGTGGCCGTAGAGG-3′. The mutation was confirmed by sequencing. Purification and characterization of the mutant were carried out as described for the wild-type.
2.2.4 Protein Expression and Purification
Recombinant KKSG9 was overexpressed using Rosetta-gami B (DE3) as follows. About 10 ng of recombinant plasmid was transformed into Rosseta-gami-B(DE3) chemically competent cells. The transformation reaction was then plated on Luria–Bertani (LB) media plates containing 100 µg/mL ampicillin, 15 µg/mL kanamycin, 34 µg/mL chloramphenicol and 12.5 µg/mL tetracycline. The positive transformant was selected and cultured into 10 mL LB broth containing the combination of the above-mentioned antibiotics and cultured overnight at 37 °C temperature. 500 mL of LB containing the above-mentioned antibiotics was then inoculated with the entire 10 mL from the overnight culture. The culture was grown at 37 °C with shaking (200 rpm) until the optical density (OD)600 reaches about 0.5. Cells were grown for 5 h post IPTG addition. The cells were harvested by centrifugation at 6000 rpm for 12 min at 4 °C. Lysis buffer containing protease inhibitor and lysozyme was added to the pellets to facilitate the breakdown of the cell wall. The cells were lysed by sonication and centrifuged at 8000 rpm for 60 min. The supernatant was collected for GST purification. Protein purification was carried out using an ÄKTA Purifier FPLC equipped with UNICORN software Version 5.1 and a fraction collector (Frac900) for greater automation of the purification process. The clear crude enzyme was applied to a Glutathione Sepharose™ High Performance column (GSTrap™ HP, 5 mL) and pre-equilibrated with 25 mM sodium phosphate buffer pH 7.4 maintained at a flow rate of 0.5 mL/min. The column was thoroughly washed with buffer A to remove non-specifically bound proteins. The bound GSTs were eluted with 10 mM glutathione in buffer A. The collected fractions with activities towards CDNB were pooled and concentrated using centrifugal concentrator (Vivaspin 20: 10 000 WMCO).
2.2.5 Glutathione S-transferase Assay and Determination of Kinetic Parameters
The GST activity towards CDNB and ethacrynic acid was carried out as described by Habig et al., [23]. The peroxidase activity of the enzyme towards hydrogen peroxide and cumene hydroperoxide was carried out according to the method described by Di Ilio et al. [24]. The kinetic parameters for wild type and mutants of KKSG9 were determined using nitro-benzyl chloride (NBC) and GSH as substrates. This was performed by varying the NBC concentration (0.02–0.2 mM) while keeping the GSH concentration constant (1.0 mM). For the GSH, it was determined by keeping the concentration of NBC constant (1.0 mM) while varying the GSH concentration (0.4–1.2). All the measurements were carried out using Cary 60 UV–Visible spectrophotometer. The data was fitted to the Michealis-menten equation V = Vmax[S]/Km + [S] and was analyzed using non-linear regression analysis with GraphPad Prism 7 software. Kinetic parameters (Km and Vmax) were determined from the graph while Kcat was calculated using the equation Kcat = Vmax/[E]t.
2.2.6 Dehalogenation Function of KKSG9
The activity of purified of KKSG9 toward DCA, permethrin, and dieldrin was measured by its ability to release chloride ions from the substrates. The amount of chloride ion released was quantified based on the method as previously described [25].
2.2.7 Protein Concentration
The protein concentration was determined using Bradford assay by employing bovine serum albumin as a standard [26].
2.2.8 Sodium Deodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
SDS-PAGE was performed in 12% polyacrylamide gel at 120V using the method as previously described [27]. The gels were stained with commassie blue. The stained gels were scanned with Image Scanner III (GE Healthcare), thereafter, it was visualized and analyzed with Image Master software.
2.3 Molecular Docking Studies
In order to understand the binding pattern and the nature of the interaction between KKSG9 and the organochlorine substrates; permethrin and dieldrin, a molecular docking study was carried out using autodock 4.2 software [28]. The modeled protein was built by the Swiss model server using a crystal structure of maleyl pyruvate isomerase in complex with glutathione (PDB code: 2jl4.1.A) which shared 21.95% sequence similarity with KKSG9 [29]. The model quality assessment was done using Global QMEAN scoring function (http://swissmodel.expasy.org/qmean/cgi/index.cgi). Chemsketch software was used to draw the structure of all the ligands which were stored in the form of a mol2 file [30]. Finally, the protein data bank file (PDB) of all the ligands was constructed using Open Babel software. The PDB files of both the ligands and the proteins were used as input file in autodock tools. A blind docking analysis was set up using an autogrid size of 126, 126 and 126 for the x, y, and z-axis respectively. Default parameters were used in all the molecular docking process. A total of 100 Lamarckian genetic algorithm docking runs were performed for each ligand. Docked conformations clustered within a root mean square deviation of 2 Å were visualized using discovery studio software (http://www.accelrys.com).
3 Results
3.1 Bioinformatic Analysis
In-silico analysis of GSTs from Acidovorax sp. KKS102 revealed the presence of at least eleven putative GSTs. A putative sequence with NCBI accession number (WP015014376) which displayed a motif closely related to that of Zeta class GSTs was designated as KKSG9 and selected for further studies. In order to examine the genetic relationship between this sequence and all known GST sequences, a phylogenetic tree was constructed using representatives from all GST classes. Phylogenetic analysis showed that KKSG9 is closely related to zeta class GSTs, having its clade emanate from Zeta groups (Fig. 1a). However, the protein displayed very low sequence similarity of only 23.30% with eukaryotic zeta GST of Capsicum annum. GSTs that share greater than 40% sequence similarity are grouped into the same class while those with less than 25% sequence similarity are grouped into a different class [4]. The gene has an open reading frame of 657 bp, coding for 219 amino acid residues with predicted molecular weight and theoretical isoelectric point of 24.4 kDa and 6.12 respectively.
a Evolutionary history of Acidovorax sp. KKS102 GSTs. The phylogenetic tree was constructed using a representative from all GST classes. Numbers after the underscore are the NCBI accession numbers. Bold arrows represent KKSG9. b Multiple sequence alignment of KKSG9 with representatives from zeta class; WP015016207 is KKSG9 from Acidovorax sp. KKS102, AAO61856 is from eukaryotic Malva pusilla, AAN39918 is from eukaryotic Capsicum annuum, Q3S4B4 is from Polaromonas naphthalenivorans CJ2, O86043 is from Ralstonia sp., Q5K5T6 is from Escherichia coli and Q5EXK2 is from Klebsiella pneumonia. The boxed letters indicate the motif observed in KKSG9 and other zeta class GSTs. The small box inside the motif indicates the variation in amino acid between different zeta class GSTs
3.2 Molecular Docking Studies
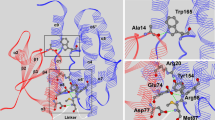
Molecular docking study was carried out in order to explore the binding pattern and the amino acids involved in the interaction of permethrin and dieldrin within the catalytic cavity of KKSG9. It was found that GSH, dieldrin and permethrin interact with Ser10, Ser11, Tyr12, His40, Lys52, Pro52, Phe53, Glu65, Ser66, Gln110, His111, Ala112, and Asn 114. Cluster analysis of docked result in permethrin using root mean square tolerance (RMSD) of 2 Å revealed 46 different conformations. The lowest binding energy obtained was − 10.55 kJ/mol which occurred in the 7th run of the most populated cluster containing 12 members. In dieldrin, the cluster analysis revealed 4 different conformations. The lowest binding energy was − 8.83 kJ/mol, which occurred in the 13th run of the most populated cluster containing 37 members.
3.3 Substrate Specificity and the Kinetic Parameters
In order to determine the substrate specificity of KKSG9, the full-length sequence was cloned into pET101-D-TOPO vector. Site-directed mutagenesis was also carried out on the Tyr12 residue which is converted to cysteine as described in the methodology section. Both the wild-type and mutant proteins were expressed and purified using GSH affinity column where a single band was obtained on SDS-PAGE (Fig. 2). The enzyme displayed a typical behavior of zeta class GSTs as reported by Board et al. [16], as it binds to the GST-affinity column slowly before it was eluted using 10 mM GSH. Substrate specificity of the enzyme was determined using a range of known GST substrates. The enzyme displayed wide substrate specificity compared to most zeta class GSTs by reacting with (CDNB), (NBC), (EA), hydrogen peroxide (H2O2) and cumene hydroperoxide (CuOOH) (Table 1).
3.4 Dehalogenase Activity of KKSG9 Towards DCA, Permethrin, and Dieldrin
The dehalogenating activity of the wild-type and mutant KKSG9 was further determined using dichloroacetate (DCA), permethrin, and dieldrin as substrates. Dichloroacetate (DCA) is a water contaminant that was believed to be carcinogenic and hepatotoxic [10, 31]. Permethrin is one of the widely used pyrethroids insecticides considered to have low toxicity, however, increasing evidence suggests that it has hepatotoxic, neurotoxic and genotoxic effects to animals and humans alike [32]. Dieldrin is a persistent organochlorine pesticide produced as a by-product of microbial biodegradation of aldrin, it is more persistent and more toxic to humans and animals than the aldrin itself [33]. KKSG9 was found to exhibit dehalogenase activities against these substrates as determined by its ability to release chloride ions from the substrates (Fig. 3). Furthermore, the wild-type enzyme displayed higher dehalogenase activity against all the substrates when compared with the mutant (Y12C) enzyme.
4 Discussion
Zeta class GSTs were known to contained a motif at the N-terminal part of the protein; SSCX(W/H)RVRIAL and RSSASYRVRIAL for eukaryotic and prokaryotic sequences respectively [6, 11]. Comparative analysis of the motif observed in KKSG9 showed that it contained SSYTQKVLIAL, which matches with the established sequence as shown in box (Fig. 1b). The first serine residue in these motifs is highly conserved and is analogous to the catalytic serine found in Theta, Phi and Delta classes [11]. In the prokaryotic sequences, the residues RS----RVRIAL were also shown to be > 85% conserved with the first arginine functions in substrate recognition. Furthermore, while the eukaryotic sequences were found to contain Cys residue in the motif, the prokaryotic sequence was found to replace the Cys with (Ser, Thr or Ala) [6]. In KKSG9, the commonly found Ser, Thr or Ala in prokaryotic sequences was found to be replaced by Tyr. In addition, the motif in KKSG9 completely lacks arginine residues including the first arginine which functions in substrate recognition.
The presence of several polar amino acid side chains in both the G-site and H-site of the binding cleft suggest that both hydrogen bonding and hydrophobic interactions could be stabilizing the complexes formed. In permethrin, in addition to the hydrophobic interactions observed, at least two hydrogen bonds were predicted between Asn 114 and His 111 and the ligand oxygen (Fig. 4a). In dieldrin, at least one hydrogen bonding was also predicted between the chloride atom of dieldrin and His111 (Fig. 4a). The validity of the interactions was further confirmed experimentally in the chloride ion detection assay.
The dehalogenase activity of wild-type KKSG9 against DCA, permethrin, and dieldrin is 1.41, 1.51 and 1.44-fold higher when compared with the mutant. This could also be attributed to the stabilization of the protein structure by the Tyr12 residue in the wild type when compared with the Cys residue as reflected in the specific activity of the enzyme. Dichloroacetate is one of the substrates of zeta class GSTs which is eventually converted into glyoxylic acid and chloride ions leading to its detoxification. Organochloride pollutants were shown to be more recalcitrant as the number of chlorine atoms attached to the parent compound is increased. Dehalogenation of these compounds was shown to decrease their recalcitrance and eventually helps in their rapid biodegradation. These findings suggest that KKSG9 could be useful in the biodegradation of wide variety of organochlorine compounds in addition to the dichloroacetate.
We examined the differences that may occur in the catalytic activity of the protein when Tyr12 is mutated to originally known cysteine residue. While mammalian zeta class GST largely maintained cysteine residue at that position, bacterial zeta class GST contained either Cys, Ser, Thr or Ala with Ala containing > 50% of the residue. Several studies have shown that the Cys, Ser, Thr and Ala located in the glutathione binding site played a significant role in the binding of the co-substrate GSH and hence, affecting the catalytic activity of zeta class GSTs [10, 12]. However, while the catalytic activity of the enzyme could be affected, the overall biotransformation pathways are not affected by the differences in the amino acid residue located at the glutathione binding site. This is because the usual metabolites are still detected after the biotransformation. The result of the specific activity of (Y12C) mutant showed a general decrease in the catalytic activity of the enzyme towards all the substrates with CDNB showed the highest decrease of 1.75-fold when compared with the wild type while para nitro benzyl chloride displayed 1.33-fold decrease in the catalytic activity of the protein. The peroxidase activity towards hydrogen peroxide and cumene hydroperoxide also showed 1.43 and 1.56-fold decrease respectively while activity with ethacrynic acid indicates 1.51-fold decrease.
Kinetic analysis using NBC and GSH as substrates further revealed the importance of the residue Tyr12 in the binding of the substrates. Data from the kinetic studies (Table 2), revealed that the binding of the substrate NBC was affected by the mutation of Tyr12 to Cys as there was a 2.23-fold decrease in the Km value of the mutant. This was also reflected in the catalytic efficiency and Vmax of the mutant which decreases by 1.48 and 4.07 respectively. However, the affinity of GSH was not much affected as there was no significant change in the Km value between the wild-type and the mutant. This is probably because while the wild type containing the tyrosine residue could provide a hydrogen bonding with the nitrogen or thiol group of the cysteinyl moiety, the mutant containing the cysteine residue could also provide disulfide linkage with the thiol group of the cysteinyl moiety. The improved catalytic activity of the wild-type, when compared with the mutant might be as a result of the presence of aromatic ring in the tyrosine residue, the aromatic ring was shown to form a stack with other aromatic amino acids in some proteins thereby stabilizing the protein structure and improve its catalytic activity [34]. The tyrosine residue in KKSG9 is lying close to a phenylalanine residue (Phe9) which could form a stack with the aromatic ring of Tyr12 and thereby further enhances the stabilization of the protein. These findings suggest that the variation in the equivalent residues at that position in other bacterial zeta class GSTs and the presence of tyrosine in KKSG9 might represent an evolutionary step for enhancing the catalytic activity of the protein. Furthermore, the finding also corroborates with the suggestion by Fang et al. [5], that this residue may be important in the rational design of an effective and competitive enzyme in zeta class GSTs.
In conclusion, the biochemical properties of a novel zeta-like glutathione S-transferase, KKSG9, from Acidovorax sp. KKS102 were studied. The functional role of a Tyr12 residue was also investigated by site-directed mutagenesis. The wider substrate specificity displayed by KKSG9 compared to other zeta class GSTs suggested an evolutionary trend toward enhancing the catalytic activity of this cGST. Despite the fact that KKSG9 showed very low sequence similarity with other zeta class GSTs, however, several of its biochemical properties coupled with the lack of cysteine residue in its motif pointed to the fact that it probably belongs to maleylpyruvate isomerase (MPI) class. Furthermore, the enzyme also displayed dehalogenation function against a pyrethroid insecticide (Permethrin) and dieldrin, in addition to dichloroacetate. This indicates that it could potentially be useful in the bioremediation of diverse classes of organochlorine pollutants.
Abbreviations
- GST:
-
Glutathione S-transferase
- CDNB:
-
1-Chloro-2,4-dinitrobenzene
- NBC:
-
p-Nitrobenzyl chloride
- EA:
-
Ethacrynic acid
- CuOOH:
-
Cumene hydroperoxide
References
Alias Z, Clark AG (2010) Adult Drosophila melanogaster glutathione S-transferases: effects of acute treatment with methyl parathion. Pestic Biochem Phys 98:94–98
Yamamoto K, Suzuki M, Higashiura A, Nakagawa A (2013) Three-dimensional structure of a Bombyx mori Omega-class glutathione transferase. Biochem Biophys Res Commun 438:588–593
Gopal GJ, Kumar A (2013) Strategies for the production of recombinant protein in Escherichia coli. Protein J 32:419–425
Allocati N, Federici L, Masulli M, Di Ilio C (2009) Glutathione transferases in bacteria. FEBS J 276:58–75
Chrysostomou C, Quandt EM, Marshall NM, Stone E, Georgiou G (2015) An alternate pathway of arsenate resistance in E. coli mediated by the glutathione S-transferase GstB. ACS Chem Biol 10:875–882
Zhang W, Yin K, Li B, Chen L (2013) A glutathione S-transferase from Proteus mirabilis involved in heavy metal resistance and its potential application in removal of Hg2+. J Hazard Mater 261:646–652
Zhang W, Yin K, Chen L (2013) Bacteria-mediated bisphenol A degradation. Appl Microbiol Biotechnol 97:5681–5689
Yin K, Lv M, Wang Q, Wu Y, Liao C, Zhang W, Chen L (2016) Simultaneous bioremediation and biodetection of mercury ion through surface display of carboxylesterase E2 from Pseudomonas aeruginosa PA1. Water Res 103:383–390
Rahman MA, Culsum U, Tang W, Zhang SW, Wu G, Liu Z (2016) Characterization of a novel cold active and salt tolerant esterase from Zunongwangia profunda. Enzyme Microb Technol 85:1–11
Pandey T, Chhetri G, Chinta R, Kumar B, Singh DB, Tripathi T, Singh AK (2015) Functional classification and biochemical characterization of a novel rho class glutathione S-transferase in Synechocystis PCC 6803. FEBS Open Bio 5:1–7
Skopelitou K, Dhavala P, Papageorgiou AC,. Labrou NE (2012) A glutathione transferase from Agrobacterium tumefaciens reveals a novel class of bacterial GST superfamily. PLoS ONE 7:342–353
Stourman NV, Branch MC, Schaab MR, Harp JM, Ladner JE, Armstrong RN (2011) Structure and function of YghU, a nu-class glutathione transferase related to YfcG from Escherichia coli. Biochemistry 50:1274–1280
Fang T, Li D-F, Zhou N-Y (2011) Identification and clarification of the role of key active site residues in bacterial glutathione S-transferase zeta/maleylpyruvate isomerase. Biochem Biophys Res Commun 410:452–456
Marsh M, Shoemark DK, Jacob A, Robinson C, Cahill B, Zhou N-Y, Williams PA, Hadfield AT (2008) Structure of bacterial glutathione-S-transferase maleyl pyruvate isomerase and implications for mechanism of isomerisation. J Mol Biol 384:165–177
Thom R, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ (2001) The structure of a zeta class glutathione S-transferase from Arabidopsis thaliana: characterisation of a GST with novel active-site architecture and a putative role in tyrosine catabolism. J Mol Biol 308:949–962
Board GP, Baker TR, Chelvanayagam G, Jermiin SL (1997) Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem J 328:929–935
Yamamoto K, Shigeoka Y, Aso Y, Banno Y, Kimura M, Nakashima T (2009) Molecular and biochemical characterization of a zeta-class glutathione S-transferase of the silkmoth. Pestic Biochem Phys 94:30–35
Ohtsubo Y, Maruyama F, Mitsui H, Nagata Y, Tsuda M (2012) Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J Bacteriol 194:6970–6971
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Saitou N. Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Liu H. Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8:1–6
Habig WH, Pabst MJ, Jakoby WB (1974) Glutathione S-transferases the first enzymatic step in mercapturic acid formation. J Biol Chem 249:7130–7139
Di Ilio C, Sacchetta P, Bello ML, Caccuri AM, Federici G (1986) Selenium independent glutathione peroxidase activity associated with cationic forms of glutathione transferase in human heart. J Mol Cel Cardiol 18:983–991
McGuinness M, Mazurkiewicz V, Brennan E, Dowling D (2007) Dechlorination of pesticides by a specific bacterial glutathione S-transferase, BphKLB400: potential for bioremediation. Eng Life Sci 7:611–615
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nat 227:680–685
Goodsell DS, Morris GM, Olson AJ (1996) Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 9:1–5
Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1–13
Mills N (2006) ChemDraw ultra 10.0 CambridgeSoft. 100 CambridgePark Drive, Cambridge
Zeen T, Philip G, Anders M (1998) Glutathione transferase zeta catalyses the oxygenation of the carcinogen dichloroacetic acid to glyoxylic acid. Biochem J 331:371–374
Wang X, Martínez M-A, Dai AM, Chen D, Ares I, Romero A, Castellano V, Martínez M, Rodríguez JL, Martínez-Larrañaga M-R (2016) Permethrin-induced oxidative stress and toxicity and metabolism: a review. Environ Res 149:86–104
Birolli WG, Yamamoto KY, de Oliveira JR, Nitschke M, Seleghim MH, Porto AL Biotransformation of dieldrin by the marine fungus Penicillium miczynskii CBMAI 930. Biocatal Agric Biotech 4:39–43
Brennan E, McGuinness M, Dowling DN (2009) Bioinformatic analysis and in vitro site-directed mutagenesis of conserved amino acids in BphK LB400, a specific bacterial glutathione transferase. Int Biodeter Biodegrad 63:928–932
Acknowledgements
We wish to thank Dr. Yuji Nagata of the graduate school of life sciences, Tohoku University, Japan, for granting us the permission to use the strain. We also wish to thank Japan collection of microorganism (JCM) for providing us with the strain. This work was supported by the University of Malaya IPPP [PG170-2016A]. One of the authors (DS) would like to thank Bayero University, Nigeria, for the financial assistance.
Funding
This study was funded by University of Malaya IPPP Grant [PG170-2016A].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Shehu, D., Alias, Z. Functional Role of Tyr12 in the Catalytic Activity of Novel Zeta-like Glutathione S-transferase from Acidovorax sp. KKS102. Protein J 37, 261–269 (2018). https://doi.org/10.1007/s10930-018-9774-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-018-9774-x